Abstract
Internal ribosome entry site (IRES) elements consist of highly structured RNA regions that determine internal initiation of translation. We have previously shown that the foot-and-mouth disease virus (FMDV) IRES contains a GNRA tetraloop spanning residues G178UAA181. Here we show that tertiary RNA interactions dependent on the GNRA motif determine the structural organization of the central domain. By using mutational analysis in combination with RNA probing, we have identified distant reciprocal interactions between the GNRA motif and the invariant region G240CACG244, termed motif A. Mutations in motif A caused a decrease in IRES activity as severe as the GUAG substitution in the GNRA motif. Substitutions in either GNRA or motif A sequences induced a common reorganization around the conserved R199AAA202 stem–loop, suggesting that the latter contributes to stabilize the GNRA–motif A interaction. This finding was also consistent with a significant increase in the efficiency of RNA–RNA interactions determined in gel shift assays using as probe the hairpin that contains the GNRA motif compared to a transcript encompassing the entire apical region of the central domain. Thus, we propose that the central domain of the FMDV IRES contains a structural conformation essential for IRES activity stabilized by a tertiary contact involving residues in the GNRA tetraloop and motif A conserved sequences.
Keywords: internal initiation of translation, IRES, RNA structure, RNA tertiary interactions, GNRA motif
INTRODUCTION
Internal ribosome entry site (IRES) elements are relatively long cis-acting sequences that recruit the translational machinery to an internal initiation codon (Hellen and Sarnow 2001; Martínez-Salas et al. 2001). Despite considerable research efforts, including identification of IRES-binding proteins (Belsham and Jackson 2000; Stoneley and Willis 2004), the specific mechanisms by which these complex structures recruit the ribosome are unknown. The secondary structure of the picornavirus foot-and-mouth disease virus (FMDV) and encephalomiocarditis virus (EMCV) IRES elements is phylogenetically conserved, indicating that the overall RNA spatial organization needs to be maintained in order to retain activity. However, only a few conserved IRES motifs have been demonstrated to interact with essential proteins, including the translation initiation factor eIF4G (Kolupaeva et al. 1998; López de Quinto and Martínez-Salas 2000; Clark et al. 2003) and the auxiliary proteins PTB or ITAF45 (Pilipenko et al. 2000). A precise role for other conserved motifs has not been established yet.
The element in the viral RNA that constitutes the IRES adopts a complex RNA structure, likely stabilized by RNA-binding proteins (Belsham and Jackson 2000; Hellen and Sarnow 2001; Bedard et al. 2004; Martínez-Salas and Fernández-Miragall 2004). Experimental evidence for tertiary structure generated by RNA–RNA interactions is available for portions of IRES elements from FMDV (Ramos and Martínez-Salas 1999; Fernández-Miragall and Martínez-Salas 2003), HCV (Kieft et al. 1999; Jubin et al. 2000; Lyons et al. 2001; Spahn et al. 2001; Kieft et al. 2002; Vos et al. 2002; Lukavsky et al. 2003; Rijnbrand et al. 2004), and the intergenic region of insect picornavirus-like viruses (Jan and Sarnow 2002; Costantino and Kieft 2005). These elements appear to have a dynamic structure, suggesting that translation efficiency may be regulated at the level of RNA structure.
The FMDV IRES consists of ~450 nt, predicted to fold in stem–loops organized in domains (Martínez-Salas and Fernández-Miragall 2004). Little is known about the role performed by domain 3 (also termed I), the largest and central domain that spans 215 residues of the FMDV IRES (see Fig. 1 ▶). To our knowledge, none of the essential motifs present in the central domain constitute the binding sites of proteins whose interaction is a key determinant of IRES activity (Walter et al. 1999; Pilipenko et al. 2000; López de Quinto et al. 2001; Fernández-Miragall and Martínez-Salas 2003). The apical region of domain 3 contains two purine-rich motifs, GNRA and RAAA, located in distal loops (López de Quinto and Martínez-Salas 1997) (N stands for any nucleotide and R for purine). These motifs do not tolerate nucleotide substitutions, deletions, or insertions, in support of its essential contribution to IRES activity (Drew and Belsham 1994; López de Quinto and Martínez-Salas 1997; Robertson et al. 1999). A combination of functional analysis and RNA probing has shown that the GUAA motif adopts a tetra-loop conformation (Fernández-Miragall and Martínez-Salas 2003) that is responsible for the organization of the adjacent stem–loops. The study of substitution mutants bearing GUAG or UNCG motifs revealed a lack of protection to chemical and enzymatic attack in native RNA at distant nucleotides in the secondary structure of the central domain, suggesting that the GNRA motif dictates the organization and stability of domain 3 (Fernández-Miragall and Martínez-Salas 2003). This feature prompted us to undertake studies aimed at defining the role of the GNRA-dependent interactions in the FMDV IRES activity.
