Abstract
RNA–RNA interactions play a crucial role at many different levels of the cellular metabolism such as plasmid replication control, viral encapsidation, or transcriptional and translational regulation. Therefore, methods are necessary to investigate the molecular determinants of given interactions, including their stabilities, or to screen for new interacting partners. We designed an RNA-hybrid system in S. cerevisiae, based on the yeast three-hybrid system. In this setup, the activation of a reporter gene is dependent on the interaction of two RNAs. A loop–loop interaction similar to the dimerization initiation site of the HIV genome was used as a model system, demonstrating that in this novel RNA-hybrid system only cognate RNAs promote the activation of the reporter gene. Levels of reporter activation correlate well with interaction stabilities determined in vitro by UV melting analyses, suggesting that conditions used for the analysis of in vitro structural stabilities translate well into the intracellular environment. Furthermore, the system was applicable for a screen against a test library. Nine out of ten selected clones were identified as predicted interaction partners for the bait RNA. In summary, we present a yeast reporter system depending on RNA–RNA interactions, which can be used alternatively for analysis of known interactions or for screening libraries in search for new interaction partners.
Keywords: in vivo RNA–RNA interaction, loop–loop interaction, yeast three-hybrid system, intracellular RNA structural stability
INTRODUCTION
RNA–RNA interactions in cellular metabolism are gaining in prominence with the discovery that a large number of transcripts in higher eukaryotes are noncoding RNAs (ncRNAs). For example, in mouse cDNA libraries, almost half of the transcripts are ncRNAs (Mattick 2003). It is therefore anticipated that the functional RNAs already identified constitute only the tip of the iceberg. Frequently, the interaction partners of ncRNAs are not only proteins, but also other RNA molecules. Classic examples of interactions involving two RNA molecules are snRNAs (Forne et al. 1996); snoRNAs with their targets (Bachellerie et al. 2002); micro-RNAs from the RNAi pathway with their target mRNA(s) (Ambros 2004; Murchison and Hannon 2004); ncRNAs from Escherichia coli, also called sRNAs (Hershberg et al. 2003; Repoila et al. 2003); and loop–loop interactions (Brunel et al. 2002).
There is a need for efficient methods allowing the detection of RNA–RNA interactions and the screening of genomes for new targets of RNAs. In the past, the yeast two-and three-hybrid systems (Fields and Song 1989; SenGupta et al. 1996) proved essential in the examination of the networks of protein–protein and RNA–protein interactions. However, the building blocks necessary for the design of a similar system applicable to RNA–RNA interactions were not available until RNA by itself was demonstrated to be able to promote transcription (Sengupta et al. 1999; Buskirk et al. 2003; Saha et al. 2003). We reasoned that, as for their protein counterparts, it should be possible to separate the signal responsible for promoter localization (in this case, a MS2-RNA binding domain to a lexA-MS2 coat-protein fusion) from the activation domain.
Using this strategy, we designed a new system termed the “RNA-hybrid system.” With this new method, it should be possible to investigate the determinants of known RNA–RNA interactions in vivo. Furthermore, this system would also be suited for library screenings and the identification of unknown interaction partners of RNAs. To demonstrate the feasibility of this system, we applied it to the HIV-1 type dimerization signal loop–loop interactions.
Intermolecular loop–loop interactions, also called kissing complexes, are widely spread and serve a diverse range of biological functions. Loop–loop interactions are implicated in plasmid replication control and involve, for example, the RNAs CopA–CopT or RNA I–RNA II (Eguchi and Tomizawa 1990). Another well-known instance of kissing interaction is the dimerization initiation signal (DIS) loop responsible for initiation of the dimerization of the genomic HIV RNA (Paillart et al. 1996). This kissing complex between two DIS-loops is the initial transitory structure, precluding a refolding of the two molecules leading to the formation of an extended duplex between the two copies of the HIV genome (Laughrea and Jette 1996; Brunel et al. 2002; Windbichler et al. 2003). Our laboratory previously determined the thermodynamic parameters governing the DIS kissing complex (Weixlbaumer et al. 2004).
In this article, we show that the RNA-hybrid system can efficiently distinguish between cognate and noncognate RNAs from two different kissing complexes differing only in the interacting loops. The β-galactosidase activity obtained with the RNA-hybrid system correlates with the stability of the interaction as determined by physicochemical methods in vitro. This demonstrates that the nonphysiological conditions (1 M NaCl) used to quantify the stability of RNA structures in vitro translate well to the intracellular conditions. Furthermore, we confirm the potential of the RNA-hybrid system for the screening of new interaction partners of known RNAs.
RESULTS
Design of plasmids and transcripts
The RNA-based transcriptional activator (m26-11) was evolved by Buskirk et al. (2003) from a random library cloned in pIIIa MS2-2. This RNA molecule activates transcription in a yet unknown mechanism when tethered to the promoter of a reporter gene through the binding of the MS2-RNA to the LEXA-MS2 coat-protein fusion. In order to monitor RNA–RNA interactions, the simplest design imaginable was to separate the RNA transcriptional activator from the DNA tethering sequence (MS2 binding domain) and fuse both domains to interacting RNAs, allowing the formation of an activation complex only via the interaction of the two hybrid RNA molecules.
