Abstract
The signature DNA lesion induced by ionizing radiation is clustered DNA damage. Gamma radiation-induced clustered DNA damage containing base lesions was investigated in plasmid DNA under cell mimetic conditions and in two cell lines, V79-4 (hamster) and HF19 (human), using bacterial endonucleases Nth (endonuclease III) and Fpg (formamidopyrimidine DNA glycosylase). Following irradiation with 60Co γ-rays, induction of double-strand breaks (DSB) and clustered DNA damage, revealed as DSB by the proteins, was determined in plasmid using the plasmid-nicking assay and in cells by either conventional pulsed field gel electrophoresis or a hybridization assay, in which a 3 Mb restriction fragment of the X chromosome is used as a radioactive labeled probe. Enzyme concentrations (30–60 ng/µg DNA) were optimized to minimize visualization of background levels of endogenous DNA damage and DSB produced by non-specific cutting by Fpg and Nth in cellular DNA. 60Co γ- radiation produces a 1.8-fold increase in the yields of both types of enzyme sensitive sites, visualized as DSB compared with that of prompt DSB in plasmid DNA. In mammalian cells, the increase in yields of clustered DNA damage containing either Fpg or Nth sensitive sites compared with that of prompt DSB is 1.4–2.0- and 1.8-fold, respectively. Therefore, clustered DNA damage is induced in cells by sparsely ionizing radiation and their yield is significantly greater than that of prompt DSB.
INTRODUCTION
Ionizing radiation causes a spectrum of biological effects in cells. Damage produced within DNA may lead to mutations and cancer in mammals. Ionizing radiation produces a variety of DNA lesions through direct and indirect effects, such as single-strand breaks (SSB), double-strand breaks (DSB), AP sites (either apyrimidinic or apurinic), DNA–DNA and DNA–protein cross-links along with a plethora of base modifications (1–3). Many endogenous DNA lesions, formed as by-products of oxygen metabolism in cells, are chemically identical to the individual lesions induced by ionizing radiation (4). These endogenously induced lesions are mainly isolated. Although many of the lesions induced by ionizing radiation are isolated, radiation also induces clustered DNA damage, which represent two or more lesions formed within one or two helical turns of DNA and is in part responsible for the biological effects of ionizing radiation. This type of damage includes not only prompt DSB, but also non-DSB clustered damage such as a SSB formed in close proximity to additional breaks or base lesions on both strands. It has been shown that as the ionizing density of radiation and hence the yield and complexity of clustered DNA damage increases, the repairability of DSB in cells decreases (5–7). From biophysical modeling (8), it was hypothesized that clustered DNA damage induced by radiation is less readily repaired than individual lesions. The cytotoxic, mutagenic and carcinogenic effects of ionizing radiation may be caused by clustered DNA damage.
The majority of studies have focused on DSB and less so on non-DSB clustered DNA damage containing a combination of base lesions or SSB. It is important to consider the contribution that base lesions within clustered DNA damage make to the biological severity of damage induced by ionizing radiation. The majority of cells maintain the integrity of the genome by repairing isolated lesions in DNA using base excision repair and strand break repair pathways (4,9,10). However, there is evidence which shows that non-DSB clustered DNA damage containing base lesions and/or SSB may compromise the efficiency of eukaryotic DNA damage repair by reducing the ability of glycosylases to excise base lesions and AP endonucleases to incise AP sites within a clustered damage (11–19). There is also evidence in prokaryote systems that non-DSB clustered damage is present and may, in part, be converted into DSB post-irradiation (20).
It is also possible to detect non-DSB clustered lesions by converting them into DSB using bacterial endonucleases, which recognize and remove modified DNA bases leaving AP sites that are then converted into strand breaks by an independent or associated glycosylase activity. The resulting DSB can be detected in cells using pulsed field gel electrophoresis (PFGE) and in plasmid by constant field gel-electrophoresis. Previous studies have used bacterial enzymes to detect base lesions induced in naked DNA (21,22). In this study, Nth (endonuclease III) and Fpg (formamidopyrimidine DNA glycosylase) were used to cleave oxidized pyrimidines or oxidized purines, respectively, and convert the resulting AP site into a SSB since each enzyme has associated AP lyase activity (23). Both Nth and Fpg enzymes caused non-specific cutting of non-irradiated DNA at high concentrations. Therefore it was important to carry out titrations of enzyme concentration versus DSB yield for each enzyme to find the optimum concentration for maximal specific cutting of base lesions and minimal non-specific cutting. The plasmid-nicking assay permits detection of SSB, DSB, base damage and non-DSB clustered lesions in plasmid DNA. PFGE allows detection of DSB and non-DSB clustered damage within the whole genome of mammalian cells and the hybridization assay allows detection of these types of damage in a smaller, gene sized, fragment of the genome (24).
To date, non-DSB clustered DNA damage has been shown to be induced in cells by heavy ion radiation (25). In this study, the first evidence that non-DSB clustered DNA damage is induced in high abundance relative to prompt DSB in cells even by sparsely ionizing (low LET) radiation is presented. The yields of clustered DNA damage induced in cells has been compared with that in plasmid DNA under conditions where the mean diffusion distance of the water radicals are comparable with that in cells (cell mimetic conditions). For reasons stated earlier, it is important to quantify the relative yields of radiation induced lesions to the endogenous levels within mammalian cells.
MATERIALS AND METHODS
Plasmid
Plasmid pUC18 (2686 bp) was prepared in its mainly (>95%) supercoiled form using the caesium chloride preparation method (26).
Cell culture
HF19 cells are non-transformed human fibroblasts derived from the lung of a female fetus (27). V79-4 cells are fibroblasts derived from the Chinese hamster ovary (CHO) cell lines. Both cell types were grown as monolayers as described previously (5). Briefly, cells were maintained at 37°C in 95% air, 5% CO2, in Eagle’s minimal essential medium (MEM, Sigma), supplemented with 10% fetal calf serum (Mycoplex/PAA), 0.1% penicillin/streptomycin (Gibco) and 0.1% l-glutamine (Gibco).