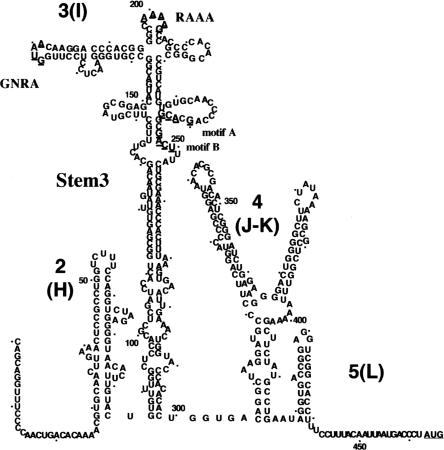
FIGURE 1.
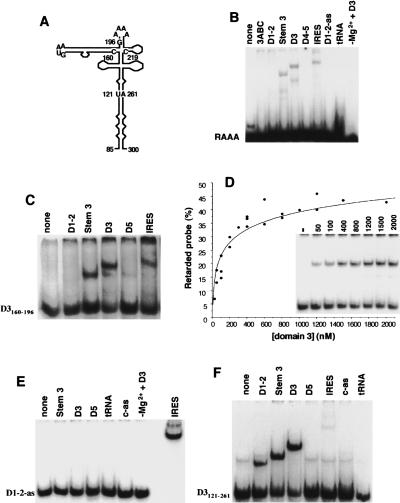
Secondary structure of FMDV IRES. Domains are numbered 1–5 or H–L; a dot is positioned every 10 residues. RNA structure of the central domain is depicted according to RNA probing analysis (Fernández-Miragall and Martínez-Salas 2003). Underlined symbols depict the substitutions in RAAA motif (A199AAAG203) to CGCCC, in GNRA motif (G178UAA181) to GUAG or UACG; motif A (G240CACG244) to ACGGC; and motif B (G247ACU250) to CGGG. Transcript stem 3 encompasses residues 86–299 with 151–227 deleted.
GNRA tetraloops are often involved in RNA folding; they bring together distant regions by providing sites for RNA tertiary contacts (Correll and Swinger 2003). GNRA-dependent tertiary interactions have been previously documented in natural RNAs (Doherty et al. 2001; Nissen et al. 2001), as well as in high affinity ligands selected in vitro (Costa and Michel 1997). The RNA recognition motif in GNRA tetra-loops constitutes A-minor contacts that involve tandem adenosine docking into the minor groove of an RNA helix. In particular, the receptor region of a GUAA tetra-loop is often composed of two C·G pairs (Nissen et al. 2001; Correll and Swinger 2003).
Here we have studied conserved motifs in the central domain that showed a common alteration in the folding pattern of GNRA mutants. Mutational and structural analysis have allowed us to determine the relation between changes in RNA folding in vitro and activity of the mutated IRES elements in vivo. Changes in distant residues around residue G240 were observed in T2 RNase probing in vitro of GNRA mutants, in agreement with previous RNase T1 and DMS probing results (Fernández-Miragall and Martínez-Salas 2003). Mutational analysis of the distant G240CACG244 invariant residues to ACGGC showed its involvement in IRES activity in vivo. This substitution was carried out to destroy C·G pairs that may act as a potential binding site of the GUAA tetraloop (Correll and Swinger 2003). Analysis of the RNA folding pattern of the GCACG motif mutant indicated a lack of protection at the distant GNRA motif, demonstrating a reciprocal protection induced by RNA–RNA interaction. Specific binding of the GNRA hairpin to the central domain was also observed in gel shift assays. This interaction may be stabilized by sequences within the apical region, as suggested by the change in local RNA conformation around the RAAA motif. We propose that the G240CACG244 invariant motif is a strong candidate to be the functional receptor of the GNRA motif.
RESULTS
Identification of long-distance interactions of the GNRA motif with a conserved region within the central domain
Domain 3, the largest and central domain in the FMDV IRES, shows two distinct regions in terms of secondary structure. The apical region is organized as a cruciform structure (Fig. 1 ▶), whereas the proximal region has been characterized as a stem interrupted by several bulges. Two conserved and essential motifs, GNRA and RAAA (with sequences G178UAA181 and A199AAA202, respectively), are present in the apical region. Structural probing of UNCG or GUAG substitution mutants carried out with DMS and RNase T1 revealed that the GNRA motif adopts a tetraloop conformation and dictates the organization and stability of domain 3 (Fernández-Miragall and Martínez-Salas 2003).
