Abstract
RNA interference (RNAi) is a process by which short interfering RNAs (siRNAs) direct the degradation of complementary single-strand RNAs. In this study, we investigated the effects of full-strand phosphorothioate (PS) backbone and 2′-O-methyl (2′-OMe) sugar modifications on RNAi-mediated silencing. In contrast to previous reports, we have identified active siRNA duplexes containing full 2′-OMe-modified sense strands that display comparable activity to the unmodified analog of similar sequence. The structure of these modified siRNAs is the predominant determinant of their activity, with sequence and backbone composition being secondary. We further show, by using biotin-tagged siRNAs and affinity-tagged hAgo2/eIF2C2, that activity of siRNA duplexes containing full 2′-OMe substitutions in the sense strand is mediated by the RNA-induced silencing complex (RISC) and that strand-specific loading (or binding) to hAgo2 may be modulated through selective incorporation of these modifications.
Keywords: 2′-OMe RNA, anti-sense, RNA, small interfering
INTRODUCTION
Double-stranded short interfering RNAs (siRNAs) direct the degradation of complementary target RNAs in a process known as RNA interference (RNAi) (Fire et al. 1998; Hammond et al. 2001b; Hannon 2002; McManus and Sharp 2002). Identification of siRNAs as intermediates of the RNAi pathway has led to the demonstration that chemically synthesized siRNAs are sufficient substrates for reduction of target mRNA in mammalian systems (Caplen et al. 2001; Elbashir et al. 2001a). Further application of medicinal chemistry to these synthetic oligoribonucleotides has in turn enabled usage and identification of modifications that can be incorporated into siRNAs for further mechanistic analysis (Manoharan 2004), as well as development of siRNAs with improved pharmacokinetic and pharmacodynamic properties for in vivo applications (Layzer et al. 2004; Soutschek et al. 2004; de Fougerolles et al. 2005).
Two modifications that have been extensively evaluated, for both anti-sense oligonucleotides (ASO) and siRNAs, are the phosphorothioate (PS) backbone modification (Eckstein 2000) and the 2′-O-methyl (2′-OMe) sugar modification (Lubini et al. 1994; Braasch and Corey 2002; Grunweller et al. 2003; Harborth et al. 2003). The replacement of a nonbridging oxygen with a sulfur atom creates a PS linkage that may afford an increased resistance to nuclease degradation and raise the in vivo bioavailability of the modified siRNA, as has been demonstrated previously for single-stranded ASOs that operate via an RNase H-based mechanism (Crooke 1999; Eckstein 2000; Geary et al. 2001). Incorporation of the PS modification, however, also lowers thermal duplex stability for complementary sequences, promotes nonspecific protein binding, and—upon extensive incorporation—may result in cytotoxicity (Amarzguioui et al. 2003; Harborth et al. 2003).
The 2′-OMe sugar modification, although more bulky than the 2′-OH, preserves the ribose sugar pucker and retains the canonical right-handed A-form helical geometry (Lubini et al. 1994; Cummins et al. 1995), which is required for siRNA activity (Chiu and Rana 2003). As with the PS modification, this modification has also been shown to increase the nuclease resistance of oligoribonucleotides (Lubini et al. 1994; Cummins et al. 1995) and siRNA duplexes (Chiu and Rana 2003; Czauderna et al. 2003).
The effect of the PS and 2′-OMe modifications on siRNA activity has been found to be dependent on both position and extent of incorporation. With respect to the latter, a significant number of reports in the literature have demonstrated that siRNA constructs containing full 2′-OMe substitutions in either sense, anti-sense, or both strands display little to no RNAi activity (Elbashir et al. 2001b; Amarzguioui et al. 2003; Braasch et al. 2003; Chiu and Rana 2003; Czauderna et al. 2003). For example, an siRNA that had either or both strands 2′-OMe modified was shown to completely lose RNAi activity against a luciferase mRNA in Drosophila cell lysates (Elbashir et al. 2001b). In agreement with this, Braasch and coworkers (Braasch et al. 2003) failed to observe inhibition of hCAV expression in HeLa cells with fully substituted 2′-OMe RNA in one or both strands of the duplex. In further studies using 19-bp 3′-dTdT duplexes in HeLa cells, Chiu and Rana (2003) found that 2′-OMe-modified siRNA greatly diminished EGFP silencing, achieving 16%–25% RNAi activity when either the anti-sense or the sense strand was fully modified, and no activity was seen when both strands were modified, where 93% activity was achieved with the respective unmodified siRNA. Czauderna and coworkers also demonstrated that blunt-end 21-bp duplexes targeting human PTEN with either one or both strands fully substituted with 2′-OMe were relatively inactive compared to the unmodified parent construct in their mammalian cell culture system (Czauderna et al. 2003).
In contrast to prior observations, this report demonstrates that siRNA duplexes containing full 2′-O-methyl-modified sense strands, targeted to sites within the PTEN mRNA, mediate mRNA target reduction with similar efficacy and potency as the unmodified parent siRNA in HeLa cells. To determine the basis of the differences between our observations and those of other investigators, we evaluated the effects of construct design, target site sequence, and backbone composition on 2′-OMe-modified siRNA activity in cell culture.
We also sought to determine (or confirm) the underlying mechanism of target mRNA reduction by the 2′-OMe-modified siRNA complexes. Argonaute2/eIF2C2 (hAgo2) has been identified as an endonuclease activity associated with the RNA-induced silencing complex (RISC) (Hammond et al. 2000, 2001a), which provides the minimal scaffold for catalytic RISC cleavage (Liu et al. 2004; Meister et al. 2004; Rand et al. 2004; Song et al. 2004; Rivas et al. 2005). We found by using biotin-tagged siRNAs, an affinity-tagged hAgo2 overexpression vector, an RNase H-based ASO knock-down assay, and an in vitro hAgo2-based RNA cleavage assay that hAgo2 is a mediator of target mRNA knock-down in cell culture and RNA substrate cleavage in solution for this class of modified siRNAs.
RESULTS
Target site selection and siRNA design
We selected four active siRNA sites (Table 1 ▶, Sites I–IV), identified previously from a screen of 36 siRNAs that target the endogenous tumor suppressor PTEN mRNA (Vickers et al. 2003), to examine the effects of full-strand sugar-phosphate backbone modifications on siRNA activity. Full-strand PS and 2′-OMe modifications were evaluated in a combinatorial fashion at each site, in conjunction with two types of siRNA construct designs, the canonical 19-bp duplex with 3′-dTdT overhangs (19-bp 3′-dTdT) and a 20-bp duplex with blunt ends (blunt-end 20 bp).
TABLE 1.
Location of PTEN target sites
| Site | Start | Sequence 5′–3′ (sense mRNA) |
| I | 1505 | A.G.U.A.A.G.G.A.C.C.A.G.A.G.A.C.A.A.A |
| IB | 1504 | A.A.G.U.A.A.G.G.A.C.C.A.G.A.G.A.C.A.A.A |
| II | 1541 | C.A.G.U.C.A.G.A.G.G.C.G.C.U.A.U.G.U.G |
| IIB | 1541 | C.A.G.U.C.A.G.A.G.G.C.G.C.U.A.U.G.U.G.U |
| III | 1855 | G.G.G.U.A.A.A.U.A.C.A.U.U.C.U.U.C.A.U |
| IIIB | 1855 | G.G.G.U.A.A.A.U.A.C.A.U.U.C.U.U.C.A.U.A |
| IV | 2097 | C.A.A.A.U.C.C.A.G.A.G.G.C.U.A.G.C.A.G |
| IVB | 2096 | U.C.A.A.A.U.C.C.A.G.A.G.G.C.U.A.G.C.A.G |
A comparative analysis of 36 siRNA sequences identified as active siRNA sites within human phosphatase and tensin homolog PTEN (Accession No. U92436) message (Vickers et al. 2003). The location and sequence of the active target sites used in this study are shown above. The PTEN coding sequence (CDS) starts at 1035 and terminates at 2246. The sites shown in gray were used for generating 19-mers with 2-nt deoxythymidine overhangs at the 3′–end. The sites shown in white were used for the generation of 20-mer blunt siRNAs. The sequence of the antisense strand is an exact complement to the sense strand shown in the table.