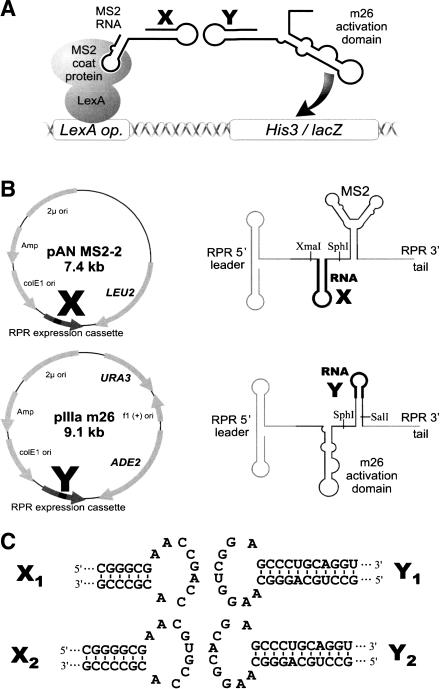
The basic strategy for the analysis of RNA–RNA interactions using the RNA-hybrid system is shown in Figure 1A ▶. Yeast strain YBZ-1, from the three-hybrid system (Bernstein et al. 2002; Hook et al. 2005), carries both lacZ and HIS3 reporter genes under the control of several lexA operators, and expresses constitutively the fusion protein LEXA-MS2 coat-protein. The bait RNA (RNA X–MS2-RNA) binds to the LEXA-MS2 coat-protein hybrid protein. If the prey RNA (RNA Y–m26) interacts with the bait, the m26 transcriptional activator is tethered to the promoter of the reporter gene and induces gene expression.
FIGURE 1.
Outline of the RNA hybrid system. (A) Schematic representation of the system. Yeast strain YBZ-1 expresses constitutively the fusion protein LexA-MS2 coat-protein. Both lacZ and HIS3 are under the control of multiple lexA operators. When two interacting hybrid RNAs are coexpressed, the MS2–RNA X hybrid binds to the MS2 coat-protein fusion and tethers the RNA Y–m26 fusion RNA to the promoter of lacZ and HIS3, thereby activating gene expression. (B) Overview of plasmids pAN MS2-2 and pIIIa-m26 used in the RNA hybrid system. RNA X and RNA Y can be cloned into pAN MS2-2 (respectively, pIIIa-m26) between the restriction sites XmaI and SphI (SphI and SalI). The schematic structure of the hybrid RNA is shown next to the corresponding plasmid with the positions of the respective restriction sites indicated. (C) Sequence and secondary structure of two pairs of RNAs used in this study.
Our principal concerns during the design of the RNA-hybrid vectors were RNA localization and folding. To promote colocalization of both RNAs in the nucleus, they are expressed from the same RNase P RNA (RPR) promoter used in the classical three-hybrid system. This ensures that both RNAs have identical promoter, leader, and 3′ tail sequences. Thus, we constructed plasmid pIIIa-m26 and plasmid pAN MS2-2 (Fig. 1B ▶). Plasmid pIIIa-m26 is a derivative of m26-11 in which the MS2-RNA binding sequence was deleted via simple excision of the SalI fragments containing the MS2-RNA tandem repeats. To construct pAN MS2-2, we cloned the RNA expression cassette from pIIIa MS2-2 in the shuttle vector pACTII. When inserting potential interaction partners in both plasmids, special care was taken to preserve the secondary structure of the m26 activation domain, as described below.
Loop–loop interactions
We used the HIV-1 type DIS-loop as a model for the analysis of RNA–RNA interactions using the RNA-hybrid system. The DIS-loop is a hairpin structure that contains a 6-nt self-complementary loop sequence flanked by a double and a single adenine (Paillart et al. 1996). Self-complementarity would be detrimental in this case, as it would lead to the formation of homodimers (X–X or Y–Y) instead of heterodimers (X–Y). We therefore use nonpalindromic loop sequences throughout this study. First, two different stem-loop pairs were cloned into pAN MS2-2 and pIIIa-m26. To guarantee that the transcribed RNAs fold correctly, we analyzed the secondary structure of different constructs using the mFOLD algorithm (Zuker 2003). This is of particular importance in the case of the activator domain m26, which is located at a three-way junction.