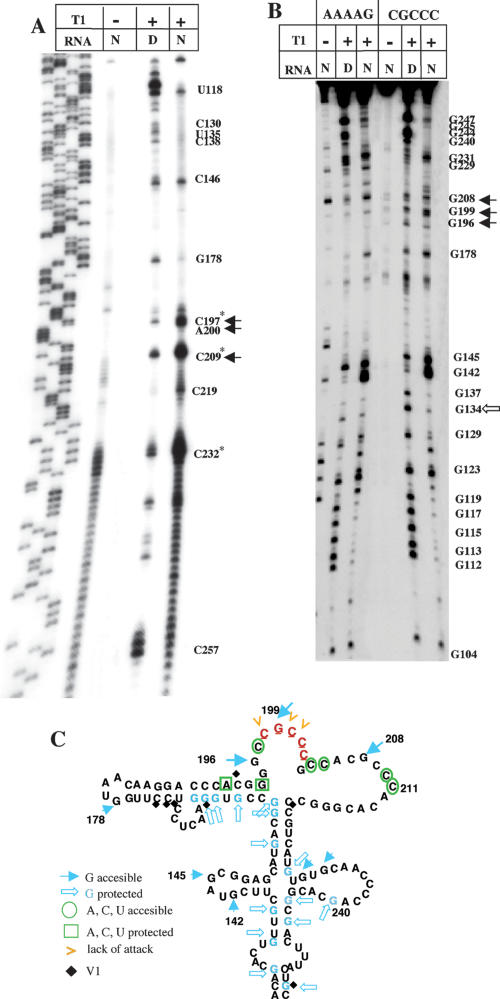
Because of the essential role played by the adenine residues of the GNRA motif in IRES activity (López de Quinto and Martínez-Salas 1997; Robertson et al. 1999), the structural organization of the central domain was further analyzed here using RNase T2. This RNase exhibits a preferential attack to adenine residues (Irie and Oghi 2001). Following incubation with calibrated amounts of RNase T2 in native (N) and denaturing (D) conditions, partially digested RNA was analyzed by primer extension using specific 5′-end-labeled oligonucleotides (Fig. 2 ▶). In this assay, the RNase cleavage products give rise to a specific reverse transcriptase (RT) extension stop (Brunel and Romby 2000; Mitchell et al. 2003; Terenin et al. 2005). Thus, new bands detected in the comparison of untreated (−) to treated (+) RNA indicate accessibility of RNAse T2 to the template RNA. Moreover, lack of RT stop in the comparison of RNA incubated in native to denaturing conditions indicated protection to RNase digestion induced by RNA folding. A representative example of the primer extension analysis of the wild-type FMDV IRES, carrying the GUAA sequence at the GNRA motif, is shown in Figure2 ▶, lanes 1–4. Prominent RT stops in residues U230–C235 in the wild-type GUAA RNA and strong protection at positions A199–A202 were readily observed.
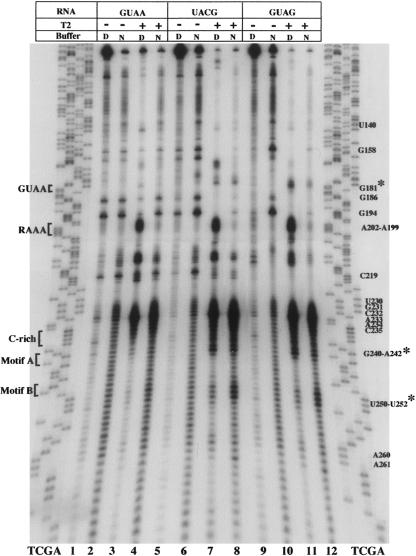
FIGURE 2.
RNase T2 probing of domain 3. RNA harboring the wild-type GUAA or UACG and GUAG substitution in the GNRA motif was incubated with RNase T2 in native (N) or denaturing (D) buffer conditions. A 5′-labeled primer was then used in a reverse transcriptase reaction, and cDNA products were subsequently analyzed on 6% acrylamide 7 M urea gels. A wild-type DNA sequence, prepared with the same oligonucleotide, was run in parallel to identify the RT stops. Residues are marked as the complementary sense sequence. Asterisks denote new attacks observed in the mutant RNAs relative to the GUAA sequence.
We have shown previously that the central domain of the FMDV IRES constitutes an independently folded region. Chemical probing of transcripts encompassing the full-length FMDV IRES, with either GUAA or GUAG sequences, led to the same pattern of accessibility as transcripts encoding only the central domain (Fernández-Miragall and Martínez-Salas 2003). This result was also in agreement with the finding that the apical region of domain 3 interacts exclusively with the central region of the FMDV IRES in gel shift assays (Ramos and Martínez-Salas 1999; Fernández-Miragall and Martínez-Salas 2003).
