Abstract
MicroRNAs (miRNAs) are ~22-nt RNA segments that are involved in the regulation of protein expression primarily by binding to one or more target sites on an mRNA transcript and inhibiting translation. MicroRNAs are likely to factor into multiple developmental pathways, multiple mechanisms of gene regulation, and underlie an array of inherited disease processes and phenotypic determinants. Several computational programs exist to predict miRNA targets in mammals, fruit flies, worms, and plants. However, to date, there is no systematic collection and description of miRNA targets with experimental support. We describe a database, TarBase, which houses a manually curated collection of experimentally tested miRNA targets, in human/mouse, fruit fly, worm, and zebrafish, distinguishing between those that tested positive and those that tested negative. Each positive target site is described by the miRNA that binds it, the gene in which it occurs, the nature of the experiments that were conducted to test it, the sufficiency of the site to induce translational repression and/or cleavage, and the paper from which all these data were extracted. Additionally, the database is functionally linked to several other useful databases such as Gene Ontology (GO) and UCSC Genome Browser. TarBase reveals significantly more experimentally supported targets than even recent reviews claim, thereby providing a comprehensive data set from which to assess features of miRNA targeting that will be useful for the next generation of target prediction programs. TarBase can be accessed at http://www.diana.pcbi.upenn.edu/tarbase.
Keywords: microRNA, microRNA target, database, experimentally supported, computational
INTRODUCTION
MicroRNAs (miRNAs) are ~22-nt RNA segments that are involved in the regulation of protein expression primarily by binding to one or more target sites on an mRNA transcript and inhibiting translation. The first miRNA gene, lin-4, was identified more than a decade ago in Caenorhabditis elegans (Lee et al. 1993). It is now reported that miRNAs constitute anywhere from ~1% (Lim et al. 2003; Bartel 2004) to ~3% (Bentwich et al. 2005; Berezikov et al. 2005) of the known genes in eukaryotes. The Sanger Institute’s miRBase reports 326 human miRNAs. Although computational studies have suggested that thousands of human genes are targets of these 326 miRNAs, the lack of a high-throughput experimental technique has impeded a large-scale confirmation of these targets (Lewis et al. 2005). Notwithstanding this limitation, the last 2 yr have witnessed a significant increase in the number of experimentally supported miRNA targets, primarily due to an increase in the number of laboratories interested in miRNA targeting.
The first target prediction programs for mammals (Lewis et al. 2003; Kiriakidou et al. 2004) and for fruit fly (Enright et al. 2003; Stark et al. 2003; Rajewsky and Socci 2004) were published in late 2003/early 2004. Since then, at least four others for mammals (John et al. 2004; Lewis et al. 2005; Krek et al. 2005; Rusinov et al. 2005) and five others for fruit fly (Rehmsmeier et al. 2004; Burgler and Macdonald 2005; Grun et al. 2005; Robins et al. 2005; Saetrom et al. 2005) have been published. A majority of these 14 programs are available for public use (Table 1 ▶). Several of these programs search for potential miRNA targets by scanning the genome for short segments that have features very similar to a set of experimentally supported miRNA targets (Enright et al. 2003; Stark et al. 2003; John et al. 2004; Kiriakidou et al. 2004; Rajewsky and Socci 2004). Examples of these sets are: Rajewsky et al.: target sites of C. elegans lin-4 and let-7; Enright et al.: target sites of Drosophila lin-4 and let-7; Stark et al.: target sites of C. elegans lin-4, let-7, and Drosophila bantam; and Kiriakidou et al.: target sites of C. elegans lin-4, let-7, and variants of a let-7 target site on C. elegans lin-41 obtained from extensive mutational analyses. A common limitation of these programs is the very small sets of data from which they decipher rules/steps for genome-wide predictions. Since the first generation of programs in 2003/2004, many more miRNA targets have gained experimental support in human cells, fruit flies, worms, and even zebrafish. However, due to the lack of any up-to-date collection of experimentally supported targets, subsequent target prediction programs that relied on training sets still harbored the same limitation mentioned above (Robins et al. 2005; Saetrom et al. 2005). For example: Robins et al.: target sites for C. elegans lin-4, let-7, and Drosophila bantam, and Saetrom et al.: target sites for C. elegans lin-4, let-7, and Drosophila mir-13a, bantam. The target sites in these sets account for a small percentage of experimentally verified miRNA targets in mammals, fruit flies, worms, and zebrafish. A comprehensive collection of experimentally supported targets could improve the performance of the programs mentioned above and even introduce a new class of more sophisticated machine-learning approaches. However, it must be noted that there exists a distinct class of target prediction programs that do not rely on training sets, but rather, rely heavily on conservation (Lewis et al. 2003, 2005; Krek et al. 2005; Xie et al. 2005). The utility of these programs reveals that training sets are optional and merely provide a means for future machine-learning approaches that may supplement existing approaches.