Activity of 19-bp 3′-dTdT siRNA duplexes with full 2′-O-methyl sense strands is sequence dependent
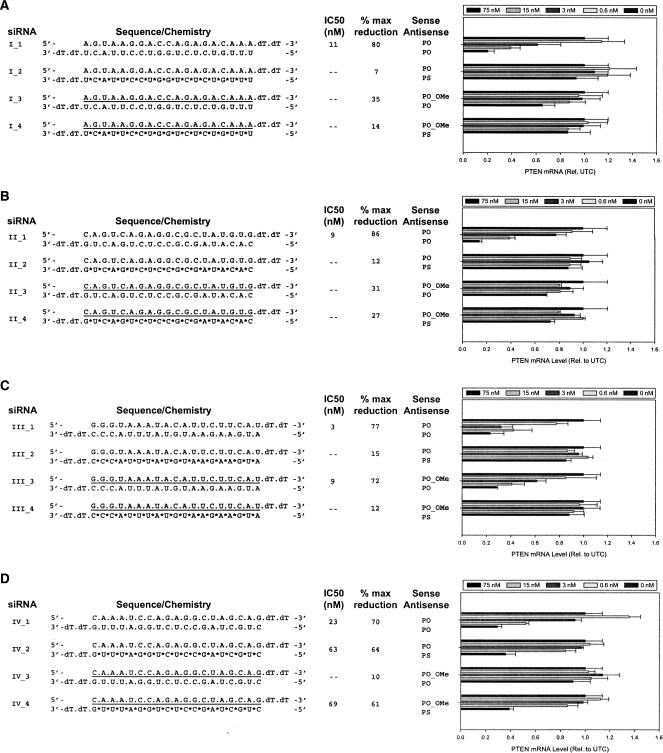
In our initial analysis, we evaluated combinations of a PS-modified anti-sense strand, a 2′-OMe sense strand, and the respective unmodified strands in the context of the canonical 19-bp 3′-dTdT overhang construct. Each of the unmodified siRNA molecules (I_1, II_1, III_1, and IV_1) directed against the four PTEN target sites displayed a dose-dependent reduction in PTEN mRNA expression levels (Fig. 1 ▶). The siRNA duplex targeting Site III (III_1) was the most potent with an IC50 of 3 nM. Constructs targeting Sites I and II (I_1 and II_1) were less potent, yet displayed significant target reduction with IC50s between 9 and 10 nM. The least potent siRNA was directed against the Site IV (IV_1) with an IC50 of 22 nM. The efficacy of all four sites was similar, achieving ~70%–80% reduction of target in each case.
FIGURE 1.
Activity of 19-bp 3′-dTdT siRNA duplexes with full 2′-O-methyl sense strands is sequence-dependent. Dose response analyses of 19-bp 3′-dTdT siRNAs targeting Site I (A), Site II (B), Site III (C), and Site IV (D) of the PTEN mRNA in HeLa cells. The sequence and structure of siRNAs are shown to the left of each graph and are unmodified oligoribonucleotides unless indicated otherwise. (*) A phosphorothioate (PS) modification, (underline) 2′-OMe-modified nucleotides, (dT) 2′-deoxythymidine. HeLa cells were treated with siRNAs at 0.6, 3, 15, and 75 nM, as detailed in Materials and Methods. PTEN mRNA levels were determined relative to c-raf mRNA levels by real-time RT-PCR (TaqMan). Results shown represent the percentage of untreated control expression (UTC). Each bar represents the mean of three biological replicates (±standard deviation).
In comparison, the 19-bp 3′-dTdT duplexes containing a fully modified 2′-OMe sense strand designed against Site I (Fig. 1A ▶, siRNAs I_3 and I_4), and Site II (Fig. 1B ▶, siRNAs II_3 and II_4) showed no reduction in PTEN mRNA levels when tested in combination with either a PO or PS anti-sense strand. At Site III, however, the duplex consisting of a PO anti-sense strand duplexed to a fully modified PO 2′-OMe sense strand showed efficient reduction of PTEN mRNA expression levels with an IC50 of 9 nM (Fig. 1C ▶, III_3). The corresponding PO:PS_2′- OMe duplex was slightly more potent with an IC50 of 6 nM (data not shown). In addition, we noted that if a modified PS anti-sense strand was inactive (Fig. 1C ▶, III_2), activity could not be restored by incorporating a fully modified 2′-OMe sense strand into the duplex (Fig. 1C ▶, III_4). Constructs containing a full 2′-OMe sense strand that targeted Site IV displayed very limited activity when duplexed to the PS anti-sense strand (Fig. 1D ▶, cf. IV_2 and IV_4).
Taken together, these results suggest that a fully modified 2′-OMe sense strand can elicit an effective siRNA response with similar potency and efficacy as the unmodified parent duplex. However, target mRNA reduction with fully modified 2′-OMe sense strands is impacted by strong sequence-dependent effects as only one active site out of the four examined was able to reduce target RNA in the context of the 19-bp 3′-dTdT construct design (Fig. 1 ▶).
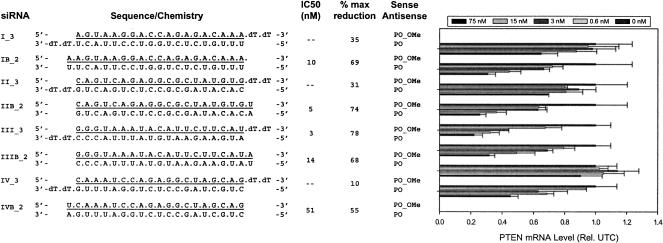
Activity of siRNAs with full 2′-O-methyl sense strands is dependent on construct design
We next investigated the effects of construct design on activity of siRNAs with full 2′-OMe-modified sense strands. Blunt-end 20-bp constructs were designed against the PTEN Sites I, II, III, and IV (Table 1 ▶), and their activity was compared to the analogous 19-bp 3′-dTdT constructs by dose response analysis in HeLa cells. In contrast to the 19-bp 3′-dTdT overhang constructs, target mRNA reduction was observed at all four sites with the blunt-end 20-bp duplexes containing a full 2′-OMe sense strand (Fig. 2 ▶). Constructs IB_2, IIB_2, and IIIB_2 achieved ~70%–75% PTEN mRNA reduction at the 75 nM dose with IC50s of 10, 5, and 14 nM, respectively. Activity of the blunt-end construct at Site IV (IVB_2) displayed ~50% reduction at the highest dose tested (75 nM). This relatively low level of activity is in accordance with the activity of the unmodified siRNA at Site IV, which was also the least active in its class. The 19-bp 3′-dTdT construct corresponding to Site III remained the most potent and efficacious design with an IC50 of 3 nM and 78% total reduction of target RNA.
FIGURE 2.
Activity of siRNAs with full 2′-O-methyl sense strands is dependent on construct design. Dose response analysis of two types of siRNA constructs (19-bp 3′-dTdT vs. blunt-end 20 bp) targeting each of the four PTEN mRNA target sites in HeLa cells. The construct structure and sequence are shown to the left of each graph and are unmodified oligoribonucleotides unless indicated otherwise. (Underline) 2′-OMe-modified nucleotides, (dT) 2′-deoxythymidine residues. HeLa cells were dosed at 0.6–75 nM with siRNA, as described in Materials and Methods. The results shown represent the percentage of untreated control expression (UTC). Each bar represents the mean of three biological replicates (±standard deviation).
These data suggest that activity of siRNAs with fully modified 2′-OMe sense strands is dependent on construct design. Furthermore, the blunt-end 20-bp design appears to be more broadly applicable when incorporating a full 2′-OMe sense strand, as it has a broader range of activity across the four sites in comparison to the 19-bp 3′-dTdT overhang constructs.