Determination of radiation-induced DNA damage in plasmid DNA
Plasmid nicking assay
Plasmid DNA (1.5 µg in 5 µl) was irradiated with 60Co γ-rays at 4°C, pH 7.5 in aqueous aerated solution containing 0.2 M Tris as a scavenger to provide cell mimetic conditions. The irradiations were carried out at a dose rate of ∼50 Gy min–1. After irradiation, the DNA was precipitated with ice cold ethanol and resuspended in 40 µl endonuclease reaction buffer (40 mM HEPES, pH 7.9, 100 mM potassium chloride, 0.5 mM EDTA and 0.2 mg/ml BSA). Control, non-irradiated plasmid was treated in the same way. Each sample was split into two 20 µl volumes, one for treatment with enzyme and one for mock treatment without enzyme. Optimal concentrations of Nth and Fpg enzymes were determined by titration (data not shown) so that there is no significant non-specific cutting of non-irradiated plasmid DNA. Nth and Fpg enzymes (stored in buffer containing 50% glycerol, 100 mM potassium phosphate, pH 6.6, 100 mM DTT and 0.005% Triton X-100) were used at a final concentration of 2.6 and 42 ng/µg DNA, respectively. Mixtures were incubated at 37°C for 30 min (Nth) or 60 min (Fpg) and the reaction was stopped by the addition of 5 µl 0.5 M EDTA, pH 8.0. Samples were stored on ice before addition of 10 µl of loading buffer [0.1% bromophenol blue, 30% sucrose in 1× TBE (89 mM Tris, 89 mM boric acid and 2 mM Na2EDTA)]. Samples (20 µl) were loaded onto a 1% agarose (Sigma type 1A) gel in 1× TBE at pH 7.1 and run at 74 mV/cm, 6 mA for 17 h at 4°C. The resulting gel was subsequently stained in 600 ml 0.5 µg/ml ethidium bromide in 1× TBE for 1 h at 4°C. DNA was visualized on a UV transilluminator. An image of the gel was captured through a CCD linked to a computer supporting Quantity One (Bio-Rad laboratories) software and the relative proportions of supercoiled, relaxed and linear plasmid were determined. Ethidium bromide has a lower binding efficiency to supercoiled form than to linear or relaxed DNA; intensities of the supercoiled band were therefore multiplied by a factor of 1.4 (28).
Determination of strand break yield. The SSB yield (SSB/Gy/Da) was calculated using the following equation.
SSB yield = 1/(D37 × 2686 × 650)1
Where D37 is the dose (Gy) required to give, on average, 1 SSB per plasmid molecule (assuming a Poisson distribution of breaks), 2686 is the number of base pairs per plasmid molecule and 650 is the molar mass of 1 base pair.
The yield of DSB was determined from the linear dependence of the percentage linear plasmid on dose. The number of DSB present at a dose corresponding to the D37 was calculated after fitting the D37 value to the linear regression analysis. The yield of base lesions was calculated by subtraction of the yield of SSB in plasmid that had received only mock enzyme treatment from the yield of SSB in plasmid following enzyme treatment. The yields of non-DSB clustered damage were determined from the difference in yields of DSB determined in the presence and absence of enzymatic treatment of irradiated DNA.
Determination of radiation-induced DNA damage in mammalian cells
The fraction of activity released (FAR) assay
The FAR assay allows detection of induced DSB in the whole genome of cells. A 24 h subculture of cells (2.5 × 105 cells) in T25 flasks (Falcon) was labeled with tritiated [3H]thymidine. Briefly, 5 ml of normal growth medium was replaced with 5 ml medium containing 92.5 µl PBS, 2.5 µl (1 mg/ml) thymidine and 5 µl (37 MBq/ml) [3H]thymidine (Amersham International). One hour before γ-irradiation, the cells were ‘chased’ with unlabeled thymidine (0.1 mM) at 37°C and then equilibrated at 4°C prior to γ-irradiation. Aerated flasks were irradiated with ∼1 Gy/min 60Co γ-radiation at 4°C as described in (5). After irradiation, cells were trypzinized on ice to minimize DSB rejoining and centrifuged at 150 g for 10 min at 4°C. The pellet was resuspended in 0.8% low gelling temperature agarose (Sigma, type VII) (at 35–40°C) at a cell density of ∼0.25 × 106 cells/ml. A total of 100 µl of agarose–cell mixture was immediately transferred into moulds to form cell plugs that were placed on ice to set the mixture. In this way, repair of damage was minimized during preparation of the cell plugs.
Two different lysis conditions, namely proteinase-K lysis and salt lysis, were used for determination of DNA damage using the FAR assay. During proteinase-K lysis, cells are held at 37°C for a prolonged period of time, whereas during salt lysis, cells remain at 4°C. It should be noted that the percent DNA extracted following salt lysis is lower than that following proteinase-K lysis.
Proteinase-K lyse. Plugs were placed in 10 ml of buffer A containing proteinase-K (0.05%), N-Lauroyl sarcosine (2%) and 0.5 M EDTA at pH 7.5 for 1 h at 4°C, followed by 24 h at 37°C.
Salt lyse. Plugs were transferred into 30 ml ice cold buffer B (10 mM Tris–HCl, pH 7.6, 140 mM NaCl, 1 mM MgCl2). After 10 min on ice, the buffer was replaced with fresh buffer B supplemented with 0.5% Triton X-100. After 20–30 min the plugs were washed three times in buffer B to remove residual detergent. Plugs were incubated at 4°C for 1 h in buffer B containing 2 M NaCl and this was replaced with fresh buffer B containing 2 M NaCl for continued incubation overnight. This method of lysis of the cells maintained the DNA at 4°C throughout.
Treatment of cells with Fpg and Nth. Plugs were washed at least six times for 45 min each in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 7.5) at room temperature. Prior to treatment with Nth or Fpg proteins, plugs were suspended in 400 µl 1.25× endonuclease reaction buffer for 1 h at room temperature. For treatment and mock-treatments, plugs were suspended in fresh 1.25× endonuclease reaction buffer containing the optimum concentration of Nth or Fpg enzymes (see Results). Reactions were incubated at 37°C for 60 min.
The yields of DNA DSB were measured using PFGE (Bio-Rad model CHEF DR11). A 0.8% agarose (Ultrapure, Gibco) gel was prepared in 0.5× TBE buffer. The yeast molecular weight markers Schizosaccharomyces pombe (3.5–5.7 Mbp) and Saccharomyces cerevisiae (0.2–2.2 Mbp) were used to determine the exclusion size of the DNA. The conditions of the PFGE run were 45 V with 60-min pulse times for 96 h at pH 8.3 and 16°C. After electrophoresis, the gel was stained for 1 h with 0.5 µg/ml of ethidium bromide. DNA within the gel was viewed over a UV transilluminator and the lanes of gel containing the DNA were sectioned into 3–4 mm pieces. Sections were placed into individual glass scintillation vials with 200 µl of 1 M HCl to prevent re-gelling of the agarose, and the mixture was heated to melt the gel. When the vials had cooled, 3 ml of liquid scintillant (Optiphase Hisafe 3) was added to each vial and mixed thoroughly by vortexing. The activity of 3H in each vial was determined using a scintillation counter (Beckman LS6500).
Determination of FAR. The yield of DSB after irradiation was determined from the amount of DNA extracted from the well.
