Abstract
We describe a novel approach to characterize the functional domains of a protein in vivo. This involves the use of a custom-built Tn5-based transposon that causes the expression of a target gene as two contiguous polypeptides. When used as a genetic screen to dissect the budding yeast PRP8 gene, this showed that Prp8 protein could be dissected into three distinct pairs of functional polypeptides. Thus, four functional domains can be defined in the 2413-residue Prp8 protein, with boundaries in the regions of amino acids 394–443, 770, and 2170–2179. The central region of the protein was resistant to dissection by this approach, suggesting that it represents one large functional unit. The dissected constructs allowed investigation of factors that associate strongly with the N- or the C-terminal Prp8 protein fragments. Thus, the U5 snRNP protein Snu114p associates with Prp8p in the region 437–770, whereas fragmenting Prp8p at residue 2173 destabilizes its association with Aar2p.
Keywords: protein domains, protein interaction, RNA splicing, snRNP, transposon mutagenesis
INTRODUCTION
Studies of the structure, function, and molecular interactions of very large proteins are often hindered by the difficulty in obtaining sufficient quantities of the intact protein. On the other hand, many proteins appear to have been assembled from several independently folding domains, each of which has a distinct function, such as catalysis, ligand-binding, protein or nucleic acid interaction, or membrane-spanning properties (Doolittle 1995). Thus, the ability to identify and isolate individual protein domains offers a reductionist approach to facilitate protein characterization. However, in the absence of clear structural features it may be difficult to recognize domain structure in a protein. We describe here a novel approach to characterize the functional domains of a protein in vivo, using a custom-built Tn5-based transposon that causes the expression of a target gene as two contiguous polypeptides must function in trans. We used this approach to gate the domain structure of the large (280 kDa in yeast) spliceosomal protein, Prp8p. Prp8p is known to multiple interactions in ribonucleoprotein (RNP) plexes, including the spliceosome (for review, see and Beggs 2005); however, despite the high level of servation of this essential splicing factor, informatic has provided few clues as to its likely domain structure.
The spliceosome is a dynamic complex that nuclear pre-mRNA splicing. It assembles on the mRNA in a series of steps, in which the U1, U2, U4, and U6 snRNAs in the form of small RNP (snRNPs) and a variety of non-snRNP proteins with the pre-mRNA. Prp8p is the largest and most conserved spliceosomal protein. Biochemical, genetic, two-hybrid interaction data indicate that Prp8p physically and/or functionally with a large number of ing factors. It is a component of the U5 snRNP, a snRNP/Prp19p complex, the U5·U4/U6 tri-snRNP, the spliceosome (Grainger and Beggs 2005). It the catalytic center of the spliceosome, as detected photochemical cross-linking to functionally regions of the U5 and U6 snRNAs and to the 5′ splice the 3′ splice site, and the branch point of the pre-(for review, see Grainger and Beggs 2005). In Prp8p has been implicated as a regulator of several conformational rearrangements in the spliceosome (Newman 1997; Staley and Guthrie 1998; Collins and Guthrie 1999; Brow 2002; Kuhn et al. 2002). Achsel et al. (1998) showed that human Prp8p (hPrp8p) forms a stable RNA-free core complex with h116K (ySnu114p), h200K (yBrr2p), and h40K (no known yeast homolog). Thus Prp8p may function as a large protein scaffold in the spliceosome and play a key role at its catalytic center.
Further characterization of Prp8 protein function(s) would be facilitated by knowledge of the domain structure of this very large 2413-residue protein. However, the sequence of Prp8p is largely novel and reveals little about the protein’s domain structure or its possible biochemical activities. We show here, using a novel random transposon insertion strategy, that the PRP8 gene of Saccharomyces cerevisiae can be dissected at any one of three positions (amino acids 394–443, 770, and 2170–2179), such that the yeast Prp8p can function as pairs of separate, interacting polypeptides. These dissected constructs allowed us to identify amino acids 437–770 of Prp8p as being associated with Snu114p, whereas disruption of Prp8p at 2173 interferes with its association with Aar2p. The central region of Prp8p, from amino acids 770 to 2173, was resistant to dissection by this approach, suggesting that it represents one large functional unit.
RESULTS
Transposon insertion to dissect PRP8
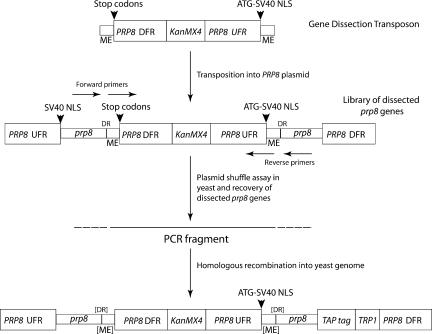
Our approach involves the random insertion of a custom-built Tn5-based transposon into the PRP8 gene to create a library of thousands of prp8 genes, each with a transposon sequence interrupting the open reading frame. The transposon sequence is designed to terminate translation of the upstream reading frame and at the same time drive expression of the prp8 sequence downstream of the site of insertion. The transposon consists of four elements (Fig. 1 ▶) inserted between the “Mosaic End” recognition sequences for Tn5 Transposase: (1) PRP8 gene downstream flanking region; (2) Kan or KanMX4 cassette to mark the transposon in bacteria or yeast, respectively; (3) PRP8 gene upstream flanking region, including the promoter; and (4) translation start site upstream of an SV40 nuclear localization signal (NLS). Insertion of this transposon will generally be lethal for five out of the six possible reading frames because in functional terms the gene will be truncated. However, in the remaining frame the transposon will direct the synthesis of an NLS-tagged downstream polypeptide fragment, resulting in two separate polypeptides encoded by distinct mRNAs.