TABLE 1.
Current target prediction programs that are available for public use
| Paper | Program name | URL | Organisma |
| Lewis et al. 2003 | TargetScan | http://genes.mit.edu/targetscan/ | Mammals |
| Kiriakidou et al. 2004 | DIANA-microT | http://www.diana.pcbi.upenn.edu/cgi-bin/micro_t.cgi | Mammals |
| John et al. 2004 | MiRanda | http://www.microrna.org/ | Mammals |
| Krek et al. 2005 | PicTar | http://pictar.bio.nyu.edu | Mammals |
| Lewis et al. 2005 | TargetScanS | http://genes.mit.edu/targetscan/ | Mammals |
| Enright et al. 2003 | MiRanda | http://www.microrna.org/ | Fruitfly |
| Rehmsmeier et al. 2004 | RNAhybrid | http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/ | Fruitfly |
| Grun et al. 2005 | PicTar | http://pictar.bio.nyu.edu | Fruitfly |
| Rusinov et al. 2005 | MicroInspector | http://mirna.imbb.forth.gr/microinspector/ | Any |
| Saetrom et al. 2005 | TargetBoost | https://demo1.interagon.com/demo/ | Worm and Fruitfly |
aOrganism(s) for which the program is best suited.
Here we present a database, TarBase, that provides a collection of all experimentally tested miRNA targets. Tar-Base describes at least 45 human/mouse genes, 28 fruit fly genes, 7 worm genes, and 1 zebrafish gene that have gained experimental support as translationally repressed miRNA targets. TarBase also describes ~350 human/mouse genes and 3 worm genes that have gained experimental support as cleaved miRNA targets. The total number of target sites recorded in TarBase for human/mouse, fruit fly, worm, and zebrafish exceeds 550—a much larger data set for the next generation of target prediction programs. Furthermore, TarBase describes each supported target site by the miRNA that binds it, the gene in which it occurs, the location within the 3′ UTR where it occurs, the nature of the experiments that were conducted to validate it, and the sufficiency of the site to induce translational repression and/or cleavage. Such a comprehensive description of each target site will be useful for focused bioinformatic and experimental studies to further understand the features of miRNA targeting, the mechanisms of miRNA-based translational repression and/or cleavage, and the roles of miRNAs in various biological networks. TarBase can be accessed at http://www.diana.pcbi.upenn.edu/tarbase.
RESULTS
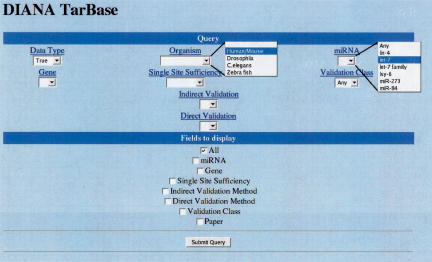
A snapshot of the graphical user interface to the database is provided in Figure 1 ▶. Users can query the database of either true or false miRNA targets as determined by experimental testing. Furthermore, users can query for target sites according to the organism to which it belongs, the gene in which it occurs, the miRNA that it binds, the technique used for its experimental support, its single-site sufficiency status, or any combination thereof. The schema of the database is depicted in Figure 2 ▶. Users are provided the option of choosing the fields from the schema that they would like to view for each of their queries (Fig. 1 ▶).
FIGURE 1.
A snapshot of the graphical user interface to the database.
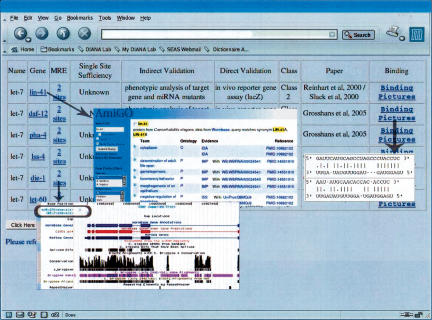
FIGURE 2.
A snapshot of the results page showing the various functionalities of the database. Target genes are linked to Gene Ontology (GO); miRNA Recognition Elements (MREs) are linked to UCSC Genome Browser via custom tracks; and binding pictures are viewable in a new window.