FAR = c.p.m. (lane)/[c.p.m. (lane) + c.p.m. (well)]2
The percentage of activity released is given by FAR × 100. The value of FAR was converted into the yield of DSB/cell/Gy using the following equation:
FRETAINED = exp[–ηD(k/m)] {1 + –ηD(k/m) · [1 – (k/m)]}3
The experimentally determined FRETAINED = 1 – FE, η is the mean frequency of DSB per chromosome per unit dose, k is the exclusion size of DNA able to leave the well (≥5.7 Mbp), m is the mean size of a chromosome. The value of m was taken to be 245 Mbp for V79-4 cells and 130 Mbp for HF19 cells and the number of chromosomes to be 22 for V79-4 cells and 46 for HF19 cells.
Hybridization assay
In this assay, induction of DSB is measured in a defined large fragment of the genome (a 3 Mbp section of the X chromosome) (24). Cells were irradiated as above and suspended in agarose plugs as described for the FAR assay (above), except that the cell density was ∼2.5 × 106 cells/ml.
Cells were lysed in the plug with a solution of proteinase-K (1 mg/ml), N-Lauroyl sarcosine (1%) and 0.5 M EDTA at pH 8.0 for 1 h at 4°C, followed by 48 h at 50°C. Fpg and Nth treatments were performed as described in the FAR assay. Plugs were then washed twice in TE buffer and were suspended in 1.25× NotI restriction enzyme buffer (7.5 mM Tris–HCl, pH 7.9, 187.5 M NaCl, 7.5 mM MgCl2, 1.25 mM DTT and 0.1 mg/ml BSA) for 1 h at room temperature. Each plug was placed in fresh buffer containing 20 U NotI restriction enzyme (Promega) and incubated at 37°C overnight. The reaction was stopped by addition of 20 µl 0.5 M EDTA, pH 8.0 and DSB yield was determined using PFGE and Southern blotting as described by Löbrich et al. (24). An agarose (Ultrapure, Gibco) gel was prepared at a concentration of 0.8% in 1× TAE (0.04 M Tris–acetate, 0.001 M EDTA) buffer. The yeast molecular weight markers S.pombe (3.5–5.7 Mbp) and S.cerevisiae (0.2–2.2 Mbp) were also run on each gel. The conditions of the PFGE run were 2 V/cm in 1× TAE buffer, pH 8.3 with 50–5000 s switch times for 72 h at 12°C. After electrophoresis, the gel was stained for 1 h with 0.5 µg/ml ethidium bromide in TAE. DNA within the gel was viewed over a UV transilluminator and photographed. The gel was treated with 0.25 M HCl for 10 min to partially depurinate the DNA therein. After rinsing in water, the gel was transferred onto nylon membrane (Hybond-N+, Amersham) with denaturing/transfer solution (0.5 M NaOH) using a vacuum blotter for 90 min. The membrane was washed in 2× SSPE (150 mM NaCl, 10 mM sodium phosphate and 1 mM sodium–EDTA, pH 7.4). Pre-hybridization was for at least 5 h at 65°C, rotating in a hybridization oven (Hybaid) in buffer containing 5× SSPE, 7.5× Denhardts, 3% SDS and 4 mg/ml salmon sperm DNA. The probe specific for the 3 Mbp NotI fragment of interest (ATCC 95474) was radioactively labeled according to the manufacturer's recommendations (Amersham rediprime II random primer labeling system) with 1850 KBq [α-32P]dCTP, specific activity 222 TBq/mmol. Hybridization of the probe was performed overnight at 65°C in pre-hybridization buffer. The membrane was washed, wrapped in saran wrap and placed with a phosphorimager screen for 1–5 days. The image was captured by phosphorimager (Bio-Rad molecular imager FX) and analyzed using Quantity One (Bio-Rad) software. The yield of DSB (DSB/fragment) was determined using equation 4, assuming a Poisson distribution of DSB in the DNA. The equation depends on the intensity of activity associated with the specific 3 Mbp band compared with the intensity of the whole lane, relative to that in the control, non-irradiated DNA lane.
DSB yield = –ln[(IBand/ILane)D/(IBand/ILane)control]4
Where IBand is band intensity, ILane is intensity in the entire lane, D is dose and control is non-irradiated DNA.
RESULTS
Plasmid
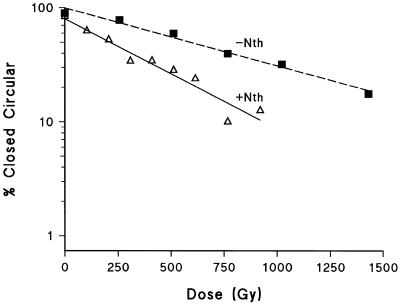
Figure 1 shows data from one experiment of the dependence of the loss of closed circular plasmid DNA on radiation dose (at 4°C) after subsequent treatment with buffer at 37°C in the absence and presence of Nth. The yields of SSB and base damage, revealed as SSB following Nth treatment were determined from the relative slopes of these dependencies. The yield of base damage revealed by Fpg treatment was also determined from dose dependencies (data not shown). The yield of base damage (revealed as additional SSB by either Nth or Fpg treatment) per SSB for plasmid DNA irradiated under cell mimetic conditions is presented in Table 1 (column 2). Therefore substantial numbers of base lesions are induced by Co60 γ-radiation that are recognized by Nth and Fpg enzymes. Treatment with both enzymes combined gave a yield of base damage equivalent to that obtained by summation of individual yields of Fpg and Nth sensitive sites (Table 1). Therefore these enzymes are effectively acting additively and recognize only a minimum of the same damaged sites [in accordance with Milligan et al. (29)]. The number of strand breaks revealed by incubation with buffer at 37°C includes heat labile sites, consequently at 37°C an additional 23% of SSB were observed compared with the yield of SSB measured at 4°C.
Figure 1.
Dependence of the loss of closed circular plasmid DNA on γ- radiation dose (at 4°C) after subsequent treatment with buffer at 37°C in the absence (closed squares) and presence (open triangles) of Nth. This is an example of data from one experiment.
Table 1. Yields of base damage per SSB and non-DSB clustered damage per DSB, measured in plasmid DNA.
| Treatment | Base damage | Non-DSB clustered damage |
|---|---|---|
| Nth | 2.0 ± 0.3a | 0.8 ± 0.3a |
| Fpg | 0.9 ± 0.2a | 0.8 ± 0.2a |
| Nth + Fpg | 2.9 ± 0.5a | 1.6 ± 0.5a |
aMean of at least three repeat experiments.