FIGURE 1.
Structure of the gene dissection transposon and dissected prp8. The transposon was custom-built from three main components: (1) PRP8 gene downstream flanking region (DFR) containing signals for transcription termination, polyadenylation, and stop codons in all three reading frames; (2) KanMX4 cassette (Wach et al. 1994) conferring Kanamycin-resistance in Escherichia coli and G418-resistance in yeast (KanMX4); (3) PRP8 gene upstream flanking region (UFR) containing signals for initiation of transcription and translation. The transposon is flanked by 19 bp “Mosaic End” (ME) sequences recognized by Tn5 transposase. An example of the structure of one of a library of transposon-dissected prp8 genes is shown (not to scale). The transposon terminates transcription and translation of the upstream prp8 gene sequences and drives expression of the prp8 gene sequences downstream, producing two contiguous protein fragments via translation of two separate mRNAs. Each fragment is tagged at its N terminus with a nuclear localization signal (NLS). The disrupted alleles were introduced into the TAP-tagged chromosomal copy of PRP8 in yeast strain RG8T by homologous recombination using PCR-amplified DNA; the outer primer set was for partition after codon 436, and the inner primer set was for partition after codons 770 and 2173 to avoid generating the 9-bp direct repeat (DR) generated during transposition.
We reasoned that if Prp8p does indeed have a modular structure, a small fraction of transposon-dissected genes might be functional. For example, if the transposon inserted in a region between essential functional modules, the two Prp8p fragments might fold independently and associate with each other and/or with other spliceosome components to perform the protein’s essential functions. We screened for functional transposon-dissected alleles of prp8 via plasmid shuffle in a yeast strain carrying a deletion of the chromosomal copy of PRP8. An estimated 60,000 transposition events occurred, of which at least 40,000 were located within the PRP8 open reading frame (i.e., an average of five to six transposition events per nucleotide of coding sequence), and DNA digests showed a broad spread of insertion points (data not shown). As expected, the majority of prp8 alleles carrying a transposon were unable to support growth, but a small number did survive the selection. The prp8::Tn plasmids from these strains were isolated and retested, and the transposon locations were determined by restriction mapping and DNA sequencing.
Several transposons mapped within the first 80 residues of Prp8p, a region containing multiple Proline-rich tracts that are found only in plant and fungal Prp8 proteins (Grainger and Beggs 2005) and are not essential in yeast (K.-L. Boon and J.D. Beggs, unpubl.); these alleles were not analyzed further. Of the remaining gene dissection points, several mapped close together in the vicinity of amino acids 394–443 (specifically, after codons 394, 413, 416, 428, 436, and 443), and there were functional alleles with transposons inserted after codons 770, 2170, and 2179. Significantly, we screened through many thousands of transposons in the interval between 770 and 2170 in Prp8p in an effort to identify additional viable alleles, but none emerged. Western blotting confirmed that Prp8p fragments of the expected sizes were produced in place of full-length Prp8p, and sequence analysis showed that the transposon was inserted in the correct frame to drive expression of an NLS-tagged downstream Prp8p fragment (data not shown). Finally we found by subcloning fragments of dissected prp8 genes to separate plasmids and transforming them individually into yeast cells that for each dissected prp8 allele, expression of both the N- and the C-terminal Prp8p fragments was required for growth (data not shown).
TAP-tag purification of Prp8p complexes
Yeast strains were constructed in which PRP8 is C-terminally TAP-tagged (Prp8T) and dissected immediately after codon 436 (strain KL8TG/436 produces Prp8T/436p, consisting of polypeptides 1–439 plus 437–2413), codon 770 (strain KL8T/770 produces Prp8T/770p, consisting of poly-peptides 1–770 plus 771–2413), or codon 2173 (strain RG8T/2173 produces Prp8T/2173p, consisting of polypeptides 1–2173 plus 2174–2413). Western blotting confirmed that the Prp8p C-terminal fragments produced in the wild-type and the dissected strains were the expected sizes (data not shown, but see below). When tested for growth at various temperatures (18°, 23°, 30°, and 37°C) the strains producing full-length Prp8p (BMA38a and RG8T) or Prp8/ 2173p grew at all the temperatures, although the Prp8T/ 2173 strain grew more slowly, and the strains producing Prp8T/436p or Prp8T/770p were slow-growing and heat-sensitive, failing to grow at 37°C (data not shown).
