Abstract
It has been reported that some double-stranded RNA (dsRNA) binding proteins interact with small RNA biogenesis-related RNase III enzymes. However, their biological significance is poorly understood. Here we examine the relationship between the Arabidopsis microRNA- (miRNA) producing enzyme DCL1 and the dsRNA binding protein HYL1. In the hyl1-2 mutant, the processing steps of miR163 biogenesis were partially impaired; increased accumulation of pri-miR163 and reduced accumulation of short pre-miR163 and mature miR163 as well as misplaced cleavages in the stem structure of pri-miR163 were detected. These misplaced cleavages were similar to those previously observed in the dcl1-9 mutant, in which the second double-stranded RNA binding domain of the protein was disrupted. An immunoprecipitation assay using Agrobacterium-mediated transient expression in Nicotiana benthamiana showed that HYL1 was able to form a complex with wild-type DCL1 protein, but not with the dcl1-9 mutant protein. We also examined miR164b and miR166a biogenesis in hyl1-2 and dcl1-9. Increased accumulation of pri-miRNAs and reduced accumulation of pre-miRNAs and mature miRNAs were detected. Misplaced cleavage on pri-miR164b was observed only in dcl1-9 but not in hyl1-2, whereas not on pri-miR166a in either mutant. These results indicate that HYL1 has a function in assisting efficient and precise cleavage of pri-miRNA through interaction with DCL1.
Keywords: microRNA, plant, biogenesis, DCL1, HYL1
In animals, small RNA (miRNA and siRNA) biogenesis requires the RNase III enzymes Drosha and Dicer. It has been reported that these enzymes interact with certain double-stranded RNA (dsRNA) binding proteins to function. DGCR8 or Pasha, together with RNase III Drosha, are essential in pri-miRNA processing in mammals and Drosophila (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004b; Landthaler et al. 2004). The Dicer-1-Loqs complex is required for specific pre-miRNA processing activity in Drosophila (Forstemann et al. 2005; Saito et al. 2005). R2D2 is required for loading of siRNA into the RNA-induced silencing complex (RISC) by interacting with Dicer-2 in Drosophila (Liu et al. 2003; Tomari et al. 2004).
We previously reported that Arabidopsis miR163 biogenesis requires three cleavage steps by RNase III-like enzymes (Kurihara and Watanabe 2004). The first step is from pri-miR163 into a long miR163 precursor (pre-miR163); the second step is cleavage of the long pre-miR163 into short pre-miR163 and small RNA UL; the last step is from short pre-miR163 into the mature miR163 and a remnant. It was also shown that DCL1 is involved in processing of both pri-miRNA and pre-miRNA.
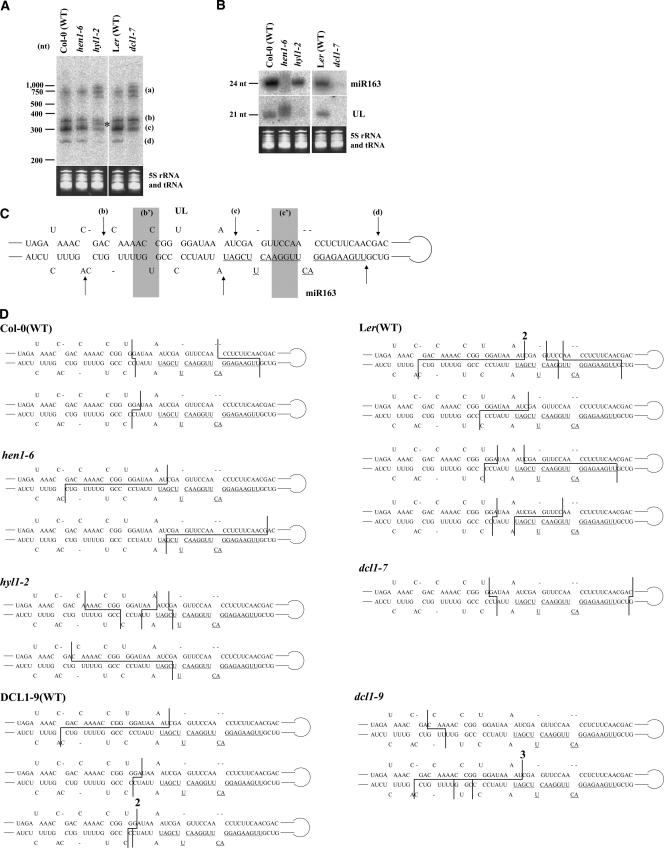