It is important to note that not all target sites are tested using the same experimental technique. For example, Krek et al. (2005) use a traditional in vitro reporter gene assay to test the ability of a target 3′ UTR to induce translational repression, whereas Zhao et al. (2005) use transgenic mice overexpressing an miRNA of interest to test changes in target protein levels. Also, not all target sites are individually tested. Kiriakidou et al. (2004) use a reporter gene assay to individually test a single target site (STS), whereas Lewis et al. (2003) use a reporter gene assay to test multiple target sites (MTS) simultaneously. The former is useful for constructing the class of target sites that are independently sufficient for translational repression, whereas the latter is useful for constructing the class of target sites that may require the presence of other target sites on the same 3′ UTR to induce translational repression. As mentioned previously, TarBase includes the experimental techniques used for support and the sts/mts status of each target site. Users of TarBase can search and compare target sites supported by different experimental techniques. TarBase does not provide a direct comparison of the techniques or the full details of the advantages and limitations of each, but we encourage any user of TarBase to use and understand the data within the context of the experimental techniques that are employed.
TarBase also includes functional links to other databases such as Gene Ontology (GO) and UCSC Genome Browser. A target gene with experimental support is linked to GO in order to provide a clearer picture of what kinds of biological pathways the targeting miRNA may regulate. The specific target sites within the target gene are linked to UCSC Genome Browser via custom tracks for facile viewing of their genic location and sequence composition. Finally, each target site is also linked to an in-house database of all known miRNA:target site base-pairing diagrams. All of these centralized data are valuable either for inquiries regarding a specific gene/miRNA/target site/experimental technique or for extracting general features of miRNA targeting or miRNA-based mechanisms for translational repression and/or cleavage. We used TarBase to study the locations of target sites within their 3′ UTRs.
Location of experimentally verified target sites within their 3′ UTRs
A recent paper suggests that miRNPs impede 5′ cap recognition, thereby interfering with translation (Pillai et al. 2005). However, details of the mechanism of miRNA-based translational repression are not yet well understood. It has been proposed that miRNA/RISC complexes may prefer to bind at certain locations on the 3′ UTR, perhaps to facilitate miRNP binding at the 5′ cap. We took 60 human/mouse experimentally supported target sites from TarBase and mapped their locations within their 3′ UTRs. We then determined the distance of each target site from the start of the host 3′ UTR and normalized this distance by the length of the host 3′ UTR. It appears that there is a marked tendency for experimentally supported target sites to occur closer to the start of the 3′ UTR than the end of the 3′ UTR. However, we repeated this test for 15 of the human/mouse target sites that were shown to be false positives and we noticed a similar distribution. Therefore, it would seem that the location within the 3′ UTR is not a feature that would increase the specificity (i.e., discriminating between true and false positives) of target prediction programs.
Since both the true and the false set of target sites have a similar distribution of locations within the 3′ UTR, it is possible that target prediction programs are implicitly biased toward locations nearer to the start of the 3′ UTR. As a final point, it must be noted that it is also possible that the current sets of true and false target sites are still too small for such an analysis as described above.
DISCUSSION
We describe a database, TarBase, which houses a manually curated collection of experimentally tested miRNA targets, in human/mouse, fruit fly, worm, and zebrafish. Simple analyses of the data have raised interesting considerations about the features used for target prediction and the experimental techniques used for support (see Results, above). We believe that TarBase will not only be useful for biologists interested in miRNA function, but also for bioinformaticians interested in using the most comprehensive set of supported targets currently available to train and test a new cohort of machine-learning methods for target prediction. Machine-learning methods are inherently biased toward predicting target sites that bear a significant resemblance to sites in the training set. Small training sets are likely to have a highly homogenous composition, restricting the types of sites that can be predicted. However, as the number of sites with experimental support increases, we expect the training set to provide a more accurate representation of target site diversity. We must emphasize, however, that machine-learning methods for target prediction are not necessarily superior to approaches that do not employ training sets (Lewis et al. 2003, 2005; Krek et al. 2005; Xie et al. 2005). But as machine-learning methods become more popular with an increasing set of experimentally supported targets, it will be interesting to observe how they compare with and how they can supplement these other approaches.
To ensure that TarBase is up to date, we have provided two semiautomated means of submission. We kindly request that future experimentally supported targets are submitted in a timely manner so that we can maintain TarBase for the growing miRNA research community. Authors are directed to http://www.diana.pcbi.upenn.edu/cgi-bin/AddTarBase.cgi for submission instructions.