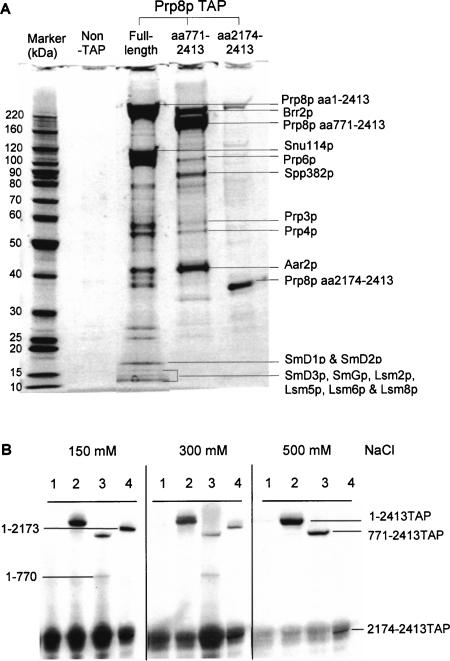
To determine the extent to which Prp8p complexes were intact or were disrupted by the dissection, extracts from parent (RG8T) and dissected strains, KL8T/770 and RG8T/ 2173, were subjected to the TAP affinity-selection procedure (Rigaut et al. 1999) and the coselected proteins were separated by SDS-PAGE and analyzed by mass spectrometry. Proteins that were coselected with full-length Prp8p included Aar2p, Brr2p, Prp3p, Prp4p, Prp6p, and Snu114p, and several Sm and Lsm proteins (Fig. 2A ▶). These represent a subset of the proteins that were previously shown to be components of the U5 snRNP or U4/U6.U5 tri-snRNP (Gottschalk et al. 1999; Stevens et al. 2001, 2002). The Prp8T/770p C-terminal fragment (771–2413) coselected Aar2p, Brr2p, Prp3p, Prp4p, Prp6p, and Spp382p. The Prp8T/2173p C-terminal (2174–2413) coselected a protein similar in size to Brr2p, whereas other coselected proteins were in very low abundance due to the reduced stability of this complex. Association of Brr2p with the extreme C terminus of Prp8p was observed previously in two-hybrid interaction studies (van Nues and Beggs 2001).
FIGURE 2.
TAP purification of Prp8-associated proteins. (A) TAP-purified proteins were fractionated in a 4%–12% PAGE gel and analyzed by mass spectrometry. (B) Effect of salt on the association of the pairs of dissected Prp8 polypeptides. Splicing extracts were incubated with IgG-agarose in the presence of 150, 300, or 500 mM NaCl. Proteins were fractionated, blotted, and probed with anti-8.6 antibodies. (Lanes 1) Nontagged full-length Prp8p, (lanes 2) Tap-tagged full-length Prp8p, (lanes 3) TAP-tagged Prp8p dissected at residue 770, (lanes 4) TAP-tagged Prp8p dissected at residue 2173.
Interestingly, Snu114p was hardly detectable in association with Prp8T/770p, whereas the amount of Aar2p associated with Prp8T/770p was increased compared to the full-length Prp8p. Aar2p was previously found associated with a 16S form of yeast U5 snRNP that contained Prp8p, Snu114p, and the Sm proteins but lacked other known Prp8-associated proteins, including Brr2p. It was suggested that the Aar2-U5 snRNP complex represents an intermediate particle in U5 snRNP biogenesis (Gottschalk et al. 2001). Also, Spp382p was associated with Prp8T/770p but was not detectable with full-length Prp8p or Prp8T/2173p. Spp382p has been affinity-selected previously with Prp8p and other splicing factors (Gavin et al. 2002) but is not known to be a stable component of U5 snRNPs. Thus, Prp8T/770p produced a different profile of coselected proteins compared to full-length Prp8p and this likely includes an Aar2p-U5 snRNP precursor complex related to that described by Gottschalk et al. (2001).
As Western blotting indicated that the N-terminal fragments (1–770 and 1–2173) were detectable in the TAP-purified fraction, albeit at a low level (data not shown), the effects of different salt concentrations were tested in order to establish conditions that gave complete separation of the N- and the C-terminal Prp8p fragments. The Prp8p C-terminal TAP-tagged fragment was affinity-selected using IgG agarose beads, and the presence of the Prp8p N-terminal fragment in coprecipitates was investigated by Western blotting with anti-8.6 antibodies (which recognize the N-terminal fragments of Prp8p but also interact with the protein A moiety of the TAP-tag on the C-terminal fragments). This single-step affinity selection in the presence of 150 mM or 300 mM salt did not disrupt the association of the N- and the C-terminal Prp8p fragments; however, with 500 mM salt, only C-terminal TAP-tagged Prp8p polypeptides were selected (Fig. 2B ▶). It should be noted that 500 mM salt also disrupted the association of the U5 snRNA with Prp8p (data not shown).
Association of Snu114p and Aar2p with Prp8p
The association of N- and C-terminal fragments of Prp8p with other splicing factors was investigated in the presence of 500 mM NaCl. TAP-tagged C-terminal Prp8p fragments were pulled down with IgG-agarose, and the supernates were then incubated with anti-8.6 antibodies to pull down the N-terminal Prp8 polypeptides (Fig. 3A ▶), and these immune precipitates were analyzed by Western blotting for the presence of Prp8p N-terminal fragments, Snu114p, or Aar2p (Fig. 3 ▶).