In this paper, we investigated the function of Arabidopsis HYPONASTIC LEAVES 1 (HYL1), a dsRNA binding protein. It was reported that the HYL1 protein is a part of a macromolecular complex and is involved in miRNA maturation (Han et al. 2004a; Vazquez et al. 2004). But, how HYL1 is involved is not well characterized. To investigate how HYL1 participates in miR163 processing, we used a T-DNA insertion mutant, hyl1-2 (SALK_064863). We also used two mutants, hen1-6 (T-DNA insertion mutant; SALK_090960) and dcl1-7. It was shown that HEN1 functions after the processing step and is a methyl-transferase protein able to methylate miRNA/miRNA* duplexes (Yu et al. 2005). dcl1–7 protein has a point mutation (P415S) in the helicase domain (Schauer 2002). We have shown that miR163 processing is impaired in this dcl1-7 mutant (Kurihara and Watanabe 2004). RNA samples were extracted from respective plants and fractionated, and subjected to Northern blot analysis using a miR163 precursor-specific probe covering the regions encoding stem– loop structures. In the hyl1-2 mutant, as in the dcl1-7 mutant, the accumulation of pri-miR163 (bands a) increased and the accumulation of short pre-miR163 (band c) was reduced compared with the wild-type (Fig. 1A ▶). The result revealed that miR163 processing was partially impaired in hyl1-2 but not hen1-6 mutants. This pattern indicated that HYL1 is involved in the miRNA processing step of miRNA biogenesis. However, the impairment in miR163 processing in hyl1-2 was different from that in the dcl1-7 mutant, in that another band appeared between the long pre-miR163 (band b) and short premiR163 (band c) (Fig. 1A ▶, asterisk). The amounts of small RNA UL and mature miR163 were reduced in hyl1-2 as in dcl1-7 (Fig. 1B ▶). In hen1-6, although a processing pattern similar to that in wild-type plants was observed as shown in Figure 1A ▶, smeared signals derived from small RNAs (Fig. 1B ▶) were observed as reported previously (Han et al. 2004a; Xie et al. 2004).
FIGURE 1.
Impaired miR163 processing and determination of the cleavage sites on pri-miR163. (A) Northern blot analysis for detection of pri-miR163 and/or pre-miR163s. Bands a, b, c, and d correspond to pri-miR163, long pre-miR163, short pre-miR163, and the remnant, respectively. The asterisk indicates an additional band between the long pre-miR163 (b) and short pre-miR163 (c) in hyl1-2. (B) Northern blot analysis for detection of small RNA UL and miR163. 5S rRNA and tRNAs were used as loading controls. (C) The sequence of the stem region of pri-miR163. The three correct cleavage sites are indicated by arrows b–d, which correspond to the roots of structures b–d in A. The sequence of miR163 is underlined. The misplaced cleavage sites in hyl1-2 and/or dcl1-9 mutants are shown by b′ and c′, respectively. Although the miR163 gene of dcl1-9 has a 12-nt deletion and 60-nt insertion in its loop structure compared with those in Col-0 and Ler (Kurihara and Watanabe 2004), the sequence and structure of the stem region were the same as those in Col-0 and Ler. (D) Cleavage sites categorized into “others” are shown by lines. The numbers shown on the line indicate total numbers of clones at that position.