MATERIALS AND METHODS
The first step in the construction of the database was to perform an exhaustive literature search for all experimentally tested miRNA targets. We found 16 relevant papers for human/mouse (Lewis et al. 2003, 2005; Esau et al. 2004; Kiriakidou et al. 2004; Poy et al. 2004; Yekta et al. 2004; Cimmino et al. 2005; Davis et al. 2005; Kawasaki and Taira 2005; Krek et al. 2005; Lecellier et al. 2005; Lim et al. 2005; Nakamoto et al. 2005; O’Donnell et al. 2005; Wu and Belasco 2005; Zhao et al. 2005), 8 for fruit fly (Brennecke et al. 2003, 2005; Stark et al. 2003; Xu et al. 2003; Burgler and Macdonald 2005; Lai et al. 2005; Leaman et al. 2005; Robins et al. 2005), 11 for worm (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000; Slack et al. 2000; Abrahante et al. 2003; Johnston and Hobert 2003; Lin et al. 2003; Chang et al. 2004; Grosshans et al. 2005; Johnson et al. 2005), and 1 for zebrafish (Kloosterman et al. 2004). For each target site from each paper we recorded the miRNA that binds it, the gene in which it occurs, the nature of the experiments that were conducted to support it, the sufficiency of the site to induce translational repression and/or cleavage, the miRNA:target site binding/base-pairing diagram, and the paper from which all these data were extracted. It must be noted that several papers did not provide the binding diagrams. In these cases, if the authors used a target prediction program to predict these target sites prior to experimental testing, we used the same target prediction program to obtain the binding diagrams from the output of the software. If the authors did not provide binding diagrams and also did not use a target prediction program to predict the target sites, then we indicate that this binding diagram is unavailable at the current time.
The recorded data were then manually uploaded in to a SQL database with tables for each organism: human/mouse, fruit fly, worm, and zebrafish. We performed quality control on the data several times to ensure that the data were uploaded accurately. We plan to include plant and virus miRNA targets very soon.
To make the database available for public use we designed a Web site that uses Perl DBI and CGI to communicate with the database and retrieve/display data that are relevant to the user’s query. The Web site is designed so as to allow users to query the database for records involving a particular organism, miRNA(s), gene(s), sts/mts status, experimental technique(s), or any combination thereof (Fig. 1 ▶). Upon retrieval of the data relevant to the user’s query, the Web site displays the results in a table format. Target genes are provided as a link to GO, where the user can directly ascertain the molecular and cellular functions of the genes (Fig. 2 ▶). The specific target sites are provided as a link to the UCSC genome browser that displays the location of the target sites as a custom track (Fig. 2 ▶). The custom track information for each experimentally supported target site within each organism was determined by using Blat (http://genome.ucsc.edu/cgi-bin/hgBlat) to locate the target site sequence within the genome.
Acknowledgments
We thank M. Megraw for her helpful comments and discussions. We also thank P. Rajasethupathy, P. Nelson, and S.A. Liebhaber for their critical reviews of the manuscript. P.S. is supported by a predoctoral NIH training grant (5T32GM008216). B.C. and A.G.H. are supported by an NSF Career Award Grant (DBI-0238295).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2239606.
REFERENCES
- Abrahante, J.E., Daul, A.L., Li, M., Volk, M.L., Tennessen, J.M., Miller, E.A., and Rougvie, A.E. 2003. The Caenorhabditis elegans hunchback-like GENE lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Finav, U., Meiri, E., et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37: 766–770. [DOI] [PubMed] [Google Scholar]
- Berezikov, E., Guryev, V., van de Belt, J., Wienholds, E., Plasterk, R.H., and Cuppen, E. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21– 24. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cells proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Stark, A., Russell, R.B., and Cohen, S.M. 2005. Principles of microRNA-target recognition. PLoS Biol. 3: 404– 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgler, C. and Macdonald, P.M. 2005. Prediction and verification of microRNA targets by MovingTargets, a highly adaptable prediction method. BMC Genomics 6: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S., Johnston, R.J., Frakjaer-Jensen, C., Lockery, S., and Hobert, O. 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789. [DOI] [PubMed] [Google Scholar]
- Cimmino, A., Calin, G.A., Fabbri, M., Iorio, M.V., Ferracin, M., Shimizu, M., Wojcik, S.E., Aqeilan, R.I., Zupo, S., Dono, M., et al. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. 102: 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, E., Caiment, F., Tordoir, X., Cavaille, J., Ferguson-Smith, A., Cockett, N., Georges, M., and Charlier, C. 2005. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 15: 743–749. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D.S. 2003. MicroRNA targets in Drosophila. Genome Biol. 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau, C., Kang, X., Peralta, E., Hanson, E., Marcusson, E.G., Ravichandran, L.V., Sun, Y., Koo, S., Perera, R.J., Jain, R., et al. 2004. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 279: 52361–52365. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., Johnson, T., Reinert, K.L., Gerstein, M., and Slack, F.J. 2005. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell 8: 321–330. [DOI] [PubMed] [Google Scholar]