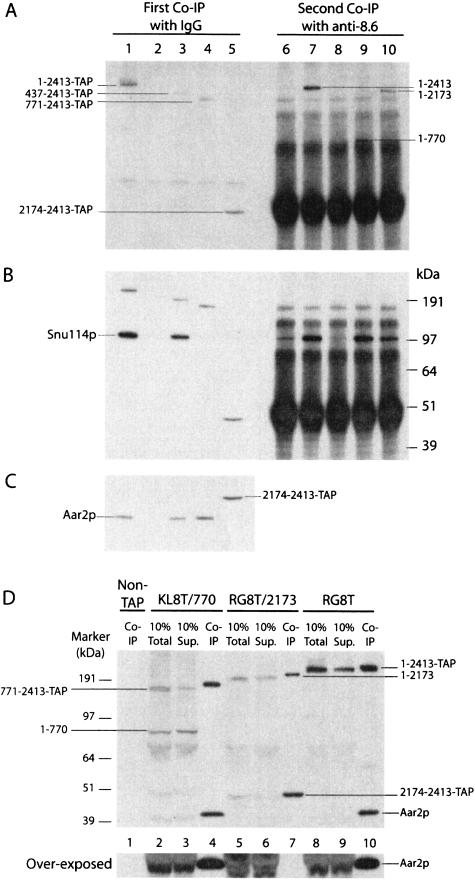
FIGURE 3.
Detection of proteins associated with the N- or C-terminal fragments of Prp8p. (A) Splicing extracts were first incubated with horse IgG agarose in 500 mM NaCl to pull down only the C-terminal Prp8p fragments (lanes 1–5). The supernates were then incubated with anti-8.6-adsorbed protein A Sepharose beads (lanes 6–10). After PAGE, Prp8p fragments that bound to the beads were detected by Western blotting, using anti-8.6 antibodies. (Lanes 1,6) full-length TAP-Prp8 protein; (lanes 2,7) full-length nontagged Prp8p; (lanes 3,8) Prp8T/436p; (lanes 4,9) Prp8T/770p; (lanes 5,10) Prp8T/2413p. The blot from A was stripped and reprobed with anti-Snu114 antibodies (B) or with anti-Aar2p antibodies (C). (D) Splicing extracts from strains producing nontagged full-length Prp8p (lane 1), Prp8T/ 770p (lanes 2–4), Prp8T/2173p (lanes 5–7), and full-length Prp8Tp (lanes 8–10) were incubated with IgG-agarose in the presence of 150 mM NaCl. The precipitates (Co-IP), 10% total, and 10% supernates (Sup) after the pulldown were analyzed by Western blotting with both anti-8.6 and anti-Aar2p antibodies.
Snu114p was detected associated with TAP-tagged full-length Prp8p (Fig. 3B ▶, lane 1) and with C-terminal Prp8p 437–2413 (Fig. 3B ▶, lane 3) but not with Prp8p 771–2413 or 2174–2413 (Fig. 3B ▶, lanes 4,5). Following the precipitation with anti-8.6 antibodies, Snu114p was found associated with non-TAP-tagged Prp8p (Fig. 3B ▶, lane 7) and with N-terminal polypeptides 1–770 and 1–2173 of Prp8p (Fig. 3B ▶, lanes 9,10) but not with full-length TAP-tagged Prp8p or with amino acids 1–439 (Fig. 3B ▶, lanes 6,8), as most Snu114p had already been pulled out of these samples during the first-round precipitation of the TAP-tagged C-terminal polypeptides (Fig. 3B ▶, lanes 1,3). Thus, Snu114p associates with the region 437–770 of Prp8p.
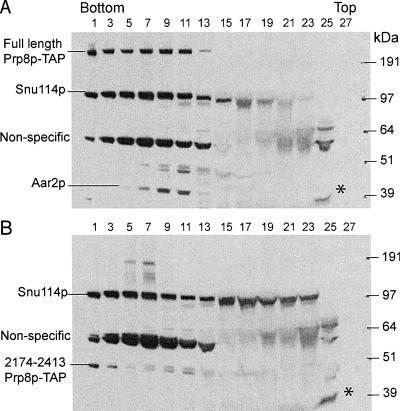
Aar2p was present in precipitates with TAP-tagged full-length Prp8p and with Prp8p residues 437–2413 and 771–2413 (Fig. 3C ▶, lanes 1,3,4) but not with untagged Prp8p, or with Prp8p amino acids 2174–2413 (Fig. 3C ▶, lanes 2,5). Even when the Prp8T/2173p immunoprecipitation was performed under conditions (150 mM NaCl) that did not disrupt the association of the N- and the C-terminal Prp8p fragments (or of the U5 snRNA), Aar2p was not coprecipitated (Fig. 3D ▶, lane 7). In addition, centrifugation through a 10%–30% glycerol gradient (100 mM KCl) showed that Aar2p mostly cofractionates with full-length Prp8p and Snu114p (Fig. 4A ▶, lanes 5–11), although some Aar2p was also detected near the top of the gradient (Fig. 4A ▶, lane 25) as was observed previously (Gottschalk et al. 2001). In contrast, the Aar2p in Prp8T/2173 extract was mostly found in the lower density fraction (Fig. 4B ▶, lane 25). Thus, the interaction of Aar2p with Prp8p was disrupted by dissecting Prp8p at position 2173.