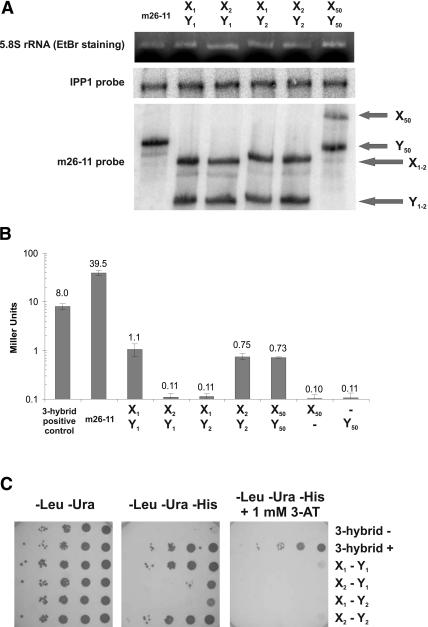
X1 and X2 stem-loops were fused to the MS2-RNA domain in pAN MS2-2. The loop of each X RNA contains 6 nt cognate to the respective Y RNA loop, preceded by two adenines and followed by one adenine (Fig. 1C ▶). The related Y1 and Y2 stem-loops were cloned in pIIIa-m26. To facilitate independent folding of the m26 domain and of the Y RNAs, we lengthened the stem originally comprising the MS2-RNA tandem repeats (Fig. 1C ▶). The stems containing X1 and X2 were chosen as to ensure correct folding. In the case of X2, the stem is 1 G–C bp longer than the X1 stem. We first tested the expression of both RNAs in yeast by Northern blot analysis with a probe hybridizing to both RNAs (Fig. 2A ▶). As expected, two bands of equal intensity are observed, indicating that both RNAs are expressed at the same level. We then tested the reporter system by using a quantitative β-galactosidase assay (Fig. 2B ▶). As can be seen, β-galactosidase activity is dependent on the coexpression of two cognate RNAs. When X1 (respectively, X2) is expressed in the presence of Y1 (Y2), the reporter gene activity is increased 10- fold over the background, while coexpression of Y2 (Y1) fails to activate lacZ expression. RNAs Y1 and Y2 only differ in the loop sequence (AAGGUCGGA and AAGGCACGA, respectively). Hence, the difference observed between lacZ expression when coexpressed with X1 and X2 can be attributed to the loop–loop interaction. In comparison to the three-hybrid system, the expression levels are low (eightfold reduction). Intermolecular reporter activation (X1–Y1) is 36-fold weaker than intramolecular activation (m26-11). This reduction is comparable with the 22-fold reduction in activity observed between the full-length GAL4 protein and the two fusion proteins interacting in the two-hybrid system (Fields and Song 1989).
FIGURE 2.
Analysis of loop–loop interactions with the RNA-hybrid system. (A) Northern blot analysis. For sample m26-11, only one RNA is expressed after cotransformation of yeast strain YBZ-1 with plasmids pIIIa-m26-11 MS2 and pACTII. In the other samples, plasmids derived from pAN MS2-2 and pIIIa-m26 containing the indicated RNA X and RNA Y were cotransformed. As controls, the band corresponding to the 5.8S rRNA after ethidium bromide staining of the gel and the band obtained after hybridization of the blot with a probe targeting the mRNA of the housekeeping gene IPP1 encoding the inorganic pyrophosphatase are shown. The probe used to observe expression of the RNAs X and Y is a PCR product from clone m26-11 containing the RNase P RNA 5′ leader and 3′tail as well as the m26 activation domain and MS2 RNA. This probe hybridizes therefore with both the X and Y RNA molecules. In lane 1 (m26-11), only one RNA is expressed (305 nt). In lanes 3–6, both RNA X and Y are detectable (X1, 283 nt; X2, 285 nt; X50, 331 nt; Y1 and Y2, 248 nt; Y50, 294 nt). (B) Quantitative analysis of β-galactosidase activity. The β-galactosidase activity is expressed in Miller units (logarithmic scale). Individual values are shown above each bar. Error bars indicate standard deviation. 3-Hybrid positive control indicates positive control of the yeast three-hybrid system using the IRE–IRP1 interaction; cells were cotransformed with plasmids pIIIa IRE MS2 and pAD IRP (Bernstein et al. 2002). m26-11 indicates MS2-RNA and m26 activation domain are on the same molecule (Buskirk et al. 2003). The other samples show expression after cotransformation of plasmids derived from pAN MS2-2 and pIIIa-m26 containing the indicated RNA X and RNA Y. When one RNA alone is indicated, the corresponding empty vector (pAN MS2-2 or pIIIa-m26) was cotransformed. (C) Growth of dilution series on selective medium. Dilution series of yeast YBZ-1 cultures were plated onto different synthetic defined media lacking leucine, uracil, and histidine, and supplemented with 1 mM 3-aminotriazole (3-AT) as indicated. 3-hybrid – indicates cells were cotransformed with plasmid pIIIa MS2-2 and pAD IRP (Bernstein et al. 2002); 3-hybrid +, same as 3-hybrid positive control in B. Other samples are as in B.