Evaluation of blunt-end 20-bp siRNA duplexes with full 2′-O-methyl and PS backbone modifications
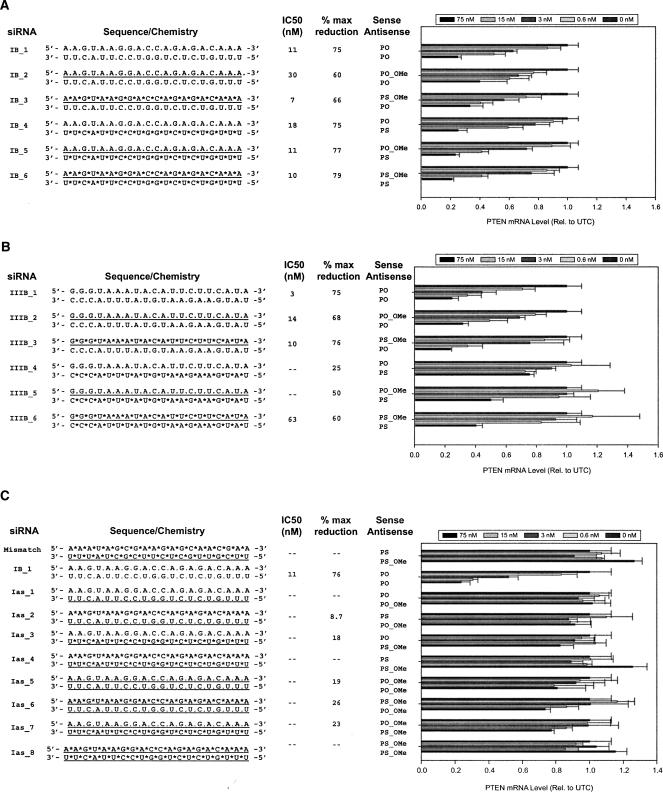
Having established that blunt-end 20-bp constructs may be the optimal design for siRNAs with 2′-OMe-modified sense strands, we next evaluated the effects of the full 2′-OMe and PS backbone modifications on RNAi activity in cell culture based on this construct design (Fig. 3 ▶).
FIGURE 3.
Blunt-end 20-bp siRNAs with full 2′-O-methyl sense strands and full PS backbone modifications mediate target mRNA reduction. Dose response analyses of modified blunt-end 20-bp siRNAs comprised of full 2′-OMe-modified sense strands to Site I (A), full 2′-OMe-modified sense strands to Site III (B), and full 2′-OMe-modified anti-sense strands to Site I (C). The siRNA constructs are shown to the left of each graph and are unmodified oligoribonucleotides unless indicated otherwise. (*) A PS modification, (underline) 2′-OMe-modified nucleotides. The results shown represent the percentage of untreated control expression (UTC). Each bar represents the mean of three biological replicates (±standard deviation).
Anti-sense strands were designed against the PTEN Sites I and III as either an unmodified PO strand or fully modified PS strand. Sense strands were duplexed to the anti-sense strands as an unmodified PO, a PO with full 2′-OMe substitutions, or a fully modified PS with full 2′-OMe modifications (Fig. 3A,B ▶). At PTEN Site I, the parent (IB_1) achieved 75% reduction of PTEN mRNA with an IC50 of 11 nM. Modified duplexes directed to this site achieved 60%–65% reduction of PTEN mRNA when either a PO-2′-OMe or a PS-2′-OMe sense strand was duplexed to a PO anti-sense strand (IB_2 and IB_3, respectively). siR-NAs containing either a PO-2′-OMe (77% reduction) or a PS-2′-OMe (79% reduction) sense strand duplexed to a PS-modified anti-sense strand were as efficacious as the parent compound (IB_4, 75%). The greatest potency was observed with the PS-2′-OMe sense strand duplexed to a PO anti-sense strand (IB_3; IC50 of 7 nM) or a PS anti-sense strand (IB_6; IC50 of 10 nM).
The unmodified blunt-end 20-bp duplex directed against Site III (IIIB_1) achieved a similar efficacy (75% reduction) as seen with the unmodified parent at Site I (IB_1) but was more potent with an IC50 of 3 nM (Fig. 3B ▶). The modified duplexes directed against Site III showed a 65%–75% reduction of PTEN mRNA when a PO anti-sense strand was duplexed to either a PO-2′-OMe (IIIB_2) or a PS-2′-OMe sense strand (IIIB_3). Modified duplexes comprised of a PS anti-sense strand and PO sense strand (IIIB_4) showed only a modest 25% reduction in target mRNA. Activity increased to 50% and 60% reduction at the 75 nM dose when the PS-modified anti-sense strand was duplexed to either a PO-2′-OMe (IIIB_5) or a PS-2′-OMe sense strand (IIIB_6), respectively. Overall, siRNAs containing a PS anti-sense strand consistently displayed lower activity at Site III in comparison to Site II, independent of construct design.
To complete evaluation of these modifications in the context of the blunt-end 20-bp duplex, we also evaluated constructs to Site I with full 2′-OMe modifications in the anti-sense strand, or both the anti-sense strand and sense strand (Fig. 3C ▶). Consistent with results reported elsewhere (Elbashir et al. 2001b; Amarzguioui et al. 2003; Braasch et al. 2003; Chiu and Rana 2003; Czauderna et al. 2003), duplexes containing either a PO-2′-OMe anti-sense strand (Ias_1, Ias_2, Ias_5, and Ias_6) or a PS-2′-OMe anti-sense strand (Ias_3, Ias_4, Ias_7, and Ias_8) were inactive and failed to reduce PTEN mRNA.
Our results indicate that 20-bp blunt-end duplexes comprised of fully modified 2′-OMe sense strands can mediate target mRNA reduction, and that complete substitutions with phosphorothioates in either or both strands is tolerated. As expected, duplexes containing a full 2′-OMe-modified sense strand displayed improved resistance against serum nucleases (Czauderna et al. 2003), as did PS-modified strands, when compared to unmodified duplexes (data not shown).
Taken together, these data suggest that activity of the modified siRNAs with a full 2′-OMe sense strand is predominantly dependent on construct design as suggested by the activity seen with the blunt-end 20 bp across the four PTEN sites (cf. Figs. 2 ▶ and 3 ▶). Sequence and backbone composition appear to be secondary determinants of activity.
Modified siRNA activity is attenuated by ASO-mediated knock-down of hAgo2
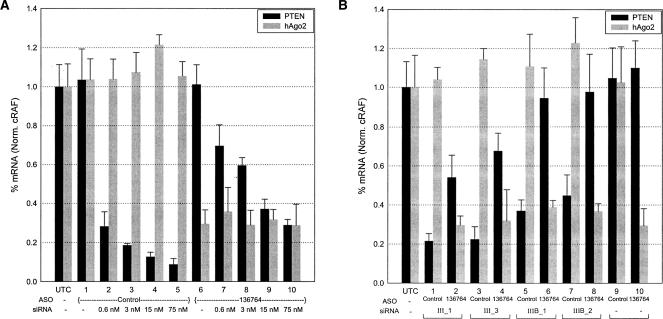
We next sought to address whether the fully modified siRNA duplexes containing 2′-OMe sense strands function through the RISC complex. As a first measure, we took advantage of the RNase H-based anti-sense oligodeoxynucleotide (ASO) technology to ascertain hAgo2’s role in 2′-OMe-modified siRNA-mediated target mRNA reduction (Bennett and Cowsert 1999; Ravichandran et al. 2004). In this assay, mRNA expression levels of hAgo2 were first reduced by treatment of cells with a complementary RNase H-based ASO (ISIS 136764). ASO-treated cells were incubated for 48 h to allow for hAgo2 protein to be eliminated through normal degradative processes, after which cells were transfected with siRNAs to determine the effect of hAgo2 knock-down on their activity.