FIGURE 4.
Glycerol gradient fractionation of Prp8p, Snu114p, and Aar2p. RG8T cell extract containing full-length Prp8Tp (A) and RG8T/2173p extract containing Prp8T/2173p (B) were fractionated in 10%–30% glycerol gradients. Alternate gradient fractions were analyzed by Western blotting with anti-Snu114p and anti-Aar2p antibodies. (*) Aar2p near the top of the gradient (lanes 25).
DISCUSSION
Yeast Prp8p is highly conserved in both its sequence (61% identical to human) and its large size. Bioinformatic approaches predicted an MPN (Mpr-1, Pad-1, N-terminal) domain, defined by amino acids 2173–2310, an RRM (RNA Recognition Motif) in the region 1059–1151, and potential NLS(nuclear localization signals, residues 96–156 (Grainger and Beggs 2005), but there is no other obvious structural domain within this huge protein. By functional, genetic classification, Kuhn et al. (Kuhn and Brow 2000; Kuhn et al. 2002) designated five regions: a (residues 233–365), b (residues 608–687), c (residues 785–864), d (residues 1091–1197), and e (residues 1621–1878).
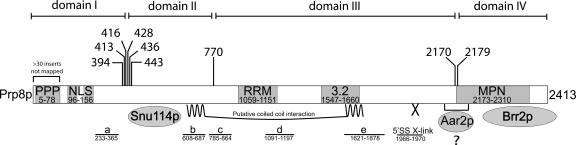
Figure 5 ▶ illustrates the sites of the viable disruptions that we identified relative to these other known features of Prp8p. Considering (1) that a very large number of disrupted prp8 alleles were screened for viability (in excess of 40,000 within the PRP8 open reading frame), (2) the broad spread of insertions observed by DNA digest analysis, and (3) the evidence that Tn5 insertions occur essentially randomly (Shevchenko et al. 2002), it seems unlikely that the three regions of PRP8 in which viable insertions were isolated might represent hotspots for Tn5 insertion. This is further supported by the fact that many viable insertions were found in the nonessential N-terminal proline-rich region of Prp8p. Therefore, the failure to obtain viable constructs with transposon insertions in the 771–2173 region of Prp8p suggests that the genetically defined regions c, d, and e and the RRM function as one indivisible unit. Thus the results presented here show that Prp8p can be dissected into four functional domains: domain I, residues 1–394; domain II, 443–770; domain III, 771–2170; and domain IV, 2179–2413. The dissection points near the C terminus of Prp8p (2170–2179) lie precisely at one boundary of the predicted MPN/JAB domain (2173–2310). This is consistent with the notion that these transposon insertions may be tolerated because they have targeted a “linker” sequence upstream of this functional module. Furthermore, the predicted NLS and the genetically defined region a lie within domain I, region b is within domain II, and the RRM and regions c, d, and e are within domain III.
FIGURE 5.
Diagram showing the positions of viable breakpoints relative to other known features in Prp8p (not to scale). Vertical lines above the protein indicate the positions at which Prp8p can be split in two and still function in trans, in vivo. The domains identified in this work are indicated. Numbers refer to amino acid residues. PPP, proline-rich region; NLS, putative nuclear localization signal; RRM, RNA recognition motif; 3.2, a region of particularly high amino acid sequence conservation; MPN, predicted Mpr-1, Pad-1, N-terminal domain; a, b, c, d, and e, genetically defined regions (Kuhn and Brow 2000); 5′SS X-link, a five-amino-acid region identified by photocross-linking to contact the 5′ splice site in pre-mRNA (Reyes et al. 1999). For further details about features in Prp8p, see Grainger and Beggs (2005).
To investigate which other splicing factors interact with the different domains of Prp8p, 500 mM salt was used to disrupt the associations of the N- and the C-terminal Prp8 polypeptides. The need for such stringent conditions to separate the pairs of dissected Prp8 polypeptides shows that strong intra- and/or intermolecular interactions hold the two ends of the protein together. Kuhn and Brow (2000) proposed the existence of a coiled-coil interaction between residues 640–670 and 1625–1650 and, if this physical interaction exists, it might be responsible for reconstituting the Prp8p molecule that is split at 770/771.
The strong association of the pairs of Prp8 polypeptides limited their utility as a tool to investigate which domains of Prp8p interact with other splicing factors, as 500 mM salt also disrupted the association of Prp8p with U5 snRNA. Nevertheless, using a two-step immunoprecipitation strategy (Fig. 3 ▶), the association of Snu114p with Prp8p was found to require Prp8p residues 437–770, which is in close agreement with two-hybrid interaction data in which Snu114p as bait selected multiple Prp8p fragments that had amino acids 420–464 in common (I. Dix and J.D. Beggs, unpubl.). The strong association between Prp8p and Snu114p is not surprising, as the human homologs (h220K and h116K) have been identified in an RNA-free complex and remain associated in 0.4 M of the chaotropic salt sodium thiocyanate (Achsel et al. 1998).