We ensured that the difference observed between cognate and noncognate RNAs with the β-galactosidase assay was reproducible by using the HIS3 reporter gene. Whereas all transformed yeast strains can grow on media lacking leucine and uracil, only cells expressing cognate RNAs are able to grow on medium also lacking histidine, indicating that HIS3 expression is turned on in these cells only (Fig. 2C ▶). When observing growth under more stringent conditions using 1 mM 3-aminotriazole (3-AT), we see a higher HIS3 expression of the three-hybrid system positive control compared with X1–Y1, confirming the results obtained with the β-galactosidase assay.
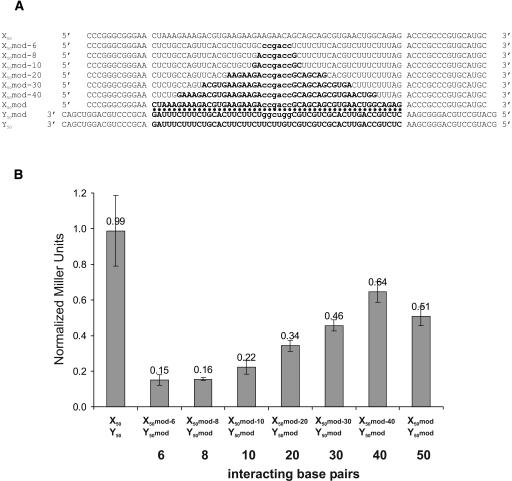
We have also measured the β-galactosidase activity of clones with a longer loop sequence. In X50 and Y50 (Fig. 4A ▶), the stem sequence is 2 bp longer, and a 50-nt sequence from the gene StpA (Zhang et al. 1995) is introduced between the flanking adenines. The arbitrarily chosen StpA coding sequence was searched for a 50-nt stretch with the lowest possible secondary structure stability. The pair was tested against its complement and against the empty RNAs transcribed from pAN MS2-2 and pIIIa-m26. As can be observed in Figure 2B ▶, lacZ expression is activated at levels comparable to the pairs X1–Y1 and X2–Y2 when both cognate RNAs are expressed (Fig. 2B ▶). We were surprised that the levels of activation resulting from an interaction over 50 bp is not higher than the signal obtained with a paring over six bases. The Northern blot (Fig. 2A ▶) reveals that the RNA containing X50 seems to be expressed at a lower level compared with X1 and X2. Other explanations for this result could be the longer distance between the promoter and the activation domain, or simply that the levels of reporter activation observed for the different pairs are already maximal.
FIGURE 4.
Influence of base-pairing length on reporter activation. (A) Sequences of the different RNAs tested. Variants of X50 and Y50 were designed to interact over different lengths with a central sequence corresponding to the six interacting base pairs of X1 and Y1. The sequence shown encompasses the cloning sites and part of the stem enclosing the interacting loops. (B) Quantitative analysis of β-galactosidase activity of the different pairs. The activity is normalized relative to the activity of X1–Y1 (1.0). Error bars indicate standard deviation. X50–Y50 indicates YBZ-1 strain transformed with plasmids expressing RNAs X50 and Y50; numbers 6–50, YBZ-1 strain transformed with plasmids expressing RNAs X50mod-6 to X50mod and Y50mod.
These results demonstrate the ability of the RNA-hybrid system to detect RNA–RNA interactions in vivo.
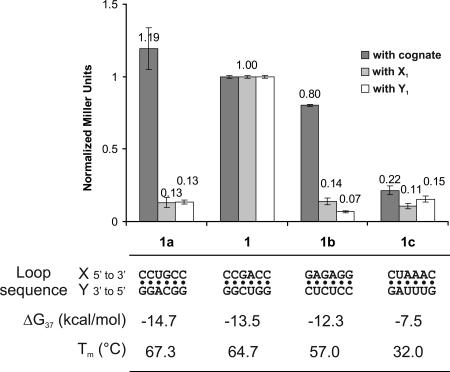
Signal intensity reflects the stability of interaction
Our laboratory previously investigated the stability of kissing loop interactions via UV melting analysis (Weixlbaumer et al. 2004). These constructs (including the pair X1–Y1) were cloned and tested in the RNA-hybrid system (Fig. 3 ▶). The only difference between the X1–Y1 pair and the pairs 1a to 1c is the sequence of the interacting loop region that was replaced in X1a to X1c and accordingly in Y1a to Y1c. We could observe a direct correlation between the β-galactosidase activity and the interaction stability measured via UV melting analysis. The most unstable pair used (X1c–Y1c), interacting with two G–C base pairs and four A–U base pairs, displays a melting temperature of 32°C and a free energy at 37°C of only −7.5 kcal/mol. This interaction can barely be detected using the RNA-hybrid system (Fig. 3 ▶).
FIGURE 3.
β-Galactosidase activity correlates with the stability of the loop–loop interaction. The β-galactosidase activity is normalized relative to the activity of X1–Y1. Error bars indicate standard deviation. The constructs X1a–X1c and Y1a–Y1c were assayed against their respective cognate partner (dark gray). As a control, the X and Y constructs were tested against the corresponding 1 construct: X1 for the Y RNAs (light gray) and Y1 for the X RNAs (white). Sequence, stability, and melting temperature at a concentration of 10−5 M of the different constructs are indicated.