Misplaced cleavage of pri-miR163 was previously observed in the dcl1-9 mutant (Kurihara and Watanabe 2004), which has a T-DNA insertion in the second dsRNA binding domain of DCL1 (Schauer 2002). It was suspected that misplaced cleavage also occurred in hyl1-2 because the band detected between the long and short pre-miR163 (Fig. 1A ▶, asterisk) seems identical to the products of misplaced cleavage in dcl1-9. We cloned processed products from wild-type plants, hyl1-2, hen1-6, dcl1-7, and dcl1-9 using ligation for connecting 5′ and 3′ ends of precursors and RT-PCR (cycle-RT-PCR) as described previously (Kurihara and Watanabe 2004) and then sequenced the clones to determine the cleavage sites. Although the miR163 gene of dcl1-9 has a 12-nt deletion and 60-nt insertion in its loop structure compared with those in Col-0 and Ler (Kurihara and Watanabe 2004), the sequence and structure of stem region were the same as those in Col-0 and Ler. The results are shown in Figure 1C,D ▶ and Table 1 ▶. Cleavage sites at positions b, c, and d in Figure 1C ▶ correspond to the stem-side of the long premiR163 (Fig. 1A ▶, b) and short pre-miR163 (c), and remnant (d), respectively. Figure 1D ▶ shows minor clones from each genotype, which are categorized into “others” in Table 1 ▶. The junctions of some clones had ~1–5 nt additional uridine and/or adenine.
TABLE 1.
Cleavage sites of the stem region of pri-miR163
| Genotype | (b) | (b′) (%) | (c) | (c′) | (d) | others | total |
| Col-0(WT) | 4 | 2 (3.9) | 21 | 2 | 19 | 3 | 51 |
| hen1-6 | 4 | 1 (2.2) | 21 | 1 | 17 | 2 | 46 |
| hyl1-2 | 4 | 14 (29.8) | 15 | 3 | 7 | 4 | 47 |
| Ler (WT) | 5 | 1 (2.1) | 18 | 2 | 12 | 9 | 47 |
| dcl1-7 | 20 | 3 (10.0) | 3 | 0 | 2 | 2 | 30 |
| DCL1-9(WT) | 5 | 2 (7.9) | 23 | 0 | 4 | 4 | 38 |
| dcl1-9 | 3 | 25 (48.1) | 3 | 17 | 0 | 4 | 52 |
The clones mapped on the stem region of pri-miR163 shown in Figure 1C ▶ were counted. (b), (b′), (c), (c′), and (d) correspond to (b), (b′), (c), (c′), and (d) in Figure 1C ▶. Cleavage differences of no more than 1 bp or 1 nt were neglected and such clones were categorized into the same group. The clones categorized into “others” are shown in Figure 1D ▶. The junctions of some clones had 1–5 nt additional uridine and/or adenine. We have not previously indicated some clones from dcl1-9, which should be categorized in “others” (Kurihara and Watanabe 2004).
In hyl1-2 plants, the first cleavage sites of some clones were mapped to zone b′ in Figure 1C ▶, as was seen in the dcl1-9 mutant, while some mapped the same as in the wild type (Table 1 ▶). The shift of the cleavage site explains the extra band between the long (b) and short pre-miR163 (c) in Figure 1A ▶. In hen1-6 and dcl1-7 mutants, positioning of the cleavage sites could be aligned as in the wild-type plants (Table 1 ▶). Taken together, the data indicate that HYL1, but not HEN1, is involved in positioning of the cleavage sites in miRNA processing.
Previous reports have suggested that the HYL1 protein is part of a macromolecular complex (Han et al. 2004a) and that the C-terminal region of the recombinant truncated DCL1 can interact with recombinant HYL1 by far Western analysis (Hiraguri et al. 2005). Besides, the results of Figure 1 ▶ and Table 1 ▶ indicate that HYL1 performs its function in collaboration with DCL1 for miRNA processing; therefore, we attempted a coimmunoprecipitation assay to detect the physical interaction between HYL1 and full-length DCL1 in vivo.