Interestingly, even at moderate salt concentrations (150 mM NaCl or 100 mM KCl), the association of Aar2p was disrupted by dissecting Prp8p at residue 2173. Previously, Gottschalk et al. (2001) purified yeast Aar2p associated with Prp8p and Snu114p. Here we show that Prp8p/771–2413p could coselect Aar2p in the absence of Snu114p (Fig. 3C ▶, lane 4), and Prp8T/ 2173p could coselect Snu114p in the absence of Aar2p (Figs. 3B ▶ [lane 10], 4B ▶). This suggests that the associations of Snu114p and Aar2p with Prp8p are not interdependent.
In summary, we describe here a novel approach to characterize the functional domains of a protein in vivo and demonstrate its utility by identifying four functional domains within the very large yeast Prp8p splicing factor, one of which—domain III—appears to require a continuous stretch of 1400 amino acids. Although Prp8p has been demonstrated to function when expressed as pairs of trans-acting polypeptides, each of these disruptions was detrimental to growth; the 2173 dissection caused slower growth and the 436 and 770 dissections resulted in heat sensitivity. For the 2173 dissection at least, the integrity of Prp8p’s interaction with another protein was destabilized. Therefore, this approach has also confirmed the functional significance of the very large size of this highly conserved protein that has been proposed to function as a scaffold for molecular interactions in the spliceosome.
MATERIALS AND METHODS
Plasmids and yeast strains used in this work are listed in Table 1 ▶ and oligonucleotides are listed in Table 2 ▶.
TABLE 1.
Plasmids and yeast strains
| Name | Details of plasmid/genotypes of yeast strains |
| pMod-Tn | PRP8 transposon plasmid. Contains PRP8 downstream flanking region, KanMX4 cassette, PRP8 upstream flanking region, SV40 NLS. |
| pBS1479 | PCR template for TAP-tagging (Rigaut et al. 1999). |
| SC261Δ8B1 | MATa, ura3-52, leu2, trp1, pep4-3, prb1-1132, prc1-407, prp8::Blasticidin [pRS316-PRP8] |
| BMA38a | MATa, his3Δ200, leu2-3,112, ura3-1, trp1Δ1, ade2-1, can1-100 |
| RG8T | PRP8:TAP-TRP1, otherwise as BMA38a |
| RG8T/2173 | PRP8(1-2173):Tn:PRP8(2174-2413):TAP-TRP1, otherwise as BMA38a |
| KL8T/770 | PRP8(1-770):Tn:PRP8(771-2413):TAP-TRP1, otherwise as BMA38a |
| KL8TG/436 | His3-PGAL1-HA3:PRP8(1-439):Tn:PRP8(437-2413): TAP-TRP1, otherwise as BMA38a |
TABLE 2.
Oligonucleotides
| AW40 and AW41 introduce an SV40 NLS at Nhel site (N terminus of Prp8p) |
| AW40 CTAGCACCTCCAAAAAGAAGAGAAAGGTAGCT |
| AW41 CTAGAGCTACCTTTCTCTTCTTTTTTGGAGGTG |
| AW21 and AW22 for prp8 3′UTR [EcoRl] |
| AW21 GCAGCAGAATTCTGACTGACTGAATATATTCC |
| AW22 TGCTGCGAATTCGATCAGTTAAATATATATAAG |
| AW25 and AW26 for Kan gene PCR [BamHl] |
| AW25 GCAGCAGGATCCAAAGCCACGTTGTGTCTCAA |
| AW26 TGCTGCGGATCCTTAGAAAAACTCATCGAGCA |
| AW17 and AW18 for prp8 5′UTR PCR [HindIII] |
| AW18 GCTGCTAAGCTTAAGCATCTTTCTTTGACGCTATT |
| AW23 and AW24 produce an SV40 type NLS when cloned into the Aflll site [CTTAAG] introduced by AW17/18PCR |
| AW23 TTAAGCCTCCAAAAAAGAAGAGAAAGGTAGCTCCC |
| AW24 TTAAGGGAGCTACCTTTCTCTTCTTTTTTGGAGGC |
| Prp8-Tap-F and Prp8-Tap-R produce the TAP construct from pBS1479 to the C-terminal of genomic PRP8 |
| Prp8-Tap-F GCGGGGGACGAAGAGTTAGAGGCCGAACAAATCGATGTATTTAGCTCCATGGAAAAGAGAAG |
| Prp8-Tap-R TATATATCTATGAAATAACAGATTCCAGTTTATTGGGGAATATATTACGACTCACTATAGGG |
| Prp8-770-F and Prp8-770-R introduce the transposon construct from pAW28 into PRP8 after codon 770 |
| Prp8-770-F TCTTGAGTTACGAAACTCGGTCATGGATGACATTTTAGAAATGATATCATCGATGAATTCTGACT |
| Prp8-770-R TGTTGCAGTATAGTTCTTGCCTTTTTCTGTCGAATACTCTCAGGCCTGAAGCTTAAGGGAGCT |
| Prp8-2173-F and Prp8-2173-R introduce the transposon construct from pAW28 into PRP8 after codon 2173 |
| Prp8-2173-F CTAATACACTATTATATTTGAGGTTAAAAAACATTTATGTTTCGGATCATCGATGAATTCTGACT |
| Prp8-2173-R TTTTTTGGAAGGACGTAAACATTCTGTTCCTCCACGAAATCATCACTGAAGCTTAAGGGAGCT |
| F-Prp8-938 and R-Prp8-1956 introduce the transposon construct from M529 into PRP8 after codon 436 |
| F-Prp8-938 GGTTATCAACGCCACTTAGAG |
| R-Prp8-1956 ACCATCTGCTAACTGGAAGG |
Transposition methods