It should be noted that the theoretical stability of the pair X2–Y2 (−12.8 kcal/mol) should lead to an activity between that of the pairs X1–Y1 and X1b–Y1b (cf. Figs. 2B ▶ and 3 ▶). However, the stem of X2 contains an additional base pair and cannot be directly compared with the other constructs. We suggest that this additional base pair changes the orientation of the activation domain on the lacZ promoter and is the cause of the lower activity observed (Fig. 2B ▶).
Influence of base-pairing length
One major application of the RNA-hybrid system would be the identification of new interaction partners through library screening. However, if 6 bp are sufficient to produce full β-galactosidase activity, many false positives would be detected during a screening for new interaction partners. To investigate this problem, we designed derivatives of X50 and Y50 in which the base-pairing between both constructs was reduced to 40, 30, 20, 10, 8, and 6 nt in the center of the loop sequence (for sequences, see Fig. 4A ▶) while the length of the loop remains constant. In the center of these constructs, six bases were exchanged to CCGACC in the X constructs and to the complementary sequence in the Y construct in order to introduce the same base-pairing as between X1 and Y1 and hence the same duplex stability. To avoid base-pairing outside of the desired region in the constructs X50 mod-6 to X50 mod-40, the nucleotides outside of the interacting region were substituted with the corresponding sequence of the Y construct (i.e., the sequence of the loop of X50-6 is the sequence of the Y50 loop outside of the central six bases). Unexpectedly, the modified constructs interacting over 50 bp (X50 mod and Y50 mod) display a lower β-galactosidase activity than do the unmodified constructs (Fig. 4 ▶). One possible explanation for this is that the loops of both X50 mod and Y50 mod can form internal secondary structures more stable than the loops of X50 and Y50 (−2.9 kcal/mol and −5.4 kcal/ mol vs. −2.4 kcal/mol and −2.3 kcal/mol, respectively) as observed by theoretical folding of the loop sequences (Zuker 2003). These structures might partially impair interaction between the two RNAs.
Nevertheless, a reduction of the number of possible base pairs between the two RNAs has a strong effect on the activation of the reporter gene (Fig. 4B ▶). Constructs interacting through 6, 8, or 10 bp do not activate β-galactosidase activity, whereas base-pairing over 30 or 40 nt fully restores activity. The pair interacting over 20 bp displays an intermediate effect, showing ~60% of the signal observed with full complementarity.
Why does a 6-bp interaction activate β-galactosidase activity in the context of the DIS-loop and not when inserted in a longer single-stranded region? We believe that the context of the DIS-loop is highly favorable for intermolecular contacts. In vivo a 6-bp complementarity does usually not lead to an interaction between two RNA molecules; in the case of the HIV genome, however, it is sufficient for initiation of dimerization. Furthermore, the interaction between two RNAs in the context of the DIS-loop is more stable than the corresponding RNA duplex (Weixlbaumer et al. 2004).
Screening a library for interaction partners
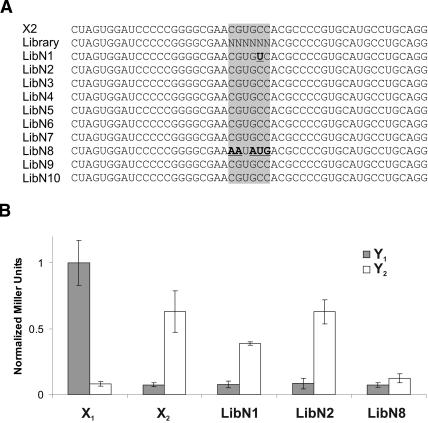
Since interacting RNAs demonstrate a growth advantage on medium lacking histidine in comparison to noncognate RNAs, the screening of a test library was performed. A library containing a 6-nt random region in place of the X2 loop was constructed and inserted into pAN MS2-2.
The number of yeast transformants after introduction of the library in YBZ-1 carrying plasmid pIIIa Y2 m26 was evaluated as 42,000 clones. Each of the 4096 possible sequences should therefore be present in ~10 copies on average. The transformants were plated on synthetic medium lacking leucine and histidine with 0.2 mM 3-AT. Of 22 growing colonies, 10 were confirmed as potential interaction partners after streaking on the same medium. Sequences of the 10 clones are shown in Figure 5A ▶. Eight clones carry the expected loop sequence from X2; the number of clones found fits nicely with the 10 expected copies of each sequence. One sequence (LibN1) displays a single point mutation C to U at the fifth position in the loop. This mutation transforms a G–C base pair in the original kissing loop complex into a G–U base pair. The sequence from clone LibN8 carries no visible similarity with the X2 loop sequence and cannot hybridize over >2 bp with Y2. The β-galactosidase activity of the different clones in the presence of Y2 and Y1 was measured (Fig. 5B ▶). As expected, none of the clones activates lacZ expression when coexpressed with Y1. When tested against Y2, clone LibN8 revealed itself as a false positive, and clone LibN1 displayed a reduced expression compared with wild-type X2 and LibN2. This result is in agreement with the fact that β-galactosidase activity correlates with the stability of interaction as this mutation transforms a G–C base pair into a G–U base pair.