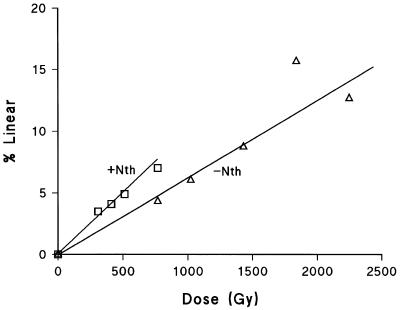
Figure 2 shows the dependence of the yield of linear plasmid DNA on radiation dose (at 4°C) after post-irradiation treatment of the DNA with buffer at 37°C in the absence and presence of Nth. The yields of DSB and non-DSB clustered damage, converted to DSB by Nth treatment, were determined from the relative slopes of these two dependencies. Similar dependencies were determined with Fpg enzyme (data not shown). Table 1 (column 3) presents the yields of DSB revealed by treatment of irradiated DNA with Nth and/or Fpg per DSB detected in the absence of enzyme treatments following the same radiation dose. In this case, Fpg and Nth treatment both reveal 0.8 extra DSB per DSB (prompt and heat labile). As these yields of DSB were measured at 37°C, heat labile sites are included in this yield, which represents an extra 94% DSB compared with the yield measured at 4°C. The combined treatment with Fpg and Nth gives a yield equal to the sum of the yields from the individual treatments, implying that the effect of the two enzymes is additive. These additional DSB represent clustered DNA damage, converted into DSB by enzyme treatment, but do not indicate the complexity of these clusters, i.e. the number of lesions in the cluster.
Figure 2.
Dependence of increase in linear plasmid DNA on γ-radiation dose (at 4°C) after subsequent treatment with buffer at 37°C in the absence (open triangles) and presence (open squares) of Nth. This is an example of data from one experiment.
The amount of damage detected in plasmid DNA has been converted into a value of lesions/cell equivalent assuming 6 × 109 bp per human diploid genome. For SSB, the value is 2217.6 SSB/cell equivalent/Gy (5.6 × 10–10 SSB/Gy/Da in pUC18). For base damage (recognized by Fpg and Nth combined) the value is 6652.8 BD/cell equivalent/Gy (1.7 × 10–9 BD/Gy/Da in pUC18). For DSB the yield is 63 DSB/cell equivalent/Gy (16 × 10–12 DSB/Gy/Da) and for non-DSB clustered DNA damage the value is 101 lesions/cell/Gy (25.5 × 10–12 BD/Gy/Da) (i.e. extra DSB after Nth and Fpg treatment).
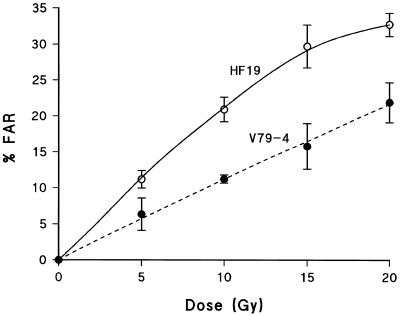
Induction of prompt DSB measured using the FAR assay
Figure 3 shows the induction of prompt DSB, presented as % FAR, in both HF19 and V79-4 cells as a function of 60Co γ-radiation dose. Based on % FAR per unit dose, almost twice as many prompt DSB are induced in HF19 cells than in V79-4 cells by this type of radiation, consistent with data presented by Foray et al. (30,31). They reported that HF19 cells irradiated by γ-radiation at 4°C produce FAR values of ∼2% FAR/Gy at low doses, whereas the majority of data (31–35) on V79-4 cells give FAR values of ∼1% FAR/Gy. Inserting experimental data into equation 3 to convert the value of % FAR into a yield of DSB, gave yields of 73.7 DSB/cell/Gy and 40.5 DSB/cell/Gy for HF19 and V79-4 cells, respectively. These values are dependent upon the exclusion size used in equation 3. If the value of the exclusion size is 10 Mbp, rather than 5.7 Mbp, the resulting DSB yield would be: 40.7 DSB/cell/Gy for HF19 and 22.6 DSB/cell/Gy for V79-4. The latter yield in HF19 cells is consistent with the reported yield of 36 DSB/cell/Gy measured by Foray et al. (29) in HF19 cells following γ-radiation, using equation 3 and an exclusion size of 10 Mbp.
Figure 3.
Dependence of the induction of prompt DSBs, presented as % FAR, in both HF19 (open circles) and V79-4 (closed circles) cells on dose of 60Co γ-radiation. Lysis of cells in the FAR assay was performed with proteinase-K (see Materials and Methods). Error bars represent the SEMs calculated from at least three experiments.
Induction of non-DSB clustered DNA damage containing Fpg sensitive sites (Fpg SS) measured using the FAR assay
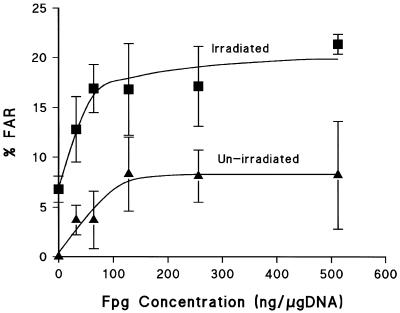
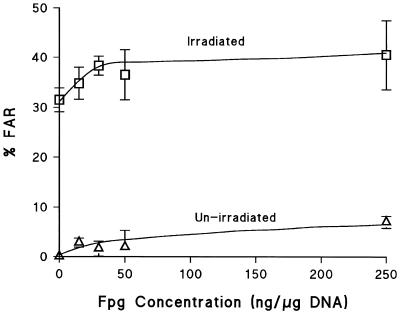
Titration curves for induction of clustered DNA damage which are converted into DSB by Fpg were determined using the FAR assay in both V79-4 and HF19 cells (Figs 4 and 5). It should be stressed that V79-4 cells in Figure 4 had undergone a salt lyse at 4°C, whereas HF19 cells in Figure 5 were lysed with proteinase-K at 37°C. In both cases there is a clear relationship between enzyme concentration and % FAR achieved with these different lysis conditions. In both cell lines, an increase in not only the yield of clustered damage containing Fpg SS in γ-irradiated DNA, but also in control, non-irradiated DNA was observed as the enzyme concentration was increased. The optimal enzyme concentration maximises inclusion of radiation induced clustered DNA damage containing Fpg SS, whilst minimizing the non-specific cutting component seen in non-irradiated DNA. This enzyme concentration was determined at the point where the difference in DSB yield between the non-irradiated and irradiated DNA was the greatest and the contribution of non-specific cutting was <5%. The optimum concentration of Fpg is ∼30 ng/µg DNA for HF19 cells (Fig. 5) and 60 ng/µg DNA for V79-4 cells (Fig. 4). Under these conditions the yield of DSB detected in un-irradiated DNA is ∼170 DSB/cell in HF19 cells and ∼355 DSB/cell in V79-4 cells; these DSB include endogenous clusters containing Fpg SS and those produced by non-specific cutting of the DNA. The FAR values were determined after treatment at 37°C in the presence or absence of Fpg. These data can be converted into actual numbers of DSB and non-DSB clustered DNA damage detected per cell per Gy and the yields are shown in Table 2.
Figure 4.
Dependence of the induction of DSBs, presented as % FAR, on Fpg concentration in V79-4 cells either un-irradiated (closed triangles) or irradiated by 20 Gy 60Co γ-radiation (closed squares). Lysis of cells in the FAR assay was performed with NaCl (see Materials and Methods). Error bars represent the SEMs calculated from at least three experiments.