The gene dissection transposon was assembled in the vector pMOD〈MCS〉 (Epicentre) by inserting three PCR products as follows: (1) PRP8 gene downstream flanking region (EcoRI site), (2) Kanamycin-resistance cassette (BamHI site), and (3) gene upstream flanking region (HindIII site). An SV40 NLS tag was then inserted into the unique AflII site near the downstream end of the transposon. The structure of the transposon in the resulting pMOD-Tn plasmid is therefore as follows: ME PRP8 gene downstream flanking region Kan PRP8 gene upstream flanking region ATG-NLS ME where “ME” denotes the Mosaic End sequences recognized by Tn5 Transposase (Fig. 1 ▶). Transpo-son DNA was released from the pMOD-Tn plasmid for in vitro transposition reactions by cleavage with PvuII and gel purification. For random insertion of the transposon into the entire PRP8 ORF the target DNA was pRS314 carrying a 9.6-kb fragment of yeast genomic DNA including the PRP8 open reading frame and flanking sequences with unique NheI and MluI sites introduced at the N and the C termini of the coding sequence, respectively. An SV40 NLS tag was also introduced at the NheI site (oligonucleotides AW40 and AW41) prior to the transposition step so that both Prp8p fragments can be localized to the nucleus after gene dissection. Target DNA (1 μg) was incubated in a 10-μL reaction with a molar equivalent of the transposon in the presence of 0.1 U/μL EZ::TN transposase (Epicentre) at 37°C for 2 h, according to the manufacturer’s instructions. The reaction was stopped by addition of 1 μL 1% SDS followed by heating to 70°C for 10 min. Aliquots (1 μL) of the transposition reaction were transformed by electroporation into XL10 Gold cells (Stratagene), and plasmids that had been targeted by the transposon were recovered by selection for ampicillin and kanamycin resistance. Plasmid DNA was prepared from the pooled transformants, and suitable fragments of the prp8 ORF containing transposons were excised, gel purified, and recloned into “gapped” pRS314-PRP8 vector, again selecting for ampicillin and kanamycin resistance. Plasmid DNA was prepared from the pooled transformants to produce a library in which each individual carries a transposon inserted in a region of the prp8 ORF. Functional copies of PRP8 were identified by plasmid shuffle after transformation into haploid yeast strain SC261ΔB1 in which the genomic copy of PRP8 is deleted and survival is dependent on a copy of PRP8 on pRS316 (marked with URA3). Cells harboring a functional transposon-tagged copy of PRP8 were isolated by growth on plates containing 5-fluoroorotic acid (5-FOA) to select for loss of the URA3-marked plasmid copy of PRP8. Plasmid DNA was isolated from these cells and retested by plasmid shuffle. The location of the transposon was determined approximately by restriction mapping and confirmed by DNA sequencing. Note that the transposition event results in the duplication of three amino acids at the site of insertion; e.g., insertion after codon 2170 results in the N-terminal polypeptide terminating with amino acid 2173 and the other starting with amino acid 2171.
Construction of yeast strains with dissected chromosomal prp8 alleles
Strain RG8T was made by integration of a linear DNA cassette, generated by PCR, that encodes the TAP tag sequence (from plasmid pBS1479) (Rigaut et al. 1999), fused in-frame with the 3′ end of the PRP8 open reading frame, plus the TRP1 gene as selectable marker. For insertion of pseudotransposons (mosaic regions omitted) into this chromosomal TAP-tagged copy of PRP8 the Kanamycin-resistance marker was replaced with a KanMX4 cassette (used for dissection after codon 436) (Wach et al. 1994), or, alternatively, a copy of the URA3 gene from Kluyveromyces lactis was inserted between the unique XbaI and PstI sites downstream of the Kanamycin-resistance marker (used for dissection after codons 770 and 2713) and PCR-generated DNA was used for yeast transformation, inserting at specific positions in the TAP-tagged chromosomal copy of PRP8 by homologous recombination. For dissection at position 436, the PCR product was generated with primers in the flanking PRP8 sequences. For dissection at positions 770 and 2173, flanking PRP8 sequences were included in the PCR primers and added directly to the transposon downstream and upstream flanking elements (Table 2 ▶). G418 (geneticin) resistant or Ura+ transformants were selected, correct integration of the cassettes was checked by PCR, and production of the TAP-tagged Prp8p was monitored by Western blotting.