FIGURE 5.
Selection of interacting partners from a random library. (A) Sequences of clones obtained after selection using Y2 as bait. Sequences of the expected result (X2) and of the library are indicated. (B) Quantitative analysis of β-galactosidase activity of the different clones. The activity is normalized relative to the activity of X1–Y1 (1.0). Error bars indicate standard deviation. The library clones were tested together with Y1 (gray) and Y2 (white).
In conclusion, in a test library screening, 90% of the selected clones were identified as interaction partners for the bait RNA. This demonstrates the accuracy of our system and the dependence for the correct interaction for signal induction.
DISCUSSION
We present in this study a new system for the investigation of RNA–RNA interactions in vivo. Using a loop–loop interaction as a model system, we show first the ability of the system to distinguish between cognate and noncognate interaction partners, demonstrating that RNA–RNA interactions can be investigated with this method. Second, we show that the interaction stability is directly correlated with the signal strength. The RNA-hybrid system can therefore be used to compare different ligands quantitatively. Third, we used the method to screen a model library and retrieved interaction partners for the bait used, indicating that this system is also suitable to search for interaction partners of RNAs in library screens.
The level of reporter activation is low compared with the three-hybrid system, which could be detrimental when screening a large library due to the low signal-background ratio. It would therefore be of interest to increase the potency of the activation domain. In this regard, an investigation of its mode of action and exact molecular requirements would be helpful. Furthermore, we plan to introduce multimers of the activation domain into a given RNA molecule to increase activity.
Expression levels of the reporter genes in cis by m26-11 are very high, and it is conceivable that further evolution of this clone was hindered by the lack of sufficient stringency (Buskirk et al. 2003). Obviously in the trans situation, we are not facing the same problem. We therefore plan to try to evolve more potent activators in the RNA-hybrid system via random mutagenesis of the m26 domain and selection on high concentrations of 3-AT in the context of the X1–Y1 interactions. Last, the low levels observed could be due to positioning effects and/or sterical hindrance at the promoter of the reporter gene. In support of this hypothesis, the pairs X1–Y1 and X2–Y2, while having comparable stabilities, display different β-galactosidase activities. A difference between the two pairs is the introduction of one additional base pair in X2. We suggest that this additional base pair is responsible for a different sterical positioning of the activation domain relative to the promoter, thereby influencing reporter activity. It would therefore be interesting to monitor the effect of the stem length on the β-galactosidase activity. Since the correct folding of the RNA molecules is critical, we are currently examining the possibility of increasing the stability of the activation domain via elongation of the flanking stems and determining the optimal scaffold of the two RNA constructs to insert RNAs X and Y in such a way to obtain strongest reporter activation by introducing longer stems at the base of the hairpins.
The correlation between in vitro stabilities and reporter activation in the RNA-hybrid system implies that the results obtained in 1 M Na+ are relevant in vivo. The stability of the kissing loop complex is highly dependent on the ionic conditions (Weixlbaumer et al. 2004). Divalent magnesium ions have a great influence on the complex stability, and a specific magnesium ion binding site has been detected in the X-ray structure of the complex (Ennifar et al. 2001). Comparison of the kissing complex stabilities in the presence of Na+ or Mg2+ in vitro showed that 1 M Na+ results in similar stabilities to 0.5–1 mM Mg2+ (Weixlbaumer et al. 2004). Our results reported here indicate that these similarities are also valid in vivo. While secondary structure formation is less dependent on divalent ions, it is still surprising that 1 M Na+ might after all be a suitable ionic condition for the prediction of secondary and tertiary interactions in vivo.
Certainly with ~20,000 putative ncRNAs already present in the mammalian ncRNA database (Pang et al. 2005), the RNA-hybrid system, while requiring some further improvements, could become a useful tool in the investigation of the role of so many transcripts with unknown function by revealing their target genes. Alternatively, the system may also be used to screen for drugs inhibiting essential RNA–RNA interactions in pathogens such as the DIS-loop interaction in HIV.