Figure 5.
Dependence of the induction of DSBs, presented as % FAR, on Fpg concentration in HF19 cells either un-irradiated (open triangles) or irradiated by 20 Gy 60Co γ-radiation (open squares). Lysis of cells in the FAR assay was performed with proteinase-K (see Materials and Methods). Error bars represent the SEMs calculated from at least three experiments.
Table 2. Yield (lesions/cell/Gy) of prompt DSB and non-DSB clustered DNA damage containing Fpg and Nth sensitive sites induced by ionizing radiation in cells.
| Cell line | Assay | DSB | Fpg | Nth |
|---|---|---|---|---|
| HF19 | FAR | 74a | 26a | – |
| HF19 | HYB | 33 | – | 25 |
| V79-4 | FAR | 40a | 44a | 32a,b |
aCalculated using equation 3, assuming a k-value of 5.7 Mb.
bJenner et al. (36).
From Table 2 the increase in the yield of DSB in HF19 cells following Fpg treatment is an extra 0.4 DSB for each prompt DSB (including heat labile sites) formed. In V79-4 cells, the maximum increase in DSB yield following treatment with Fpg enzyme reveals an extra 1.1 DSB for each prompt DSB (including heat labile sites) formed by radiation.
Induction of DSB and non-DSB clustered DNA damage containing Nth sensitive sites (Nth SS) measured using the HYB assay
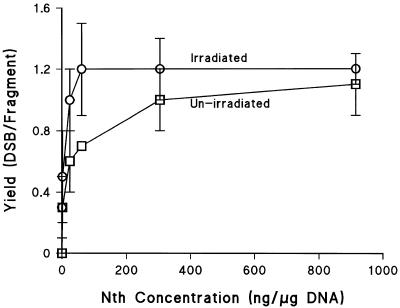
The induction of DSB was measured using the hybridization assay. Following a 20 Gy dose of γ-radiation in the absence of post-irradiation treatment with the enzyme, Nth, 0.33 prompt DSB/fragment are induced (Fig. 6), equivalent to 33 DSB/cell/Gy. This value is similar to that obtained by Löbrich et al. (24) who determined 37.8 DSB/cell/Gy in human SP3 cells irradiated with X-rays using a hybridization assay with a 1.6 Mbp fragment. However, it is lower than the value determined using the PFGE assay, equation 3 and an exclusion size of 5.7 Mbp. Figure 6 presents titration curves for induction of Nth SS in HF19 cells by γ-radiation, determined using the HYB assay. A clear relationship exists between enzyme concentration and detection of extra DSB revealed following a post-irradiation treatment with Nth. Not only does the yield of non-DSB damage containing Nth SS increase in irradiated DNA, but the yields also increase in non-irradiated DNA as the enzyme concentration is increased. It is not possible to differentiate between non-irradiated and irradiated DNA at high enzyme concentrations. The optimum enzyme concentration of Nth is between 30 and 60 ng/µg DNA, which is cognate with data obtained using the conventional FAR assay and Fpg enzyme. Under these conditions the yield of DSB detected in un-irradiated DNA is ∼500 DSB/cell in HF19 cells: these DSB include endogenous clusters containing Nth SS and those produced by non-specific cutting of the DNA. For γ-irradiation of HF19 cells, the maximum increase in yield of DSB in irradiated DNA after Nth treatment reveals an extra 0.8 DSB for each prompt DSB formed. This represents 25 non-DSB clustered damage/Gy/cell revealed by Nth treatment. This value is similar to that previously reported by us for the yield of non-DSB clustered DNA damage containing Nth SS induced by γ-radiation in V79 cells (36).
Figure 6.
Dependence of the induction of DSBs on Nth concentration in HF19 cells either un-irradiated (open squares) or irradiated by 20 Gy 60Co γ-radiation (open circles) and measured using the hybridization assay. Error bars represent the SEMs calculated from at least three experiments.
DISCUSSION
Significant yields of non-DSB clustered DNA damage have been detected in mammalian cells after exposure to sparsely ionizing, low LET γ-radiation, through the use of Fpg and Nth as probes. The observation of non-DSB clustered DNA damage induced by low LET radiation confirms qualitatively the predictions of biophysical simulations that radiation induces non-DSB clustered DNA damage. Previously it has been shown that very densely ionizing charged particles produce clustered DNA damage in cellular DNA (25).
Data obtained using plasmid DNA show that the effects of Nth and Fpg in inducing additional breaks are additive, confirming previous reports (37) that the overlap of substrate specificity of radiation-induced lesions is small. Therefore the yield of non-DSB clustered DNA damage sites in cells has been obtained from the sum of the yields of non-DSB clustered DNA damage measured following individual treatment with Fpg or Nth. The additive yields of Fpg and Nth sites predict that the yield of AP sites induced directly by γ-radiation is small. AP sites are recognized by both enzymes and converted into strand breaks. A low yield of AP sites was confirmed by the AP endonuclease HAP1, which does not cleave γ-irradiated plasmid DNA to a great extent (Siobhan Cunniffe, Medical Research Council, Harwell, personal communication). In mammalian cells, the yields of non-DSB clusters containing enzyme sensitive sites (ESS) per DSB measured at 37°C is 0.6 for HF19 cells and 0.9 for V79-4 cells. In this case, the measured number of induced DSB includes not only prompt radiation-induced DSB since 50% of the DSB measured at 37°C are due to heat labile sites (38), which may include oxidized abasic sites, for example 2-deoxyribonolactone (39). Therefore the yield of non-DSB clusters containing ESS per prompt radiation-induced DSB is 2.2 for HF19 cells and 2.8 for V79-4 cells. In other words, of the total number of clustered DNA damage sites induced by γ-radiation in mammalian cells, ∼80% are due to non-DSB clustered DNA damage. Sutherland et al. (25) report similar ratios of non-DSB clusters: DSB measured in genomic DNA following high LET charged particle radiation.
The yields above are lower limits of the number of non-DSB clustered DNA lesions induced by γ-radiation. There are several situations in which clustered DNA damage will be undetected using these enzyme probes, leading to an underestimation of their yield: (i) when a SSB or a damaged base has an associated base lesion on the same DNA strand; (ii) not every type of base lesion is recognized by—or accessible to—Nth and Fpg enzymes, for example 5-formyluracil, a major product induced by ionizing radiation (40) is excised from human DNA by a specific DNA glycosylase activity (41) and 5-(hydroxymethyl)uracil, also formed by ionizing radiation (42) is recognized by a specific 5-(hydroxymethyl)uracil DNA N-glycosylase (43); (iii) there is evidence to show that the cutting by Fpg and Nth enzymes can be inhibited by the presence of closely associated DNA lesions (14–18); and (iv) if two sites of clustered DNA damage are located close to each other, they will not be detected as distinct clustered damage sites using the assays employed, as has been discussed for high LET induced DSB (25).