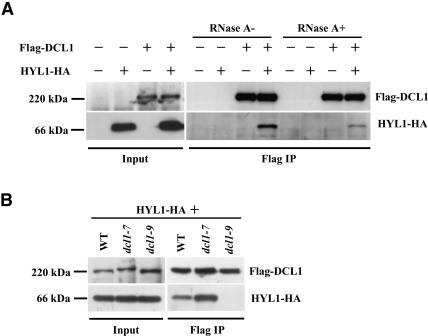
DCL1 with an N-terminal Flag epitope tag (Flag-DCL1) and HYL1 with a C-terminal HA epitope tag (HYL1-HA) were coexpressed in leaves of Nicotiana benthamiana by agroinfiltration. Leaf extracts were subsequently subjected to immunoprecipitation using anti-Flag antibody. The results showed that HYL1-HA was coimmunoprecipitated with Flag-DCL1 using anti-Flag antibody (Fig. 2A ▶). This interaction was not disturbed even in the presence of 50 μg/ mL RNase A (Fig. 2A ▶). This obviates the possibility that the coprecipitation was mediated by RNA molecules.
FIGURE 2.
Western blot analysis to detect the interaction between DCL1 and HYL1. (A) Coimmunoprecipitation of HYL1-HA with Flag-DCL1 using the anti-Flag antibody. RNase A was added (RNase A+) at a concentration of 50 μg/mL. (B) HYL1 was coimmunoprecipitated with DCL1 but not dcl1-9 protein. Each Agrobacterium culture was infiltrated, respectively, at the following densities: Flag-DCL1, FLAG-dcl1-7, FLAG-dcl1-9 OD600 = 0.5; HYL1-HA, OD600 = 0.3. (WT) wild-type, (IP) immunoprecipitate.
We also investigated whether two DCL1 mutant proteins, dcl1-7 and dcl1-9, could interact with HYL1. dcl1-7 protein has a point mutation (P415S) in the helicase domain, while dcl1-9 protein has a 74-amino-acid truncation and 8- amino-acid addition in the C-terminal second double-stranded RNA binding domain (Schauer 2002). The result showed that the dcl1-7 protein could coimmunoprecipitate HYL1, while the dcl1-9 protein could not (Fig. 2B ▶). Considering that the misplaced cleavage of pri-miR163 and premiR163s was observed in dcl1-9 mutant plants as well as in hyl1-2 plants, a close correlation between precise miR163 processing and the interaction of DCL1 with HYL1 was indicated. It prompted us to suspect that the physical interaction between DCL1 and HYL1 proteins is a requisite for adequate and precise processing of miR163 precursors (pri-miR163 and pre-miR163s).
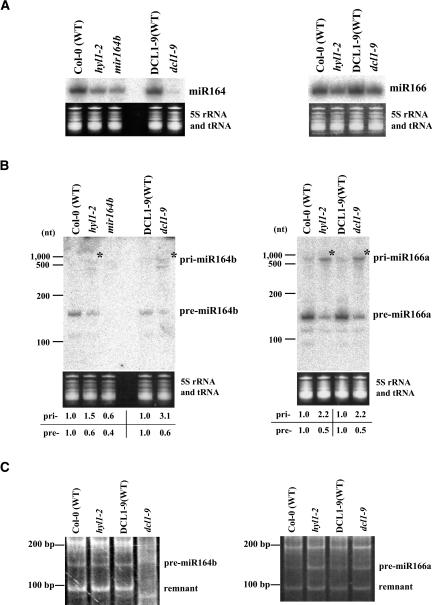
To investigate whether DCL1 and HYL1 are generally involved in processing of more typical miRNAs, we also characterized miR164b and miR166a, because it is possible to detect precursor molecules of these miRNAs by Northern blot analysis (our unpubl. observation; Fig. 3B ▶). The primary transcripts, pri-miR164b and pri-miR166a, were detected by the 5′ race method used previously (Xie et al. 2005). We used a mir164b-1 mutant (SALK_136105), where one of the three miR164 genes is disrupted, as a control (Mallory et al. 2004). Northern blot analysis showed that the accumulation of mature miR164 and miR166 was reduced in hyl1-2 and dcl1-9 (Fig. 3A ▶). The accumulation of pri-miR164b and pri-miR166a increased (Fig. 3B ▶, asterisk) and the accumulation of pre-miR164b and premiR166a was reduced in both mutants, compared with those in wild-type plants. These results indicated that the DCL1–HYL1 complex is generally involved in efficient processing from pri-miRNAs to pre-miRNAs, consistent with our previous notion that DCL1 catalyzes the first processing of miRNA biogenesis.