MATERIALS AND METHODS
Plasmid construction
To construct plasmid m26-11 (Buskirk et al. 2003) two oligonucleotides were hybridized (sequences, 5′-ATCCCCCGGGCGCGC GAGTATACTCCCCCAAGCGGATGC-3′ and 5′-GCAGGCATGC AAGAGGCTTAGGCATCCGCTTGGGGGAGTA-3′) and submitted to a PCR reaction without template to produce full-length double-stranded DNA. The product was digested with XmaI and SphI and inserted into the corresponding sites of pIIIa MS2-2 (Bernstein et al. 2002). Plasmid pIIIa-m26 (lacking the MS2-RNA binding sites) was obtained after SalI digestion of m26-11 and religation of the gel purified plasmid on itself. Vector pACT MS2-2 was constructed by insertion of the ScaI-BsrGI fragment of pIIIa MS2-2 at the same position in pACTII (Bernstein et al. 2002). In a second step, pACT MS2-2 and pACT II were digested with HindIII and SalI, respectively, and made blunt with T4 DNA polymerase. The cut plasmids were further digested with BsrGI. The 447-nt fragment of pACTII was then ligated into the 6961-nt fragment of pACT MS2-2 to create pAN MS2-2. This plasmid does not contain any of the protein expression signals present in pACTII.
The constructs containing a Y RNA were obtained by insertion of the corresponding insert at the SphI SalI sites in pIIIa-m26. X RNAs were inserted in pAN MS2-2 between XmaI and SphI. Inserts X1, X2, Y1, and Y2, respectively, were obtained via oligonucleotide hybridization with 5′-CCGGGCGAACCGACCACGCCCGTGCA TG-3′ and 5′-CACGGGCGTGGTCGGTTCGC-3′, 5′-CCGGGGCG AACGTGCCACGCCCCGTGCATG-3′ and 5′-CACGGGGCGTGG CACGTTCGCC-3′, 5′-CCTGCAGGGCAAGGTCGGAGCCCTGC AGG-3′ and 5′-TCGACCTGCA GGGCTCCGACCTTGCCCTGCA GGCATG-3′, and 5′-CCTGCAGGGCAAGGCACGAGCCCTGCAG G-3′ and 5′-TCGACCTGCAGGGCTCGTGCCTTGCCCTGCAGG CATG-3′. X1a to X1c and Y1a to Y1c were obtained with similar oligonucleotides carrying the appropriate loop mutations. X50 and Y50 were cloned after PCR of pET3A-StpA (Zhang et al. 1995) (primers, 5′-ATCCCCCGGGCGGGAACTAAAGAAAGACGTGAA GAAG-3′ and 5′-GCAGGCATGCACGGGCGGGTCTCTGCCAG TTCACGCTGCTG-3′ for X50; 5′-TCTTGCATGCCTGCAGGGC GAACTCTGCCAGTTCACGCTGCTG-3′ and 5′-TAGAGTCGAC CTGCAGGGCGTCTAAAGAAAGACGTGAAGAAG-3′ for Y50). To obtain the variants of X50 and Y50, oligonucleotides were hybridized and elongated by using Taq polymerase before digestion with the appropriate enzymes and ligation into pAN-MS2-2 or pIIIa-m26. For X50-6 the oligonucleotides used were ATCCCCCGGGCG GGAACTCTGCCAGTTCACGCTGCTGCCCGACCTCTTCT and AGGCATGCACGGGCGGGTCTAAAGAAAGACGTGAAGAAGA GGTCGGGCAGCA. The other variant were obtained with similar oligonucleotides.
Library construction
The library was obtained after ligation in pAN MS2-2 of a PCR product (primers, 5′-CTAGTGGATCCCCCGGGGCG-3′ and 5′-C TGCAGGCATGCACGGGGCG-3′; template, 5′-CTAGTGGATCC CCCGGGGCGAANNNNNNACGCCCCGTGCATGCCTGCAG-3′) digested with SphI and XmaI.
Yeast culture and transformation
YBZ-1 yeast cells were cotransformed with plasmids derived from pIIIa-m26 and plasmids derived from pACT MS2-2. pIIIa-m26–derived plasmids carry the URA3 gene, and pACT MS2-2–derived plasmids contain the LEU2 gene. Transformed cells were selected on media lacking uracil and leucine (Bernstein et al. 2002).
Quantitative determination of β-galactosidase activity was performed by using o-nitrophenyl-β-d-galactopyranoside (ONPG) as substrate as described in the Yeast Protocols Handbook from Clontech Laboratories (http://www.clontech.com).
For the comparative growth assay, serially diluted YBZ-1 strains transformed with a pACT MS2-2 and a pIIIa-m26 derivative were plated on selective medium lacking uracil, leucine, and—when indicated, histidine—and supplemented with 3-AT.
For the screening, strain YBZ-1 was transformed successively with the plasmid expressing Y2 and with the library. Transformed cells were plated on medium lacking histidine and leucine and containing 0.2 mM 3-AT. After 4 d, 22 colonies were growing. The 10 white colonies were streaked on the same medium. Colonies remaining white were streaked on medium lacking leucine and containing 5-fluorotic acid (5-FOA). Plasmids from active clones were recovered by using standard techniques (Sambrook and Russell 2001) and sequenced with primer pIIIa 4827 (5′-CTGTATCGCAAATAAGTGAA-3′).