Purification of Prp8p complexes by TAP
Yeast whole-cell extracts were produced (Umen and Guthrie 1995) from 30 L of log phase cells, and tagged proteins were isolated (Rigaut et al. 1999). The purified proteins were resolved on a Novex NuPAGE 4%–12% bis-Tris gel (Invitrogen), using mor-pholinepropanesulfonic acid (MOPS) running buffer and stained with Sypro ruby (Biorad), and the stained bands were subjected to mass spectrometric analysis by the “FingerPrints” Proteomics Facility (Wellcome Trust Biocentre, University of Dundee) and EPIC Proteomics service (Wellcome Trust Centre for Cell Biology, University of Edinburgh).
Immunoprecipitation and immunoblotting
Immunoprecipitations were performed (Teigelkamp et al. 1995) using IgG-agarose (Sigma) or anti-8.6 Prp8 polyclonal antibodies (raised against an N-terminal peptide) in 500 mM NaCl unless stated otherwise. Proteins were resolved in Novex NuPAGE 4%–12% bis-Tris gels (Invitrogen), electroblotted to a polyvinylide difluoride membrane (Hybond-P, Amersham Bioscience), probed with anti-Snu114p antibodies (Bartels et al. 2003) or anti-Aar2p antibodies (Gottschalk et al. 2001), and detected using horseradish peroxidase-conjugated goat anti-rabbit antibodies and ECL reagents (Amersham Bioscience).
Glycerol gradient analysis
Glycerol gradient analysis was performed (10%–30% glycerol gradient containing GG buffer; Bartels et al. 2003) using 80 μL splicing extract diluted with 120 μL GG buffer (20 mM HEPES at pH 7.0, 100 mM KCl, 0.2 mM EDTA; final glycerol concentration 8%). After sedimentation at 37,000 rpm for 17 h in a SW40 Ti rotor (Beckman) at 4°C, 400-μL fractions were collected and stored at −70°C. Alternate fractions were phenol/chloroform extracted, and proteins were acetone precipitated and assayed by Western blotting.
Acknowledgments
We thank Dr. Patrizia Fabrizio for the generous gift of anti-Aar2p and anti-Snu114p antibodies and Dr. Duncan Short for mass spectrometric analysis of the Aar2 and Lsm2 proteins. This work was funded by a studentship from The Darwin Trust of Edinburgh to K.-L.B., by Wellcome Trust Grant 067311 to J.D.B., and by Medical Research Council funding to A.J.N., and benefited from the facilities of the Edinburgh Protein Interaction Centre. J.D.B. is Royal Society Darwin Trust Research Professor.
Correspondence regarding the transposon-based dissection method to: Andrew J. Newman, MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK; e-mail: newman@mrc-lmb.cam.ac.uk.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2281306.
REFERENCES
- Achsel, T., Ahrens, K., Brahms, H., Teigelkamp, S., and Lührmann, R. 1998. The human U5–220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18: 6756–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels, C., Urlaub, H., Luhrmann, R., and Fabrizio, P. 2003. Muta-genesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 278: 28324–28334. [DOI] [PubMed] [Google Scholar]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Collins, C. and Guthrie, C. 1999. Allele-specific genetic interactions between Prp8p and RNA active site residues suggest a function for Prp8p at the catalytic core of the spliceosome. Genes & Dev. 13: 1970–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle, R.F. 1995. The multiplicity of domains in proteins. Annu. Rev. Biochem. 64: 287–314. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gottschalk, A., Neubauer, G., Banroques, J., Mann, M., Lührmann, R., and Fabrizio, P. 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 18: 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk, A., Kastner, B., Luhrmann, R., and Fabrizio, P. 2001. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA 7: 1554–1565. [PMC free article] [PubMed] [Google Scholar]
- Grainger, R.J. and Beggs, J.D. 2005. Prp8 protein: At the heart of the spliceosome. RNA 11: 533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A.N. and Brow, D.A. 2000. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics 155: 1667–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A.N., Reichl, E.M., and Brow, D.A. 2002. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. 99: 9145–9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A.J. 1997. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 16: 5797–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, J.L., Gustafson, E.H., Luo, H.R., Moore, M.J., and Konarska, M.M. 1999. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA 5: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Séraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotech. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Shevchenko, Y., Bouffard, G.G., Butterfield, Y.S.N., Blakesley, R.W., Hartley, J.L., Young, A.C., Marra, M.A., Jones, S.J.M., Touchman, J.W., and Green, E.D. 2002. Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 30: 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceo-some: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Stevens, S.W., Barta, I., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2001. Biochemical and genetic analysis of the U5, U6, and U4/U6.U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 7: 1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Stevens, S.W., Ryan, D.E., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9: 31–44. [DOI] [PubMed] [Google Scholar]
- Teigelkamp, S., Newman, A.J., and Beggs, J.D. 1995. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 14: 2602–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen, J.G. and Guthrie, C. 1995. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA 1: 584–597. [PMC free article] [PubMed] [Google Scholar]
- van Nues, R.W. and Beggs, J.D. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes. Genetics 157: 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsen, P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1739–1808. [DOI] [PubMed] [Google Scholar]