FIGURE 3.
(A) Northern blot analysis for detection of miR164 and miR166. (B) Northern blot analysis for detection of miR164b and miR166a precursors. mir164b-1 mutant (SALK_136105) was used as a negative control (Mallory et al. 2004). 5S rRNA and tRNAs was used as a loading control. The mean relative accumulation levels for pri-miRNAs and pre-miRNAs are indicated under the panels, respectively. The accumulation levels in wild-type plants were normalized to 1.0. (C) Cycle-RT-PCR analysis of pre-miR164b and pre-miR166a.
We performed cycle-RT-PCR analysis using premiR164b- and pre-miR166a-specific primers and obtained clear bands corresponding to pre-miRNAs and remnants containing loop structures (Fig. 3C ▶). The result showed that misplaced cleavage on pri-miR164b was observed only in dcl1-9 mutants but not in hyl1-2 mutants, whereas not on pri-miR166a in either mutants, indicating that misplaced cleavage was not necessarily accompanied by the dysfunction of HYL1 and DCL1 proteins. When we performed cycle-RT-PCR analysis with mir164b-1 mutant, where the miR164b gene was disrupted with T-DNA insertion, no such bands could be detected (data not shown).
The results above showed that miR163 processing is impaired and misplaced cleavage occurs in hyl1-2 and dcl1-9 mutants (Fig. 1 ▶; Table 1 ▶). An immunoprecipitation assay showed that HYL1 was able to form a complex with DCL1, but not dcl1-9 mutant protein (Fig. 2 ▶). Although some dsRNA binding proteins involved in small RNA biogenesis have been examined, our finding is the first example where a dsRNA binding protein, HYL1, may act in precise cleavage of a pri-miRNA by interaction with a RNase III enzyme.
Misplaced cleavage on pri-miR164b was observed only in dcl1-9, but not in hyl1-2, whereas not on pri-miR166a in either mutant (Fig. 3C ▶). It is possible that such misplaced cleavage could be observed depending on other factors such as fitness of secondary stem–loop structures of the pri-miRNAs as substrates for DCL1/HYL1.
A recent study suggested that the miR163 gene likely originated from a recent target-gene duplication event (Allen et al. 2004). Therefore, it has been unclear whether the multistep processing by DCL1, like miR163 biogenesis, is common in plant miRNA biogenesis. Analysis of two miRNAs, miR164b and miR166a, strongly suggested at least that the first typical processing step from pri-miRNA to pre-miRNA was mediated through DCL1 and HYL1 function (Fig. 3 ▶).
Previously, we described the clone numbers corresponding to each cleavage site (Kurihara and Watanabe 2004). At that time, we did not obtain the clones categorized into b′, c′, and “others” from wild-type Col-0 using 4-wk-old nonflowering young plants. In this research, 8-wk-old flowering adult plants were used. It was assumed that the processing fidelity in young plants is higher than that in adult plants. We have not previously indicated some clones from dcl1-9, which should be categorized into “others.”
DGCR8 or Pasha, together with Drosha, are essential in pri-miRNA processing in mammals and Drosophila (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004b; Landthaler et al. 2004). However, HYL1 is not fully necessary for plant miRNA processing by DCL1, because the hyl1-2 null mutant still has a small ability to accumulate miRNAs although the accumulation level is reduced (Han et al. 2004a; Vazquez et al. 2004). It is possible that other factors, which help DCL1 or supplement HYL1, are also involved in miRNA processing in vivo.