The hAgo2 knock-down assay was validated by performing a dose response analysis of the unmodified 19-bp 3′-dTdT siRNA to Site I (I_1). HeLa cells were treated with either an ASO targeting hAgo2 (ISIS 136764) or a random control ASO (ISIS 129686), prior to siRNA administration, as described in Materials and Methods. The unmodified siRNA (I_1) to Site I displayed a dose-dependent reduction of target mRNA achieving ~80%–90% knock-down following treatment with the random control ASO (ISIS 129686), while hAgo2 levels remained unchanged (Fig. 4A ▶, lanes 2–5). Treatment with an ASO targeting hAgo2 (ISIS 136764) resulted in ~65%–70% knock-down of hAgo2 mRNA and a concomitant attenuation of siRNA (I_1) activity (Fig. 4A ▶, lanes 7–10). Attenuation of siRNA activity was less pronounced at the higher dose points, suggesting that, in the presence of excess siRNA and incomplete hAgo2 knockdown, sufficient RISC complexes form to result in target PTEN mRNA reduction (70%, lane 10), but to a lesser extent than that observed in the control treated cells (92%, lane 5). These data indicate that hAgo2 is a rate-determining step for siRNA-mediated target mRNA reduction, and that the RNase H-mediated hAgo2 knock-down assay provides a useful tool for examining the mechanistic basis of activity for modified siRNA constructs.
FIGURE 4.
Modified siRNA activity is attenuated by knock-down of hAgo2. (A) Validation of ASO knock-down assay in HeLa cells. siRNA (I_1) was transfected into HeLa cells at a dose of 0.6–75 nM post-treatment with either the control ASO (ISIS 129686) or the ASO specific for hAgo2 (ISIS 136764). (B) Effect of hAgo2 knock-down on activity of siRNAs containing a full 2′-OMe-modified sense strand. Modified and unmodified siRNAs targeting PTEN Site III were transfected (75 nM) into HeLa cells previously treated with ISIS 129686 or ISIS 136764. PTEN and hAgo2 mRNA levels were determined relative to the cRAF mRNA level by real-time RT-PCR (TaqMan). The results shown represent the percentage of untreated control expression (UTC). Each bar represents the mean of three biological replicates (±standard deviation). The control ASO (ISIS 129686) is a random sequence that has no significant homology to the human genome.
We next asked whether full 2′-OMe-modified siRNA constructs require hAgo2 for target mRNA reduction. Four constructs targeted to Site III (III_1, III_3, IIIB_1, and IIIB_2) were transfected into HeLa cells following treatment with either the random control ASO (ISIS 129686) or the hAgo2-specific ASO (ISIS 136764). As shown in Figure 4B ▶, both the unmodified 19-bp 3′-dTdT (III_1) and the 20-bp blunt-end (IIIB_1) constructs displayed a loss of activity upon knock-down of hAgo2 levels. These data are in agreement with recent reports indicating that hAgo2 is the catalytic engine responsible for RNAi-mediated activity (Liu et al. 2004; Meister et al. 2004; Rivas et al. 2005).
Loss of activity was also observed with the fully modified 2′-OMe counterparts (III_3 and IIIB_2) with ASO-mediated knock-down of hAgo2 (Fig. 4B ▶). This result suggests that fully modified 2′-OMe constructs can enter the RNAi pathway and, furthermore, reduction of target mRNA is dependent on the abundance of hAgo2. The effect of hAgo2 knockdown on siRNA activity was not dependent on siRNA sequence (cf. Fig. 4, A and B ▶) or construct design (Fig. 4B ▶).
Interaction of modified siRNAs with overexpressed hAgo2
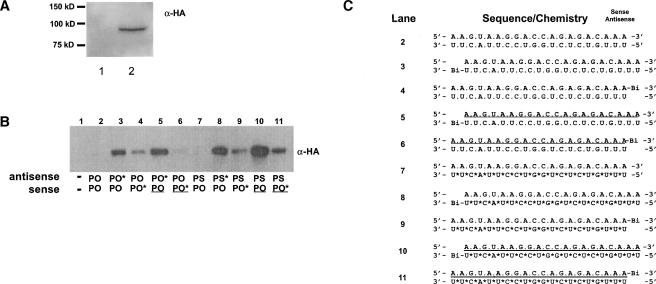
Continuing our mechanistic analysis of siRNAs containing fully modified strands, we next examined the binding properties of these duplexes with the RISC complex, specifically hAgo2. For this purpose, we generated a construct that expressed hAgo2 with an N-terminal HA epitope (pE2-N_HA). Duplexed siRNAs that contained a biotin tag on the 3′-terminus of either the anti-sense or the sense strand were then transfected into HeLa cells transiently transfected with the pE2-N_HA construct. The biotinylated strand was captured from total cell lysates under nondenaturing conditions by affinity purification, and then probed for the presence of HA-hAgo2 by Western blot. α-HA monoclonal antibodies recognized a single protein in crude HeLa extracts following transfection with the plasmid pE2-N_HA (Fig. 5A ▶).
FIGURE 5.
Modified siRNAs interact with overexpressed hAgo2. (A) Characterization of pE2-N_HA. Total HeLa lysates prepared from untreated cells (lane 1) and cells expressing HA-hAgo2 (lane 2) were separated by SDS-PAGE and immunoblotted with anti-HA antibodies. The migration of the molecular mass standards is shown on the left. (B) Western blot analysis of biotin-siRNA samples isolated from HeLa cells expressing HA-hAgo2. Biotin-tagged strands were isolated from total cell lysates, as described in Materials and Methods. Isolates were then subjected to SDS-PAGE and Western blotted with anti-HA antibody. (C) Structure and sequence of siRNA constructs included in analysis. (*) A PS modification, (underline) 2′-OMe-modified nucleotides, (Bi) biotin residue.
In the first analysis, we compared the binding ability of unmodified siRNAs to those containing a full 2′-OMe-modified sense strand (Fig. 5B,C ▶). HA-hAgo2 was not isolated from cells transfected with siRNAs lacking a biotin residue (Fig. 5B,C ▶, lanes 2,7) or the vector (pE2-N_HA) alone (data not shown). HA-hAgo2 was isolated, however, when the anti-sense strand of the duplex contained a 3′ biotin moiety, regardless of the sense strand makeup. In other words, the strand containing unmodified ribose residues was bound to hAgo2, indicating that an anti-sense strand duplexed to a full 2′-OMe sense strand can be loaded into RISC. Surprisingly, we found substantially less hAgo2 isolated when the biotin moiety was located on the 3′-end of either unmodified or modified sense strands. In addition, we found that a PS-modified AS strand bound more hAgo2 than the corresponding PO strand. This is likely a reflection of the enhanced protein-binding properties of a PS modification (Eckstein 2000).
Recent reports in the literature have shown that whichever strand is most easily unwound in a 5′–3′ direction will be preferentially loaded into RISC (Khvorova et al. 2003; Schwarz et al. 2003). The thermodynamic profile of the siRNA targeting Site I is similar for the 5′-end of the anti-sense strand and the 5′-end of the sense strand, as each end contains four A:U bp and 1 G:C bp. This would have predicted that either strand could load equally well into the RISC complex. However, as shown in Figure 5B ▶, the anti-sense strand is preferentially loaded. It is of interest to note that the sense strand is extremely purine rich (80%), and by default the anti-sense strand is pyrimidine rich.
In a second analysis, we investigated siRNAs that contained a full 2′-OMe anti-sense strand to determine if the lack of activity observed in cell culture results from a decrease in binding to hAgo2 (Fig. 6 ▶). As demonstrated in the previous analysis, HA-hAgo2 was not isolated with siRNAs lacking a biotin residue (lanes 1,4), but was isolated when the RNA anti-sense strand (either PO or PS) of the duplex contained a 3′ biotin moiety (lanes 2,5). HA-hAgo2 was also isolated when a full 2′-OMe anti-sense strand (either PO or PS) of the duplex contained a 3′ biotin moiety, although in substantially lower amounts compared to the respective parent siRNA (cf. lanes 2 and 3 and lanes 5 and 6). Therefore, as observed with the full 2′-OMe sense strand, the full 2′-OMe anti-sense strand is not effectively loaded and/or is not as tightly associated with the HA-hAgo2/RISC complex. In other words, the bulky methyl group may interfere with loading or binding to hAgo2.