- Grun, D., Wang, Y., Langenberger, D., Gunsalus, K.C., and Rajewsky, N. 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1: 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, B., Enright, A.J., Aravin, A., Tuschl, T., Sander, C., and Marks, D.S. 2004. Human microRNA targets. PLoS Biol. 2: 1862–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S.M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K.L., Brown, D., and Slack, F.J. 2005. RAS is regulated by the let-7 microRNA family. Cell 120: 635–647. [DOI] [PubMed] [Google Scholar]
- Johnston, R.J. and Hobert, O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Kawasaki, H. and Taira, K. 2005. Identification of target genes of microRNAs in retinoic acid-induced neuronal differentiation. Pure Appl. Chem. 77: 313–318. [Google Scholar]
- Kiriakidou, M., Nelson, P., Kouranov, A., Fitziev, P., Bouyioukos, C., Mourelatos, Z., and Hatzigeorgiou, A.G. 2004. A combined computational- experimental approach predicts human miRNA targets. Genes & Dev. 18: 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman, W.P., Wienholds, E., Ketting, R.F., and Plasterk, R.H.A. 2004. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 32: 6284–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, A., Grun, D., Poy, M.N., Wolf, R., Rosenberg, L., Epstein, E.J., MacMenamin, P., Piedade, I., Gunsalus, K.C., Stoffel, M., et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37: 495–500. [DOI] [PubMed] [Google Scholar]
- Lai, E.C., Tam, B., and Rubin, G.M. 2005. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes & Dev. 19: 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman, D., Chen, P.Y., Fak, J., Yalcin, A., Pearce, M., Unnerstall, U., Marks, D.S., Sander, C., Tuschl, T., and Gaul, U. 2005. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121: 1097–1108. [DOI] [PubMed] [Google Scholar]
- Lecellier, C., Dunoyer, P., Arar, K., Lehmann-Che, J., Eyquem, S., Himber, C., Saib, A., and Voinnet, O. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308: 557–560. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., and Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B., and Bartel, D.P. 2003. Vertebrate microRNA genes. Science 299: 5612. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., and Johnson, J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773. [DOI] [PubMed] [Google Scholar]
- Lin, S.Y., Johnson, S.M., Abraham, M., Vella, M.C., Pasquinelli, A., Gamberi, C., Gottlieb, E., and Slack, F.J. 2003. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639–650. [DOI] [PubMed] [Google Scholar]
- Moss, E.G., Lee, R.C., and Ambros, V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. [DOI] [PubMed] [Google Scholar]
- Nakamoto, M., Jin, P., O’Donnell, W.T., and Warren, S.T. 2005. Physiological identification of human transcripts translationally regulated by a specific microRNA. Hum. Mol. Genet. http://hmg.oxfordjournals.org/cgi/content/abstract/ddi397v1 [DOI] [PubMed]
- O’Donnell, K.A., Wentzel, E.A., Zeller, K.I., Dang, C.V., and Mendell, J.T. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., and Filipowicz, W. 2005. Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309: 573–576. [DOI] [PubMed] [Google Scholar]
- Poy, M.N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma., X., MacDonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230. [DOI] [PubMed] [Google Scholar]
- Rajewsky, N. and Socci, N.D. 2004. Computational identification of microRNA targets. Dev. Biol. 267: 529–535. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier, M., Steffen, P., Hochsmann, M., and Giegerich, R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–905. [DOI] [PubMed] [Google Scholar]
- Robins, H., Li, Y., and Padgett, R.W. 2005. Incorporating structure to predict microRNA targets. Proc. Natl. Acad. Sci. 102: 4006–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinov, V., Baev, V., Minkov, I.N., and Tabler, M. 2005. Micro-Inspector: A Web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 33: 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom, O., Snove Jr., O., and Saetrom, P. 2005. Weighted sequence motifs as an improved seeding step in microRNA target prediction algorithms. RNA 11: 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack, F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russell, R.B., and Cohen, S.M. 2003. Identification of Drosophila microRNA targets. PLoS Biol. 1: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Wu, L. and Belasco, J.G. 2005. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of the embryonal carcinoma cells. Mol. Cell. Biol. 25: 9198–9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X., Lu, J., Kulbokas, E.J., Golub, T.R., Mootha, V., Lindblad-Toh, K., Lander, E.S., and Kellis, M. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790–795. [DOI] [PubMed] [Google Scholar]
- Yekta, S., Shih, I., and Bartel, D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–596. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Samal, E., and Srivastava, D. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436: 214–220. [DOI] [PubMed] [Google Scholar]