MATERIALS AND METHODS
Plant materials
Plant growth conditions were as described previously (Kurihara and Watanabe 2004). hyl1-2 (SALK_064863), hen1-6 (SALK_090960), and mir164b-1 (SALK_136105) were described previously (Mallory et al. 2004; Vazquez et al. 2004; Li et al. 2005). DCL1-9 wild-type plants were screened as described previously (Kurihara and Watanabe 2004).
Plasmids for agroinfiltration
The coding sequence for Flag-DCL1 was amplified by overlapping PCR using cDNA from inflorescence tissue as a template and subcloned into a pCRII-TOPO vector (Invitrogen). The primers used for PCR-were as follows: FlagDCL1forward1 5′-TATAAAGATGATGATG ATAAGATGGTAATGGAGGATGAGCC-3′, FlagDCL1forward2 5′-AGGTACCTCTAGAATGGATTATAAAGAT GATGATGATAAG-3′, and FlagDCL1reverse 5′-AAACTAGTTCAAGAAAAAGTTTTATTTAAAAGCTCAAG-3′. The coding sequence of Flag-dcl1-9 was also amplified by overlapping PCR using a full-length clone of the Flag-DCL1 sequence as a template and subcloned into a pCRII-TOPO vector. The primers used for PCR were FlagDCL1forward1 Flagdcl1- 9reverse1 5′-ATCGATTTCAGCGATCAACGTTTGTCTCGTAAACG-3′ and Flagdcl1-9reverse2 5′-AACTAGTTCAACCTGGATCGATTTCAGCGATCAAC-3′.
The XbaI–SpeI fragments covering the Flag-DCL1 and Flag-dcl1-9 coding regions were inserted into a pTA7002 vector with a dexamethasone- (DEX) inducible promoter (Aoyama and Chua 1997) to obtain pTA7002-Flag-DCL1 and pTA7002-Flagdcl1-9.
Part of the Flag-dcl1-7 coding sequence was amplified by overlapping PCR using a full-length clone of the Flag-DCL1 sequence as a template and subcloned into a pCRII-TOPO vector. The primers used for PCR were as follows: Flagdcl1-7forward 5′-AACTCGAGATGGATTATAAAGATGATGATGAT-3′, Flagdcl1-7reverse1 5′-ACAATCTACTTGGCTTGAA ACACCCTTTAAATTAACAG ACGAAGC-3′, and Flagdcl1-7reverse2 5′-T CTCGAGGTTACGTATCTTTATCGCACA ATCTACTTGGCTTGAAA-3′. The XhoI– XhoI fragments from the subclone were replaced with the corresponding sites in pTA7002-Flag-DCL1 to obtain pTA7002-Flag-dcl1-7.
The coding sequence of HYL1-HA was amplified by overlapping PCR using cDNA as a template and subcloned into a pCRII-TOPOvector. The primers used for PCR were as follows: HYL1HAforward 5′-AT CTAGAATGACCTCCACTGATGTTTC-3′, HYL1HAreverse1 5′-TCCGGTACGTCGTATGGATATGCGTGGCTTGCTTCTGCT-3′, and HYL1HAreverse2 5′-GCGGCCGCTCATGCATAATCCGGTACGTCGTATGGATA-3′. The XbaI–NotI fragment covering the HYL1-HA coding region from the subclone was inserted into a pSK1 vector with a 35S promoter to obtain pSK1-HYL1-HA.