The yield of SSB induced by γ-radiation in mammalian cells is ∼1000 SSB/cell/Gy, and we have shown that Fpg treatment reveals 26 (HF19) and 44 (V79-4) non-DSB clustered damage sites and Nth treatment reveals 25 (HF19) and 32 (V79-4) non-DSB clustered damage sites. Therefore, 1 in 20 (HF19) and 1 in 13 (V79-4) SSB has an associated Fpg SS or Nth SS on the opposite DNA strand if the main type of clustered damage revealed contains a SSB opposite to a base lesion. In other words, in mammalian cells 5–8% of SSB have at least one base lesion associated with them on the opposite DNA strand following γ-radiation. If the base lesion is found on the same strand near to a SSB, this type of damage would not be seen as a DSB.
Assuming that half of base damage sites are not detected because they are on the same DNA strand as the initial lesion, then it is inferred that at least 10–16% of SSB have an associated ESS on either DNA strand. From biophysical modelling, Nikjoo et al. (8) have calculated that 37% of all SSB should have at least one base lesion on either strand following γ-radiation. In this study, 51 non-DSB clustered DNA damage sites containing ESS per human cell/Gy, revealed as an increase in the yield of DSB following γ-radiation and treatment with Nth and Fpg enzymes were detected. Pouget et al. (44) reported that there are 576 Fpg SS and 636 Nth SS per human cell/Gy γ-radiation revealed as an increase in SSB using the comet assay. These data, together with the yield of SSB induced by γ-radiation as 1000 SSB/cell/Gy, mean that 2.3% (51/2203) of the yield of individual lesions (SSB or Nth SS or Fpg SS) are non-DSB clustered DNA damage.
Plasmid DNA investigations have allowed us to look more closely at the formation of base lesions by γ-radiation under cell mimetic conditions. Many studies of base damage induction in plasmid DNA by γ-radiation have been conducted under relatively low scavenging conditions (25,45). Under these conditions, the majority of the damage comes from interactions of isolated OH radicals with DNA. There are only a few reports under cell mimetic conditions with high scavenging capacities (46–48). The yield of base lesions per SSB is 2.0 for γ-irradiated plasmid DNA under cell mimetic conditions (3 × 108 s–1). The base lesions were detected by Nth and Fpg treatment. Prise et al. (47) report a higher yield of 2.7 base damage per SSB after γ-radiation under these conditions and Nth treatment of the plasmid DNA. Milligan et al. (37) report 2.5 Nth SS per SSB and 3.0 Fpg SS per SSB following γ-irradiation of plasmid DNA under high scavenger conditions. The yield of non-DSB clustered DNA damage per DSB in plasmid DNA following γ-radiation and treatment with Nth and Fpg enzymes is 1.6. The DSB component includes heat labile sites, which represent ∼94% extra DSB to the yield of DSB detected at 4°C. Therefore, the yield of non-DSB clustered DNA damage per prompt DSB becomes 5.7.
What is the significance of radiation-induced clustered DNA damage when compared with the steady state levels of spontaneously induced DNA damage? Many endogenous oxidized base lesions in mammalian DNA are caused by cellular respiration. These individual lesions are chemically similar to those induced by ionizing radiation, but they occur in isolation. An important consideration is that radiation induced lesions are not only isolated but also occur as clusters of lesions. A wide spectrum of oxidized base lesions have been characterized in isolated DNA (49), however estimates of background levels of oxidative DNA damage reported in the literature vary widely. 8-oxoG is commonly used as a marker for oxidative DNA damage and estimates of the value of 8-oxoG per 106 guanines differ depending on the assay used to measure it. For example using HPLC to measure 8-oxoG in cells, figures between 4 and 50 per 106 guanines (4800–60 000/cell) (2,50–53) were determined. With the gas chromatography–mass spectrometry (GC/MS) method, a value of 300 per 106 guanines (360 000/cell) (54,55) was obtained and, more recently, using the comet assay or alkaline unwinding techniques together with endonucleases, values of between 0.4 and 0.6 per 106 guanines (480–640/cell) were determined (45,53). These discrepancies in yield may reflect the differences in techniques used, due to artifactual oxidation of DNA during processing. Recently, Pouget et al. (44) used a chaotropic NaI method of DNA extraction coupled with the high-performance liquid chromatography–electrochemical detection (HPLC–EC) assay to determine a background level of 0.2 8-oxoG/106 bases (2400/cell). All of these estimates are in the range of 400–60 000 8-oxoG per cell and it may be that the lower levels are more reliable for the reasons mentioned above. Values of γ-radiation-induced non-DSB clustered damage determined in this study are between 50 and 75 per cell per gray. Therefore at environmental levels of low LET radiation, the number of non-DSB clusters is low compared with the higher levels of endogenous lesions present. However, these are not individual damaged bases, but are in non-DSB clustered lesions associated with a SSB or another damaged base in close proximity. Cells contain well-defined repair enzymes for individual lesions, but it is not clear how clustered DNA damage is dealt with. Recent evidence indicates that processing of lesions within clustered DNA damage may be inhibited (11–19). Although radiation induced non-DSB clustered lesions are rare they may be biologically significant if they are converted into DSB during their repair or if their lifetime is extended within a cell through stalled processing. The biological consequences of clustered DNA damage may be significant even at low doses since there is a finite chance of a clustered DNA damage being formed in a cell by a single radiation track.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. J. Thacker, MRC, Harwell, for donation of HF19 cells. We also thank Dr D. Papworth for statistical analysis used in this study. This work was partly funded by the Commission of the European Union, contract number; FIGH-CT 1999-00005.
REFERENCES
- 1.O’Neill P. and Fielden,E.M. (1993) Primary free radical processes in DNA. Adv. Radiat. Biol., 17, 53–120. [Google Scholar]
- 2.Nakajima M., Takenchi,T., Takeshita,T. and Morimoto,K. (1996) 8-Hydroxydeoxyguanosine in human leukocyte DNA and daily health practice factors: effects of individual alcohol sensitivity Environ. Health Perspect., 104, 1336–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor and Francis, London.
- 4.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington DC.