FIGURE 6.
Reduced association of full 2′-OMe anti-sense strands with overexpressed hAgo2. (A) Western blot analysis of biotin-siRNA isolated from HeLa cells expressing HA-hAgo2. Samples were subjected to SDS-PAGE and Western blotted with anti-HA antibody, as described in Materials and Methods. (B) Structure and sequence of siRNA constructs included in analysis. (*) A PS modification, (underline) 2′-OMe-modified nucleotides, (Bi) biotin residue.
Modified siRNAs enter into the hAgo2/RISC complex to mediate target RNA cleavage
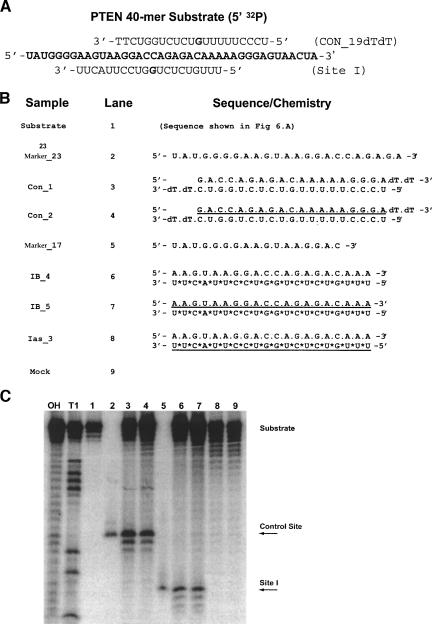
We used an hAgo2-based in vitro RNA cleavage assay to confirm that initial cleavage of the targeted RNA by the modified 2′-OMe duplexes is a direct consequence of hAgo2/RISC activity (Martinez et al. 2002; Liu et al. 2004; Song et al. 2004; Rivas et al. 2005). In brief, HeLa cells were first transfected with pE2-N_HA, to allow for expression of HA-hAgo2, and then 24 h later were transfected with the siRNAs. RISC complexes were allowed to assemble endogenously for an additional 24 h, after which cells were lysed and the HA-hAgo2 RISC complexes were immunoprecipitated with the anti-HA antibody. Following isolation of the hAgo2 RISC complex, an RNA cleavage assay was performed using a 40-nt 32P-5′-labeled RNA substrate corresponding to the region containing Site I (Fig. 7A ▶).
FIGURE 7.
Modified siRNAs enter into the hAgo2/RISC complex to mediate target RNA cleavage. (A) A 5′-[P32] end-labeled 40-mer substrate containing Site I was incubated with immunopurified RISC complexes, as described in Materials and Methods. The sequence of siRNA constructs and lane assignments are shown in B. The sequence of each RNA marker corresponds to the expected cleavage product of 23 nt (control site) or 17 nt (Site I). (*) A phosphorothioate modification, (underline) 2′-OMe-modified nucleotides. (C) Cleavage products were resolved on a 12% polyacrylamide 7 M urea gel. The 5′ cleavage products are indicated by arrows. Sequence identity was assigned according to the control digestions of the substrate by RNase T1, base (OH) hydrolysis, and synthetic RNA markers.
Constructs containing a PS anti-sense and a PO sense strand (IB_4) or a PS antisense and a PO_2′-OMe sense strand (IB_5) were selected for the analysis (Fig. 7B ▶). To confirm the sequence specificity of RNA substrate cleavage, we introduced a pair of 19-bp 3′-dTdT control siRNAs that were complementary to a different region of the RNA substrate and consequently expected to elicit cleavage at a different site.
As shown by PAGE analysis (Fig. 7C ▶), incubation of the 5′-end-labeled RNA substrate with the HA-hAgo2 RISC complex isolated from cells treated with either the unmodified control siRNA (Con_1, lane 3) or control siRNA containing a fully modified 2′-OMe sense strand (Con_2, lane 4) yielded the expected 23-nt product, where cleavage occurred between nucleotides 10 and 11 from the 5′-end of the control anti-sense strand. This assignment was verified with a 23-nt synthetic RNA marker of the same sequence as the expected product (Marker_23, lane 2). Similarly, incubation of the 5′-end-labeled RNA substrate with HA-hAgo2 RISC complexes purified from IB_4- and IB_5-treated cells also yielded the expected 17-nt cleavage product (lanes 6,7), as confirmed by a 17-nt synthetic RNA marker of equivalent sequence (Marker_17, lane 5). In this assay, we also noted that the HA-hAgo2 RISC complex isolated from cells treated with siRNA duplexes containing a full 2′-OMe anti-sense strand (Ias_3) were inactive, showing no cleavage products (lane 8). These results indicate that modified siRNAs with a full 2′-OMe sense strand are capable of cleaving mRNA in a sequence-dependent manner and that this activity is mediated by hAgo2.
DISCUSSION
Using a combination of cell culture and biochemical assays, we investigated the sequence and structure activity relationships of siRNAs containing full-strand PS backbones and 2′-OMe sugar modifications in two types of constructs, the canonical 19-bp RNA duplex with 3′-dTdT overhangs and a 20-bp RNA duplex with blunt ends. In contrast to previous reports, we found that siRNAs with a fully modified 2′-OMe sense strand can reduce target mRNA levels with similar efficacy and potency as the unmodified parent duplexes.
To better understand the mechanism of activity for si-RNAs containing fully modified 2′-OMe sense strands, we sought to determine if the mRNA knock-down seen with modified siRNAs was dependent on the RISC complex, as would be expected for an siRNA-mediated mechanism. We, and others, have found that hAgo2 is required for siRNA-mediated knock-down (Liu et al. 2004; Meister et al. 2004; Rivas et al. 2005), as inhibition of hAgo2 expression, by ASO/RNase H-mediated targeting, attenuated siRNA activity. The requirement for endogenous hAgo2 was not dependent on siRNA construct design or modifications, as attenuation of siRNA-mediated target reduction was seen with both unmodified 3′-dTdT and 20-bp blunt-end duplexes as well as with the fully modified 2′-OMe sense counterparts. Furthermore, using a siRNA biotin capture assay from HeLa cells, we demonstrated that hAgo2 is predominantly isolated with the unmodified anti-sense strand (of the duplex) containing a 3′-biotin moiety. These results suggest that siRNA duplexes containing full 2′-OMe sense strands can be recognized by the RNAi pathway and that the anti-sense strand is loaded into the RISC complex to elicit hAgo2-dependent siRNA-mediated target reduction.
Interestingly, the primary determinant of activity for these modified siRNAs turned out to be the type of construct, and not the sequence or target site. Indeed, we found that only one of the four sites of siRNA activity, previously identified in a screen for active PTEN siRNA (Vickers et al. 2003), supports activity of a 19-bp 3′-dTdT construct with a full 2′-OMe-modified sense strand, whereas all four sites supported activity in the context of the 20-bp blunt-end construct. The influence of construct design on activity of the 2′-OMe-modified siRNAs may be the basis for the lack of activity observed by those investigators who previously assessed modifications in the context of the canonical 19-bp RNA duplex with 3′-dTdT overhangs (Elbashir et al. 2001b; Amarzguioui et al. 2003; Braasch et al. 2003; Chiu and Rana 2003; Czauderna et al. 2003). Minimal activity of a 21-bp blunt-end siRNA with an unmodified anti-sense strand and a full 2′-OMe-modified sense strand was noted in one of these earlier reports (Czauderna et al. 2003). However, it is not clear as to the basis of the minimal activity observed in this structure activity relationship (SAR) study, other than the differences in siRNA construct length and target sequence.
The requirements for tolerance of full-strand 2′-OMe modifications is not immediately evident, as not all fully modified 2′-OMe duplexes display RNAi-mediated activity. We can only speculate as to the underlying causes that, in addition to construct design discussed above, may relate to unwinding of the modified duplex, loading of the unmodified anti-sense strand into RISC, sequence accessibility of the target site, or a combination, thereof.