Northern blot analysis
Total RNA was extracted from the leaves of 6–8-wk-old Arabidopsis plants using ISOGEN reagent (NIPPON GENE). Isolation of low molecular weight RNA plus (LMW RNA+) from total RNA was described previously (Kurihara and Watanabe 2004). For detection of miR163 precursors, LMW RNA+ was resolved on a denaturing 5% polyacrylamide gel (8 M urea). For detection of miR164b and miR166a precursors, LMW RNA+ was resolved on a denaturing 7.5% polyacrylamide gel (8 M urea). For detection of miRNAs, LMW RNA+ was resolved on a denaturing 15% polyacrylamide gel (7 M urea). Hybridization was carried out at 40°C for miRNA, at 65°C for miR163 and miR166a precursors, and 60°C for miR164b precursors using PerfectHyb Plus hybridization buffer (Sigma). DNA oligonucleotides probes specific for miR163 (5′-ATCGAAGTTCCAAGTCCTCTTCAA-3′), UL (5′-ATTTTACCACCGGGTTTTGG-3′), miR164 (5′-TGCACGTGCCCTGCTTCTCCA-3′), and miR166 (5′-GGGGAATGAAGCCTGGTCCGA-3′) were end labeled with γ32P-ATP using T4 polynucleotide kinase (TOYOBO). DNA probes specific for miRNA precursors were constructed by random priming reactions with α32P-dCTP using the Megaprime DNA-labeling system (Amersham Bioscience). DNA fragments used for the reactions were PCR amplified from genomic DNA using the following primers: 5′-ACACGGGGGATAATATCGAA-3′ and 5′-ACCCGGTGGATAAAATCGAG-3′ for miR163 precursors (total 331 nt), 5′-GATGGAGAAGCAGGGCACGT-3′ and 5′-GTGAAGATGGGCACATGAAG-3′ for miR164b precursors (149 nt), and 5′-AGATATATATTCAGAAACCCTAG-3′ and 5′-GGTTCATTCACTGGATCTGAAAC-3′ for miR166a precursors (246 nt). These sequences covered the regions encoding stem–loop structures specific for their respective precursors.
Cycle-RT-PCR and determination of the cleavage site sequence
Determination of the cleavage site sequence of miR163 precursors was performed as described previously (Kurihara and Watanabe 2004). The cycle-RT-PCR of miR164b and miR166a precursors was performed as described below. Aliquots of 5 μg (for miR164b precursors) or 2 μg (for miR166a precursors) of LMW RNA+ were self-ligated with T4 RNA ligase (NEB) in a 50-μL reaction volume. The reaction was performed for 2 h at 16°C. After the reaction, RNAs were precipitated with ethanol, and the pre-miRNAs and the remnants were amplified by RT-PCR using the gene-specific primers: forward 5′-GTGTGTTGAGTGTGATGATATGG-3′ and reverse 5′-CAAAATTCCGCATATATACACGC-3′ for pre-miR164b and forward 5′-GAATTGAACCTTCAGATTTCAG-3′ and reverse 5′-TTGTTAGATCGAAAGAGATCC-3′ for pre-miR166a. The reverse primers were also used for RT reactions. The PCR products were resolved on 7.5% polyacrylamide gels and detected with ethidium bromide staining.
Agroinfiltration
Three-month-old N. benthamiana plants and Agrobacterium tumefaciens strain GV3101 were used for infiltration experiments. Agrobacterium culture and infiltration were performed as described previously (Llave et al. 2000). For induction of Flag-DCL1 expression, 30 μM DEX (Wako) was sprayed on the plants twice at 24 and 45 h after agroinfiltration.
Immunoprecipitation
Forty-eight hours after agroinfiltration, leaves of N. benthamiana were ground in liquid nitrogen and homogenized in three volumes of extraction buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.5% TritonX-100, 0.2% 2-mercaptoethanol, 5% glycerol, complete proteinase inhibitor cocktail; Roche) using a mortar. Cell debris was pelleted by centrifugation for 10 min at 15,000 rpm. The supernatant (1 mL) was incubated with 1 μL of anti-Flag antibody (Sigma) and 25 μL bed volume of protein G Sepharose (Amersham) for 2 h. The immune complexes were then centrifuged and washed three times in 1 mL wash buffer A (25 mM Tris-HCl at pH 7.5, 250 mM NaCl, 2 mM EDTA, 0.05% TritonX-100, 1 mM PMSF). The volume was reduced to 60 μL using a micropipette, and 4× sample buffer was added, followed by incubation at 65°C for 10 min.