- 5.Jenner T.J., de Lara,C.M., O’Neill,P. and Stevens,D.L. (1993) Induction and rejoining of DNA double-strand breaks in V79-4 mammalian cells following gamma- and alpha-irradiation. Int. J. Radiat. Biol., 64, 264–273. [DOI] [PubMed] [Google Scholar]
- 6.Prise K.M., Ahnstrom,G., Belli,M., Carlsson,J., Frankenberg,D., Kiefer,J., Loebrich,M., Michael,B.D., Nygren,J., Simone,G. and Stenerlow,B. (1998) A review of dsb induction data for varying quality radiations. Int. J. Radiat. Biol., 74, 173–184. [DOI] [PubMed] [Google Scholar]
- 7.Blöcher D. (1988) DNA double strand break repair determines the RBE of alpha particles. Int. J. Radiat. Biol., 54, 761–771. [DOI] [PubMed] [Google Scholar]
- 8.Nikjoo H., O’Neill,P., Wilson,W.E. and Goodhead,D.T. (2001) Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res., 156, 577–583. [DOI] [PubMed] [Google Scholar]
- 9.Dianov G.L., O’Neill,P. and Goodhead,D.T. (2001) Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA. Bioessays, 23, 745–749. [DOI] [PubMed] [Google Scholar]
- 10.Schärer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhry M.A. and Weinfeld,M. (1995) The action of Escherichia coli endonuclease III on multiply damaged sites in DNA. J. Mol. Biol. 49, 914–922. [DOI] [PubMed] [Google Scholar]
- 12.Harrison L., Hatahet,Z. and Wallace,S.S.(1999) In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol., 290, 667–684. [DOI] [PubMed] [Google Scholar]
- 13.Harrison L., Hatahet,Z., Purmal,A.A. and Wallace,S.S. (1998) Multiply damaged sites in DNA: interactions with Escherichia coli endonucleases III and VIII. Nucleic Acids Res., 26, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David-Cordonnier M.-H., Laval,J. and O’Neill,P. (2000) Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem., 275, 11865–11873. [DOI] [PubMed] [Google Scholar]
- 15.David-Cordonnier M.-H., Boiteux,S. and O’Neill,P. (2001) Excision of 8-oxoguanine within clustered damage by the yeast OGG1 protein. Nucleic Acids Res., 29, 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David-Cordonnier M.-H., Laval,J. and O’Neill,P. (2001) Recognition and kinetics for excision of a base lesion within clustered DNA damage by the Escherichia coli proteins Fpg and Nth. Biochemistry, 40, 5738–5746. [DOI] [PubMed] [Google Scholar]
- 17.David-Cordonnier M.-H., Boiteux,S. and O’Neill,P. (2001) Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry, 40, 11811–11818. [DOI] [PubMed] [Google Scholar]
- 18.David-Cordonnier M.-H., Cunniffe,S.M.T., Hickson,I.D. and O’Neill,P. (2002) Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry, 41, 634–642. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhry M.A. and Weinfeld,M. (1997) Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem., 272, 15650–15655. [DOI] [PubMed] [Google Scholar]
- 20.Blaisdell J.O. and Wallace,S.S. (2001) Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli.Proc. Natl Acad. Sci. USA, 98, 7426–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epe B., Pflaum,M., Haring,M., Hegler,J. and Rüdiger,H. (1993) Use of repair endonucleases to characterize DNA damage induced by reactive oxygen species in cellular and cell-free systems. Toxicol. Lett., 67, 57–72. [DOI] [PubMed] [Google Scholar]
- 22.Dizdaroglu M., Laval,J. and Boiteux.S. (1993) Substrate specificity of the Escherichia coli endonuclease III: excision of thymine and cytosine derived lesions in DNA produced by radiation generated free radicals. Biochemistry, 32, 12105–12111. [DOI] [PubMed] [Google Scholar]
- 23.Boiteux S. (1993) Properties and biological functions of the Nth and Fpg proteins of Escherichia coli: two DNA glycosylases that repair oxidative damage in DNA. J. Photochem. Photobiol B., 19, 87–96. [DOI] [PubMed] [Google Scholar]
- 24.Löbrich M., Ikpeme,S. and Kiefer,J. (1994) DNA double-strand break measurement in mammalian cells by pulsed field gel electrophoresis: an approach using restriction enzymes and gene probing. Int. J. Radiat. Biol., 65, 623–630. [DOI] [PubMed] [Google Scholar]
- 25.Sutherland B.M., Bennett,P.V., Sidorkina,O. and Laval,J. (2000) Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl Acad. Sci. USA, 97, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fulford J., Nikjoo,H., Goodhead,D.T. and O’Neill,P. (2001) Yields of SSB and DSB induced in DNA by Al-K ultrasoft X-rays and alpha-particles: comparison of experimental and simulated yields. Int. J. Radiat. Biol. 77, 1053–1066. [DOI] [PubMed] [Google Scholar]
- 27.Cox R., Thacker,J. and Goodhead,D.T. (1977) Inactivation and mutation of cultured mammalian cells by aluminium characteristic ultrasoft X-rays. II Dose responses of Chinese hamster and human diploid cells to aluminium X-rays and radiations of different LET. Int. J. Radiat. Biol., 31, 561–576. [DOI] [PubMed] [Google Scholar]
- 28.Jones G.D.D., Milligan,J.R., Ward,J.F., Calabrojones,P.M. and Aguilera,J.A. (1993) Yield of strand breaks as a function of scavenger concentration and LET for SV40 irradiated with 4He ions. Radiat. Res., 136, 190–196. [PubMed] [Google Scholar]
- 29.Milligan J.R., Aguilera,J.A., Nguyen,T.-T.D., Ward,J.F., Kow,Y.W., He,B. and Cunningham,R.P. (1999) Yield of DNA strand breaks after base oxidation of plasmid DNA. Radiat. Res., 151, 334–342. [PubMed] [Google Scholar]
- 30.Foray N., Fertil,B., Alsbeih,M.G.A., Badie,C., Chavaudra,N., Iliakis,G. and Malaise,E.P. (1996) Dose-rate effect on radiation-induced DNA double-strand breaks in human fibroblast HF19 cell line. Int. J. Radiat. Biol., 69, 241–249. [DOI] [PubMed] [Google Scholar]
- 31.Foray N., Monroco,C., Marples,B., Hendry,J.H., Fertil,B., Goodhead,D.T., Arlett,C.F. and Malaise,E.P. (1998) Repair of radiation-induced DNA double-strand breaks in human fibroblasts is consistent with a continuous spectrum of repair probability. Int. J. Radiat. Biol., 74, 551–560. [DOI] [PubMed] [Google Scholar]
- 32.Botchway S.W., Stevens,D.L., Hill,M.A., Jenner,T.J. and O’Neill,P. (1997) Induction and rejoining of DNA double-strand breaks in Chinese hamster V79-4 cells irradiated with characteristic aluminium K and copper L ultrasoft X-rays. Radiat. Res., 148, 317–324. [PubMed] [Google Scholar]