The introduction of 2′-OMe modifications into the siRNA may introduce structural changes or alter the thermostability of the duplex that could interfere with either recognition or unwinding activity of the predominant, yet to be identified, RISC helicase. Either strand of the siRNA duplex may function as a guide that localizes RISC to the target RNA via Watson-Crick base pairing (Nykanen et al. 2001; Martinez et al. 2002, 2003). Prior to incorporation into RISC, the strands of the siRNA duplex must be unwound in a step that is thought to require an ATP-dependent helicase. Duplexes containing a full 2′-OMe sense strand may require an alternative helicase, or additional factors, to assist in unwinding the modified duplex, which, in turn, may preferentially recognize blunt-end 20-bp constructs. By example, eIF4A has been shown to bind full 2′-OMe substrates equally as well as unmodified substrates (Peck and Herschlag 1999). Its ATPase activity was, however, below detectable levels when bound to the modified substrates. This result suggests that the introduction of the 2′-OMe nucleotides interferes with the helicase ATPase-dependent activities (Peck and Herschlag 1999).
Alternatively, siRNAs containing a fully modified 2′-OMe sense strand may require additional factors to facilitate loading of the anti-sense strand into the minimal RISC complex. Multiple pathways have already been identified for loading various substrates into RISC depending on their origin and function. For example, although humans and Caenorhabditis elegans encode only one Dicer, Drosophila encodes two distinct Dicers with different functions in the RNAi pathway (Lee et al. 2004). Dcr-2, which contains a helicase domain, binds to siRNA in association with R2D2 and facilitates loading into RISC (Liu et al. 2003), whereas Dcr-1 is likely required for miRNA loading (Lee et al. 2004; Pham et al. 2004). In addition, Dcr-2 may require the canonical 19-bp 3′-dTdT overhang design for effective recognition and RISC loading in Drosophila, and likewise may be unable to unwind a 2′-OMe-modified siRNA duplex as previously demonstrated in a Drosophila lysate study (Elbashir et al. 2001b). It is likely that similar ancillary “loading” factors exist within the human genome and await identification. In this regard, a possible human homolog of R2D2 has recently been identified (TRBP) that is required for the recruitment of hAgo2 to the siRNA bound by Dicer, which may, in a similar fashion as R2D2, facilitate RISC assembly (Chendrimada et al. 2005).
Sequence accessibility of the target mRNA may account for some of the variation between the different sites due to the secondary structure present in RNA (Schubert et al. 2005). However, it is puzzling that certain sites with highly active unmodified siRNAs display very limited activity (or none at all) with full 2′-OMe-modified duplexes. In this regard, one would expect the site to be accessible because of the activity of the unmodified duplex. It is therefore unlikely that sequence accessibility of the target mRNA is a major determinant of activity with regard to fully modified 2′-OMe duplexes.
Although the mechanistic details regarding activity of 2′-OMe-modified constructs have not been completely elucidated, we can take advantage of the potential benefits of introducing a full 2′-OMe sense strand into an siRNA duplex to mediate RNAi target knock-down, as we have shown these compounds to be active. Exogenously introduced phosphodiester oligoribonucleotides are inherently sensitive to nucleases, greatly limiting their use as therapeutic agents. By incorporating a full 2′-OMe sense strand in combination with PS modifications, the siRNA duplex will benefit from an increased resistance to degradation by nucleases, as well as prolonged serum retention, and therefore may display improved pharmacokinetic properties in vivo. In addition, use of modified siRNA duplexes may lead to increased specificity by eliminating sense-strand-induced off-target gene silencing.
Certain siRNAs can discriminate between single nucleotide changes in mRNA targets (Elbashir et al. 2001a; Ding et al. 2003); however, gene expression profiling has revealed that siRNA treatment can lead to off-target silencing effects when partial complementation to unintended targets exists in either strand of the duplex (Jackson et al. 2003; Semizarov et al. 2003; Scacheri et al. 2004). We found that siRNA analogs containing the full 2′-OMe anti-sense strand, instead of the 2′-OMe sense strand, did not elicit RNAi-mediated target mRNA reduction. Nor was cleavage seen in a HeLa lysate cleavage assay using a full 2′-OMe anti-sense strand (Chiu and Rana 2003; this study). In addition, we have shown in this report that fully modified 2′-OMe strands may inhibit RISC loading, as there is a significant reduction in levels of HA-hAgo2 associated with fully modified strands. Given that a full 2′-OMe strand is not an acceptable substrate for RISC, as demonstrated by siRNAs with a full 2′-OMe-modified anti-sense strand, we propose that usage of modified sense strands will minimize or eliminate off-target gene silencing that results from this strand.
It is worth noting that siRNAs with alternating 2′-OMe and unmodified (2′-OH) nucleotides (Czauderna et al. 2003) or alternating 2′-OMe and 2′-F nucleotides (Allerson et al. 2005) have potencies similar to or exceeding that of the unmodified parent constructs. The >500-fold increase in potency, observed with the 2′-OMe_2′-F alternate constructs at certain sites, merits further analysis of the basis of target RNA reduction by this class of modified siRNAs.
In summary, we have examined the silencing efficiencies of various siRNAs with regard to construct design, target site, and sugar-phosphate backbone composition. Although the underlying basis of the effect of construct design and sequence on the activity of modified siRNAs has yet to be fully understood, one may conclude that usage of a single design and/or sequence is not sufficient when defining the scope and limitations of a given modification for usage in the RNAi pathway. Finally, for the 2′-OMe sense strand-modified siRNA constructs under study, we have shown that target RNA reduction and cleavage by this class of double-strand constructs is mediated by hAgo2/RISC.
MATERIALS AND METHODS
Preparation of synthetic siRNAs
Synthetic siRNAs, including those containing a 3′-biotin label, were manufactured by Dharmacon Research, Inc. Anti-sense strands were synthesized with a 5′-phosphate group, with the exception of the unmodified 3′-dTdT RNAs. Sequences of the oligoribonucleotides and their respective modifications are shown in Table 1 ▶. siRNA duplexes were formed according to the manufacturer’s instructions. In brief, 1.6 μL of a 250 μM anti-sense stock was combined with 1.6 μL of a 250 μM sense stock, 4 μL of 5× universal buffer (500 mM potassium acetate, 150 mM HEPES-KOH at pH 7.4, 10 mM magnesium acetate), and 12.8 μL of ultrapure water, followed by heating at 90°C for 1 min. The reaction was then allowed to cool to ambient temperature. The final concentration of the duplex was 20 μM in 1× universal buffer (100 mM potassium acetate, 30 mM HEPES-KOH at pH 7.4, 2 mM magnesium acetate).
Cell culture and transfections
The HeLa cell line (CCL-2) used in these experiments was obtained from the American Type Culture Collection (Manassas, VA) and was cultured in Dulbecco’s modified Eagle’s medium (high glucose) (DMEM; GIBCO BRL) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and Penicillin-Streptomycin (Invitrogen). Then 24-well plates (Becton Dickinson) were seeded at an initial density of 45,000 cells/well on the day prior to transfection and incubated at 37°C and 5% CO2. Synthetic siRNA was delivered to cells (typically at 65%–75% confluency) by using Lipofectin reagent (Invitrogen) according to the manufacturer’s instructions. siRNA duplexes were incubated with 6 μg/mL Lipofectin per 100 nM siRNA at indicated concentrations in serum-free and antibiotic-free OptiMEM medium (Invitrogen) for 15 min and then added to each well. After 4 h at 37°C and 5% CO2, the medium was aspirated from the cells and replaced with DMEM containing 10% FBS and antibiotics. Cells were returned to 37°C and 5% CO2 until harvested.