Western blot analysis
Protein samples were analyzed on 7.5% SDS-polyacrylamide gels and transferred to Immobilon-P Transfer Membrane (Millipore). Monoclonal anti-Flag and anti-HA (Sigma) were used as the primary antibody, and peroxidase-conjugated anti-mouse immunoglobulins antibody (Amersham) was used as the secondary antibody. Bands were visualized by enhanced chemiluminescence system (ECL) according to the manufacturer’s instructions (Amersham).
Acknowledgments
We thank the Salk Institute and Arabidopsis Biological Resource Center for providing the seeds of hyl1-2 (SALK_064863), hen1-6 (SALK_090960), mir164b-1 (SALK_136105), dcl1-7 (CS3089), and dcl1-9 (CS3828). We are also grateful to A. Takeda and T. Watanabe for helpful discussion. This work was supported by a grant-in-aid for scientific research, “Spatiotemporal Network of RNA Information Flow”, priority area (A) awarded by the Ministry of Education, Culture, Sports, Science, and Technology, Japan (No. 14035215) to Y.W. and by a grant-in-aid for scientific research (A) from the Japan Society for the Promotion of Science to Y.K.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2146906.
REFERENCES
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.H., Spatafora, J.W., and Carrington, J.C. 2004. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 12: 1282–1290. [DOI] [PubMed] [Google Scholar]
- Aoyama, T. and Chua, N.H. 1997. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605– 612. [DOI] [PubMed] [Google Scholar]
- Denli, A.M., Tops, B.B.J., Plasterk, R.H.A., Ketting, R.F., and Hannon, G.J. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235. [DOI] [PubMed] [Google Scholar]
- Forstemann, K., Tomari, Y., Du, T, Vagin, V.V., Denli, A.M., Bratu, D.P., Klattenhoff, C., Theurkauf, W.E., and Zamore, P.D. 2005. Normal microRNA maturation and germ-line stem cell maintenance requires loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, R.I., Yan, K.P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., and Shiekhattar, R. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240. [DOI] [PubMed] [Google Scholar]
- Han, M.H., Goud, S., Song, L., and Fedoroff, N. 2004a. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. 101: 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J., Lee, Y., Yeom, K.H., Kim, Y.K., Jin, H., and Kim, V.N. 2004b. The Drosha-DGCR8 complex in primary microRNA processing. Genes & Dev. 18: 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri, A., Itoh, R., Kondo, N., Nomura, Y., Aizawa, D., Murai, Y., Koiwa, H., Seki, M., Shinozaki, K., and Fukuhara, T. 2005. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 57: 173–188. [DOI] [PubMed] [Google Scholar]
- Kurihara, Y. and Watanabe, Y. 2004. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. 101: 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler, M., Yalcin, A., and Tuschl, T. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14: 2162–2167. [DOI] [PubMed] [Google Scholar]
- Li, J., Yang, Z., Yu, B., Liu, J., and Chen, X. 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., and Carrington, J.C. 2000. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. 97: 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. 2004. Micro-RNA regulation of NAC-domain targets is required for proper formation and separation of adiacent embryonic, vegetative, and floral organs. Curr. Biol. 14: 1035–1046. [DOI] [PubMed] [Google Scholar]
- Saito, K., Ishizuka, A., Siomi, H., and Siomi, M.C. 2005. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 3: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. 2002. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7: 487–491. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Matranga, C., Haley, B., Martinez, N., and Zamore, P.D. 2004. A protein sensor for siRNA asymmetry. Science 306: 1377–1380. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Gasciolli, V., Crete, P., and Vaucheret, H. 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14: 346–351. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S.A., and Carrington, J.C. 2005. Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R.W., Steward, R., and Chen, X. 2005. Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]