Northern blot
For Northern blotting, transformed yeast cells were grown to mid-log phase (OD600 0.8) and RNA was prepared by using the RNeasy Mini Protocol for Isolation of Total RNA from yeast (enzymatic lysis protocol, standard version) from Qiagen; 25 μg of the RNA samples were denatured and loaded on a 5% denaturing polyacrylamide gel before semi-dry transfer (Lee et al. 1993). The PCR probe from m26-11 (using oligonucleotides pIIIa rev 5162 5′-CTCTATACTCCCCTATAGTCTG-3′ and pIIIa 4827) was labeled by random priming. The probe was hybridized according to a standard protocol (Sambrook and Russell 2001).
Acknowledgments
We thank Prof. Marvin Wickens for the gift of the three-hybrid system strain and plasmids. We thank Stefan Ameres and Christina Lorenz for helpful discussions, and all of the Schroeder laboratory for critical review of the manuscript. This work was supported by FWF (Austrian Science Foundation) grant no. Z-72.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2105506.
REFERENCES
- Ambros, V. 2004. The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J.P., Cavaille, J., and Huttenhofer, A. 2002. The expanding snoRNA world. Biochimie 84: 775–790. [DOI] [PubMed] [Google Scholar]
- Bernstein, D.S., Buter, N., Stumpf, C., and Wickens, M. 2002. Analyzing mRNA–protein complexes using a yeast three-hybrid system. Methods 26: 123–141. [DOI] [PubMed] [Google Scholar]
- Brunel, C., arquet, R., Romby, P., and Ehresmann, C. 2002. RNA loop– loop interactions as dynamic functional motifs. Biochimie 84: 925– 944. [DOI] [PubMed] [Google Scholar]
- Buskirk, A.R., Kehayova, P.D., Landrigan, A., and Liu, D.R. 2003. In vivo evolution of an RNA-based transcriptional activator. Chem. Biol. 10: 533–540. [DOI] [PubMed] [Google Scholar]
- Eguchi, Y. and Tomizawa, J. 1990. Complex formed by complementary RNA stem-loops and its stabilization by a protein: Function of CoIE1 Rom protein. Cell 60: 199–209. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Walter, P., Ehresmann, B., Ehresmann, C., and Dumas, P. 2001. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 8: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Fields, S. and Song, O. 1989. A novel genetic system to detect protein– protein interactions. Nature 340: 245–246. [DOI] [PubMed] [Google Scholar]
- Forne, T., Labourier, E., Antoine, E., Rossi, F., Gallouzi, I., Cathala, G., Tazi, J., and Brunel, C. 1996. Structural features of U6 snRNA and dynamic interactions with other spliceosomal components leading to pre-mRNA splicing. Biochimie 78: 436–442. [DOI] [PubMed] [Google Scholar]
- Hershberg, R., Altuvia, S., and Margalit, H. 2003. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 31: 1813–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook, B., Bernstein, D., Zhang, B., and Wickens, M. 2005. RNA– protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA 11: 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughrea, M. and Jette, L. 1996. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensable for dimer formation. Biochemistry 35: 1589–1598. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. 2003. Challenging the dogma: The hidden layer of nonprotein- coding RNAs in complex organisms. Bioessays 25: 930– 939. [DOI] [PubMed] [Google Scholar]
- Murchison, E.P. and Hannon, G.J. 2004. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell. Biol. 16: 223–229. [DOI] [PubMed] [Google Scholar]
- Paillart, J.C., Marquet, R., Skripkin, E., Ehresmann, C., and Ehresmann, B. 1996. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie 78: 639–653. [DOI] [PubMed] [Google Scholar]
- Pang, K.C., Stephen, S., Engstrom, P.G., Tajul-Arifin, K., Chen, W., Wahlestedt, C., Lenhard, B., Hayashizaki, Y., and Mattick, J.S. 2005. RNAdb: A comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 33 (database issue): D125–D130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila, F., Majdalani, N., and Gottesman, S. 2003. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: The RpoS paradigm. Mol. Microbiol. 48: 855–861. [DOI] [PubMed] [Google Scholar]
- Saha, S., Ansari, A.Z., Jarrell, K.A., and Ptashne, M. 2003. RNA sequences that work as transcriptional activating regions. Nucleic Acids Res. 31: 1565–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sengupta, D.J., Zhang, B., Kraemer, B., Pochart, P., Fields, S., and Wickens, M. 1996. A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl. Acad. Sci. 93: 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, D.J., Wickens, M., and Fields, S. 1999. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. RNA 5: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer, A., Werner, A., Flamm, C., Westhof, E., and Schroeder, R. 2004. Determination of thermodynamic parameters for HIV DIS type loop–loop kissing complexes. Nucleic Acids Res. 32: 5126–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler, N., Werner, M., and Schroeder, R. 2003. Kissing complex- mediated dimerisation of HIV-1 RNA: Coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 31: 6419–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Derbyshire, V., Salvo, J.L., and Belfort, M. 1995. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA 1: 783–793. [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]