RNA isolation and RT-PCR
Total RNA was isolated 16–24 h post-transfection (unless otherwise noted) on an RNeasy 3000 BioRobot (QIAGEN). In brief, 150 μL of RLT Buffer supplemented with 1% β-mercaptoethanol was added to each well of a 24-well plate. The samples were then transferred to a 96-well plate for RNA isolation according to the manufacturer’s protocol (QIAGEN). Reduction of target mRNA expression was determined by real-time RT-PCR using an ABI Prism 7700 Sequence Detector (Applied Biosystems). Reverse transcription was performed for 30 min at 48°C followed by PCR (40 cycles of 30 sec at 94°C and 1 min at 68°C) in a total volume of 25 μL. Unless noted, PTEN mRNA expression levels were normalized to c-raf mRNA levels and/or Ribogreen (Invitrogen). The gene specific forward (F), reverse (R), and probe sequences (P) are as follows. c-raf kinase (accession no. X03484): F, 5′-AGCTTGGAAGACGATCAGCAA; R, 5′-AAACTGCTGAACTATTGTAGGAGAGATG; P, 5′-6-FAM-AGATGCCGTGTTTGATGGCTCCAGC-TAMRA. PTEN (accession no. U92436): F, 5′-AATGGCTAAGTGAAGATGACAATCAT; R, 5′-TGCACATATCATTACACCAG-TTCGT; P, 5′-6-FAM-AGATGCCGTGTTTGATGGCTCCAGCTAMRA. Primer/probe sets were synthesized by Applied Biosystems.
hAgo2 ASO knock-down assay
HeLa cells were treated with either 100 nM of a random control ASO (ISIS 129686) that had no effect on hAgo2 protein levels, or an equivalent amount targeted to hAgo2 (ISIS 136764). The PS-modified ASO used in these experiments contains 10 2′-O-(2-methoxyethyl) nucleosides (2-MOE), with five on the 5′-end and the remaining five at the 3′-end of the ASO, that serve to increase binding affinity to the target RNA and provide extended nuclease resistance (Altmann et al. 1996; Freier and Altmann 1997), while the central region contains 10 2′-deoxynucleosides required to support RNase H cleavage of the target mRNA (Wu et al. 2004). Then, 48 h later, cells were transfected with siRNA duplexes as indicated using Lipofectin. On average, hAgo2 RNA levels were reduced 60%–70% upon treatment with the hAgo2-specific ASO, ISIS 136764. Total RNA was isolated 24 h (post-siRNA treatment) to determine the effect of hAgo2 knock-down on siRNA activity.
N-terminal hAgo2 plasmid construction
cDNA encoding full-length human hAgo2 was generated by RT-PCR from HeLa-extracted total RNA. hAgo2 with an N-terminal HA epitope was generated by PCR with the following primers: phCMV2–5′_E2 (5′-TAACAATGTACCCATACGATGTTCCGGATTACGCTTACTCGGGAGCCGGCCCCGCACTTG-3′) and phCMV2–3′_E2 (5′-CCCGGGCCCGCGGTACCGTCGACTGCAGAATTAAGCAAAGTACATGGT GCGCAGAGTGT-3′). Plasmid pE2-N_HA was constructed by homologous recombination of the hAgo2 PCR product generated from the HeLa cDNA with a linear phCMV-2/Xi vector according to the manufacturer’s protocol (Gene Therapy Systems).
siRNA biotin capture assay
HeLa cells (CCL-2) from the American Type Culture Collection (Manassas, VA) were seeded at an initial density of 1,000,000 cells/100 mm plate (Becton Dickinson) on the day prior to transfection and incubated at 37°C and 5% CO2. Plasmid pE2-N_HA was delivered to cells (typically at 70%–75% confluency) by using Effectene transfection reagent (QIAGEN) according to the manufacturer’s instructions and incubated for 24 h at 37°C and 5% CO2. Synthetic siRNA (75 nM) containing a 3′-biotin modification and a 5′-phosphate group was then delivered to cells (typically at 80%–85% confluency) by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. After 4 h at 37°C and 5% CO2, the media was aspirated from the cells and replaced with DMEM containing 10% FBS and antibiotics and returned to 37°C and 5% CO2 until cells were harvested 18 h later. Briefly, cells were treated with Trypsin and harvested, then washed with 1 mL of cold PBS, and centrifuged at 1000g. The cell pellet was resuspended in 200 μL of lysis buffer (150 mM NaCl, 0.5% NP-40, 2 mM MgCl2, 2 mM CaCl2, 20 mM Tris at pH 7.5) that contained Complete Mini Protease Inhibitor Tablet (Roche) and 1 mM DTT, then passaged through a 1-cc U-100 Insulin Syringe (Becton Dickinson) 10 times, followed by a 10-min 10,000g clearing spin. The clarified lysate was normalized according to the manufacturer’s protocol for Dc Protein Assay (BioRad). Equivalent amounts of starting material were transferred to a new Eppendorf tube containing 30 μL of immobilized NeutrAvidin (Pierce), 3 μL of RNase out (Invitrogen), and lysis buffer up to the final 1-mL volume. Beads were rotated end over end for 2 h, then washed three times in wash buffer (500 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 20 mM Tris at pH 7.5). Following the final wash, the samples were eluted from the beads in 50 μL of 2% SDS at 100°C, and resolved by SDS-4%–12% Bis-Tris PAGE (NUPAGE; Invitrogen), transferred to a 0.2-μm PVDF membrane (Invitrogen), and analyzed by immunoblotting with affinity-purified monoclonal anti-HA antibodies, HA.11 (Covance).
RISC cleavage assay
HeLa cells were seeded at an initial density of 1,000,000 cells/100-mm plate (Becton Dickinson) on the day prior to the initial transfection and incubated at 37°C and 10% CO2 in DMEM containing 10% FBS. The next day, plasmid pE2-N_HA was delivered to cells (typically at 70%–75% confluency) by using Effectene transfection reagent (QIAGEN) according to the manufacturer’s instructions and incubated for 24 h at 37°C. Synthetic siRNA (75 nM) was then delivered to cells (typically at 80%–85% confluency) by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions, and incubated at 37°C for 18 h. Cells from three 100-mm plates per siRNA test were combined into a 15-mL conical tube upon harvesting with Trypsin, then washed with 1 mL of cold PBS and centrifuged at 1000g. The cell pellet was resuspended in 500 μL of lysis buffer (150 mM NaCl, 0.5% NP-40, 2 mM MgCl2, 2 mM CaCl2, 20 mM Tris at pH 7.5) that contained a Complete Mini Protease Inhibitor Tablet (Roche) and 1 mM DTT, then passed through a 1-cc U-100 Insulin Syringe (Becton Dickinson) 10 times, followed by a 10-min 10,000g clearing spin. The clarified lysate was transferred to a new Eppendorf tube containing 15 μL of immobilized HA.11 beads (Covance), 450 μL of lysate, and lysis buffer up to a final 1-mL volume. Beads were rotated end over end for 2 h, then washed three times in wash buffer (500 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 20 mM Tris at pH 7.5), followed by one wash with cleavage buffer (100 mM KCl, 2 mM MgCl2, 10 mM Tris at pH 7.5). Following the final wash, the immunoprecipitated RISC complexes were incubated for 2 h at 37°C in a heated mixer. Final reaction volume was 30 μL containing 5 U/mL RNasin Ribonuclease Inhibitor (Promega), 1 mM ATP, 0.2 mM GTP, and labeled target substrate in cleavage buffer. The reaction was stopped by the addition of TRIZOL (Invitrogen), followed by RNA extraction according to the manufacturer’s protocol and PAGE analysis.
Antibodies and immunoblotting
Cell lysates were harvested from duplicate wells of a six-well plate (Becton Dickinson) treated with 100 nM ASO at the indicated time points. Briefly, cells were scraped into 1 mL of PBS and centrifuged at 1000g. The cell pellet was resuspended in 150 μL of RIPA lysis buffer (150 mM NaCl, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris at pH 8.0) and passaged through a QIAGEN DNA Shredder column (QIAGEN), followed by a 5-min 14,000g clearing spin. The clarified lysate was normalized according to the manufacturer’s protocol for Dc Protein Assay (BioRad). Equivalent amounts of starting material (10 μg/lane) were resolved by SDS-4%–12% Bis-Tris PAGE (NUPAGE; Invitrogen), transferred to a 0.2-μm PVDF membrane (Invitrogen), and analyzed by immunoblotting with affinity-purified monoclonal anti-HA antibodies, HA.11 (Covance).