- 33.de Lara C.M., Hill,M.A., Jenner,T.J., Papworth,D. and O’Neill,P. (2001) Dependence of the yield of DNA double-strand breaks in Chinese hamster V79-4 cells on the photon energy of ultrasoft X-rays. Radiat. Res., 155, 440–448. [DOI] [PubMed] [Google Scholar]
- 34.Stenerlöw B., Carlsson,L., Blomquist,E. and Erixon,K. (1994) Clonogenic cell survival and rejoining of DNA double-strand breaks: comparison between three cell lines after photon or He ion irradiation. Int. J. Radiat. Biol., 65, 631–639. [DOI] [PubMed] [Google Scholar]
- 35.Weber K.J. and Flentje,M. (1993) Lethality of heavy ion-induced DNA double-strand breaks in mammalian cells. Int. J. Radiat. Biol., 64, 169–178. [DOI] [PubMed] [Google Scholar]
- 36.Jenner T.J., Fulford,J. and O’Neill,P. (2001) Contribution of base lesions to radiation-induced clustered DNA damage: implication for models of radiation response. Radiat. Res., 156, 590–593. [DOI] [PubMed] [Google Scholar]
- 37.Milligan J.R., Ng,J.Y.-Y., Wu,C.C.L., Aguilera,J.A., Ward,J.F., Kow,Y.W., Wallace,S.S. and Cunningham,R.P. (1996) Methylperoxyl radicals as intermediates in the damage to DNA irradiated in aqueous dimethyl sulfoxide with gamma rays. Radiat. Res., 146, 436–443. [PubMed] [Google Scholar]
- 38.Rydberg B. (1996) Clusters of DNA damage induced by ionizing radiation: formation of short DNA fragments. II. Experimental detection. Radiat. Res., 145, 200–209. [PubMed] [Google Scholar]
- 39.Kotera M., Roupioz,Y., Defrancq,E., Bourdat,A.-G., Garcia,J., Coulombeau,C. and Lhomme,J. (2000) The 7-nitroindole nucleoside as a photochemical precursor of 2′-deoxyribonolactone: access to DNA fragments containing this oxidative abasic lesion. Chem. Eur. J., 6, 4163–4169. [DOI] [PubMed] [Google Scholar]
- 40.Kasai H., Iida,A., Yamaizumi,Z., Nishimura,S. and Tanooka,H. (1990) 5-Formyldeoxyuridine: a new type of DNA damage induced by ionizing radiation and its mutagenicity to Salmonella strain TA102. Mutat. Res., 243, 249–253. [DOI] [PubMed] [Google Scholar]
- 41.Bjelland S., Eide,L., Time,R.W., Stote,R., Eftedal,I., Volden,G. and Seeberg,E. (1995) Oxidation of thymine to 5-formyluracil in DNA: mechanisms of formation, structural implications, and base excision by human cell free extracts. Biochemistry, 34, 14758–14764. [DOI] [PubMed] [Google Scholar]
- 42.Frelon S., Douki,T., Ravanat,J.-L. Pouget,J.-P., Tornabene,C. and Cadet,J. (2000) High-performance liquid chromatography-tandem mass spectrometry measurement of radiation-induced base damage to isolated and cellular DNA. Chem. Res. Toxicol., 13, 1002–1010. [DOI] [PubMed] [Google Scholar]
- 43.Boorstein R.J., Cummings,A.,Jr, Marenstein,D.R., Chan,M.C., Ma,Y., Neubert,T.A., Brown,S.M. and Teebor,G.W. (2001) Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem., 276, 41991–41997. [DOI] [PubMed] [Google Scholar]
- 44.Pouget J.-P., Douki,T., Richard,M.-J. and Cadet,J. (2000) DNA damage induced in cells by gamma and UVA radiation as measured by HPLC/GC-MS and HPLC-EC and Comet assay. Chem. Res. Toxicol., 13, 541–549. [DOI] [PubMed] [Google Scholar]
- 45.Pflaum M., Will,O. and Epe,B. (1997) Determination of steady-state levels of oxidative DNA base modifications in mammalian cells by means of repair endonucleases. Carcinogenesis, 18, 2225–2231. [DOI] [PubMed] [Google Scholar]
- 46.Milligan J.R., Aguilera,J.A., Nguyen,T.-T.D., Paglinawan,R.A. and Ward,J.F. (2000) DNA strand break yields after post-irradiation incubation with base excision repair endonucleases implicate hydroxyl radical pairs in double-strand break formation. Int. J. Radiat. Biol., 76, 1475–1483. [DOI] [PubMed] [Google Scholar]
- 47.Prise K.M., Pullar,C.H.L. and Michael,B.D. (1999) A study of endonuclease III sensitive sites in irradiated DNA: detection of alpha particle-induced oxidative damage. Carcinogenesis, 20, 905–909. [DOI] [PubMed] [Google Scholar]
- 48.Moiseenko V.V., Waker,A.J., Hamm,R.N. and Prestwich,W.V. (2001) Calculation of radiation-induced DNA damage from photons and tritium beta-particles. Part II: tritium RBE and damage complexity. Radiat. Environ. Biophys., 40, 33–38. [DOI] [PubMed] [Google Scholar]
- 49.Cadet J., Berger,M., Douki,T. and Ravanat,J.-L. (1997) Oxidative damage to DNA: formation, measurement and biological significance. Rev. Physiol. Biochem. Pharmacol., 131, 1–87. [DOI] [PubMed] [Google Scholar]
- 50.Inoue T., Mu,Z., Sumikawa,K., Adachi,K. and Okochi,T. (1993) Effect of physical exercise on the content of 8-hydroxydeoxyguanosine in nuclear DNA prepared from human lymphocytes. Jpn. J. Cancer Res., 84, 720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson V.L., Taffe,B.G., Shields,P.G., Povey,A.C. and Harris,C.C. (1993) Detection and quantification of 8-hydroxydeoxyguanosine adducts in peripheral blood of people exposed to ionizing radiation. Environ. Health Perspect., 99, 261–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degan P., Bonassi,S., de Caterina,M., Korkina,L.G. Pinto,L., Scopacasa,X., Zatterale,A., Calzone,R. and Pagano,G. (1995) In vivo accumulation of 8-hydroxy-2′-deoxyguanosine in DNA correlates with release of reactive oxygen species in Fanconi’s anaemia families. Carcinogenesis, 16, 735–742. [DOI] [PubMed] [Google Scholar]
- 53.Collins A.R., Duthie,S.J., Fillion,L., Gedek,C.M., Vaughan,N. and Wood,S.G. (1997) Oxidative DNA damage in human cells: the influence of antioxidants and DNA repair. Biochem. Soc. Trans, 25, 326–331. [DOI] [PubMed] [Google Scholar]
- 54.Olinski R., Zastawny,T.H., Foksinski,M., Windorbska,W., Jaruga,P. and Dizdaroglu,M. (1996) DNA base damage in lymphocytes of cancer patients undergoing radiation therapy. Cancer Lett., 106, 207–215. [DOI] [PubMed] [Google Scholar]
- 55.Podmore I.D., Griffiths,H.R., Herbert,K.E., Mistry,N., Mistry,P. and Lunec,J. (1998) Vitamin C exhibits pro-oxidant properties. Nature, 392, 559. [DOI] [PubMed] [Google Scholar]