Acknowledgments
We thank Christy Esau, Tim Vickers, Youngsoo Kim, Stan Crooke, and Frank Bennett for helpful discussions and critical review of the manuscript; and Tracy Reigle for assistance with graphics.
Abbreviations
RNAi, RNA interference
siRNAs, short interfering RNAs
PS, phosphorothioate
2′-OMe, 2′-O-methyl
2′-MOE, 2′-O-2-methoxyethyl
RISC, RNA-induced silencing complex
ASO, anti-sense oligonucleotides
SAR, structure activity relationship
CDS, coding sequence
PAGE, polyacrylamide gel electrophoresis
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2150806.
REFERENCES
- Allerson, C.R., Sioufi, N., Jarres, R., Prakash, T.P., Naik, N., Berdeja, A., Wanders, L., Griffey, R.H., Swayze, E.E., and Bhat, B. 2005. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 48: 901–904. [DOI] [PubMed] [Google Scholar]
- Altmann, K.H., Fabbro, D., Dean, N.M., Geiger, T., Monia, B.P., Muller, M., and Nicklin, P. 1996. Second-generation antisense oligonucleotides: Structure–activity relationships and the design of improved signal-transduction inhibitors. Biochem. Soc. Trans. 24: 630–637. [DOI] [PubMed] [Google Scholar]
- Amarzguioui, M., Holen, T., Babaie, E., and Prydz, H. 2003. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, C.F. and Cowsert, L.M. 1999. Antisense oligonucleotides as a tool for gene functionalization and target validation. Biochim. Biophys. Acta 1489: 19–30. [DOI] [PubMed] [Google Scholar]
- Braasch, D.A. and Corey, D.R. 2002. Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry 41: 4503–4510. [DOI] [PubMed] [Google Scholar]
- Braasch, D.A., Jensen, S., Liu, Y., Kaur, K., Arar, K., White, M.A., and Corey, D.R. 2003. RNA interference in mammalian cells by chemically- modified RNA. Biochemistry 42: 7967–7975. [DOI] [PubMed] [Google Scholar]
- Caplen, N.J., Parrish, S., Imani, F., Fire, A., and Morgan, R.A. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. 98: 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada, T.P., Gregory, R.I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., and Shiekhattar, R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, Y.L. and Rana, T.M. 2003. siRNA function in RNAi: A chemical modification analysis. RNA 9: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke, S.T. 1999. Molecular mechanisms of action of antisense drugs. Biochim. Biophys. Acta 1489: 31–44. [DOI] [PubMed] [Google Scholar]
- Cummins, L.L., Owens, S.R., Risen, L.M., Lesnik, E.A., Freier, S.M., McGee, D., Guinosso, C.J., and Cook, P.D. 1995. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 23: 2019–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna, F., Fechtner, M., Dames, S., Aygun, H., Klippel, A., Pronk, G.J., Giese, K., and Kaufmann, J. 2003. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 31: 2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fougerolles, A., Manoharan, M., Meyers, R., and Vornlocher, H.P. 2005. RNA interference in vivo: Toward synthetic small inhibitory RNA-based therapeutics. Methods Enzymol. 392: 278–296. [DOI] [PubMed] [Google Scholar]
- Ding, H., Schwarz, D.S., Keene, A., el Affar, B., Fenton, L., Xia, X., Shi, Y., Zamore, P.D., and Xu, Z. 2003. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell 2: 209–217. [DOI] [PubMed] [Google Scholar]
- Eckstein, F. 2000. Phosphorothioate oligodeoxynucleotides: What is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 10: 117–121. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001a. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. 2001b. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20: 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Freier, S.M. and Altmann, K.H. 1997. The ups and downs of nucleic acid duplex stability: Structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 25: 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary, R.S., Yu, R.Z., and Levin, A.A. 2001. Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr. Opin. Investig. Drugs 2: 562–573. [PubMed] [Google Scholar]
- Grunweller, A., Wyszko, E., Bieber, B., Jahnel, R., Erdmann, V.A., and Kurreck, J. 2003. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 31: 3185–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001a. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Caudy, A.A., and Hannon, G.J. 2001b. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2: 110–119. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Vandenburgh, K., Manninga, H., Scaringe, S.A., Weber, K., and Tuschl, T. 2003. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 13: 83–105. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P.S. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21: 635–637. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., and Jayasena, S.D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216. [DOI] [PubMed] [Google Scholar]
- Layzer, J.M., McCaffrey, A.P., Tanner, A.K., Huang, Z., Kay, M.A., and Sullenger, B.A. 2004. In vivo activity of nuclease-resistant siRNAs. RNA 10: 766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., and Hannon, G.J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Lubini, P., Zurcher, W., and Egli, M. 1994. Stabilizing effects of the RNA 2′-substituent: Crystal structure of an oligodeoxynucleotide duplex containing 2′-O-methylated adenosines. Chem. Biol. 1: 39–45. [DOI] [PubMed] [Google Scholar]
- Manoharan, M. 2004. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 8: 570–579. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Elbashir, S.M., Harborth, J., Hossbach, M., Urlaub, H., Meyer, J., Weber, K., Vandenburgh, K., Manninga, H., et al. 2003. Analysis of mammalian gene function using small interfering RNAs. Nucleic Acids Res. Suppl.: 333. [DOI] [PubMed]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3: 737–747. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15: 185–197. [DOI] [PubMed] [Google Scholar]
- Nykanen, A., Haley, B., and Zamore, P.D. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321. [DOI] [PubMed] [Google Scholar]
- Peck, M.L. and Herschlag, D. 1999. Effects of oligonucleotide length and atomic composition on stimulation of the ATPase activity of translation initiation factor elF4A. RNA 5: 1210–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, J.W., Pellino, J.L., Lee, Y.S., Carthew, R.W., and Sontheimer, E.J. 2004. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94. [DOI] [PubMed] [Google Scholar]
- Rand, T.A., Ginalski, K., Grishin, N.V., and Wang, X. 2004. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. 101: 14385–14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran, L.V., Dean, N.M., and Marcusson, E.G. 2004. Use of antisense oligonucleotides in functional genomics and target validation. Oligonucleotides 14: 49–64. [DOI] [PubMed] [Google Scholar]
- Rivas, F.V., Tolia, N.H., Song, J.J., Aragon, J.P., Liu, J., Hannon, G.J., and Joshua-Tor, L. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12: 340–349. [DOI] [PubMed] [Google Scholar]
- Scacheri, P.C., Rozenblatt-Rosen, O., Caplen, N.J., Wolfsberg, T.G., Umayam, L., Lee, J.C., Hughes, C.M., Shanmugam, K.S., Bhattacharjee, A., Meyerson, M., et al. 2004. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. 101: 1892–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, S., Grunweller, A., Erdmann, V.A., and Kurreck, J. 2005. Local RNA target structure influences siRNA efficacy: Systematic analysis of intentionally designed binding regions. J. Mol. Biol. 348: 883–893. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D.N., and Fesik, S.W. 2003. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. 100: 6347–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.J., Smith, S.K., Hannon, G.J., and Joshua-Tor, L. 2004. Crystal structure of Argonaute and its implications for RISC Slicer Activity. Science 305: 1434–1437. [DOI] [PubMed] [Google Scholar]
- Soutschek, J., Akinc, A., Bramlage, B., Charisse, K., Constien, R., Donoghue, M., Elbashir, S., Geick, A., Hadwiger, P., Harborth, J., et al. 2004. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432: 173–178. [DOI] [PubMed] [Google Scholar]
- Vickers, T.A., Koo, S., Bennett, C.F., Crooke, S.T., Dean, N.M., and Baker, B.F. 2003. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 278: 7108–7118. [DOI] [PubMed] [Google Scholar]
- Wu, H., Lima, W.F., Zhang, H., Fan, A., Sun, H., and Crooke, S.T. 2004. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 279: 17181–17189. [DOI] [PubMed] [Google Scholar]