To complete the study on the GNRA motif, RNAse T2 probing of UACG and GUAG mutants was carried out. Comparison of T2 accessibility with wild-type GUAA showed a new RT stop at residue 181 in each mutant, UACG and GUAG (Fig. 2 ▶, lanes 7,8,11,12), demonstrating new attacks to residues C180 and A180, respectively. Additionally, RT stops at positions 240–242, and 250–252 were more intense in both UACG and GUAG RNAs, strongly suggesting a long-distance reorganization of the central region. These mutants also showed lack of RT stops in positions G186, G194, C219, and G158 under N and D conditions in the treated RNA. This result suggested the presence of a different conformation in the mutant RNAs compared to the wild-type IRES, as previously shown in probing analysis using T1 and DMS (Fernández-Miragall and Martínez-Salas 2003). Changes around residues 135–138, potentially pairing to nt 245–248, were not observed.
Functional analysis of the conserved region interacting with the GNRA motif
Together with the change observed in residue 180, the concomitant reorganization around regions 240 and 250 pointed to a tertiary contact involving a long-range interaction with the GNRA motif. This hypothesis was further supported by the phylogenetic conservation of the FMDV IRES nucleotide sequence noticed around residues 240–244 (Martínez-Salas and Fernández-Miragall 2004; Carrillo et al. 2005). We reasoned that if the GNRA motif mediates tertiary interactions essential for IRES activity, its binding site must also be important to preserve the activity of the element. Therefore, we tested IRES activity in mutants bearing nucleotide substitutions in residues 240–244 and 247–250 (termed motifs A and B, respectively; Fig. 1 ▶) that were structurally modified in all UCCG, UACG, and GUAG mutants (Fig. 2 ▶; Fernández-Miragall and Martínez-Salas 2003). Figure3A ▶ shows the relative IRES activity of constructs with nucleotide substitutions at each of these regions. Substitutions introduced at motif A, G240CACG244 to ACGGC, were intended to destroy C·G base pairs in the secondary structure of the central domain (see Fig. 1 ▶) that may constitute a potential binding site for GYRA motifs (Costa and Michel 1997; Correll and Swinger 2003). Functional analysis in transfected cells demonstrated that substitutions in motif A diminished IRES activity ~100-fold, while substitutions in motif B (G247ACU250 to CGGG) only reduced activity fivefold (Fig. 3A ▶). A single point mutation, G240 to C, was then introduced to analyze the contribution of this invariant nucleotide to IRES activity, leading to threefold reduction in IRES activity. It appeared that not just G240 but the whole motif was important for IRES function. These results pointed out the biological relevance of nucleotides substituted in motif A for IRES activity.
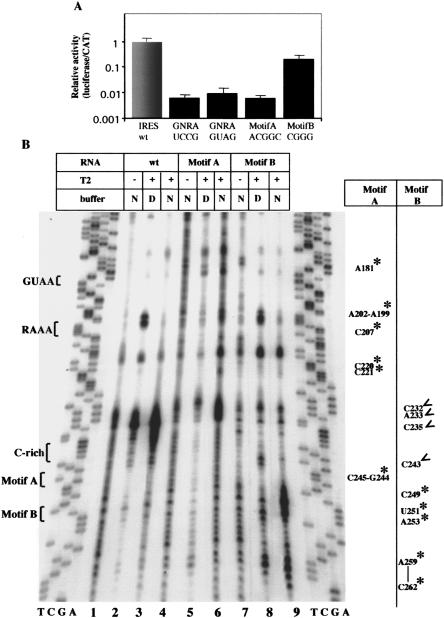
FIGURE 3.
(A) Relative activity of IRES mutants. Bicistronic constructs of the form CAT-IRES-luciferase carrying the indicated substitutions in motifs A or B were transfected in BHK-21 cells. GUAG and UCCG substitutions in the GNRA motif have been included for comparison. Relative IRES activity was calculated as the luciferase/CAT activity in each extract, made relative to that observed in the wild-type IRES. (B) Enzymatic probing of mutants in motifs A and B. RNA harboring the wild-type sequence or substitution in motifs A or B shown in A was incubated with RNase T2 in native (N) or denaturing (D) buffer and then analyzed by primer extension. Residues marked with asterisks denote new attacks relative to the wild-type sequence; open arrows denote lack of attack in the mutant RNAs.
Bidirectional interaction between the GUAA tetraloop and the invariant motif A detected by enzymatic and chemical probing
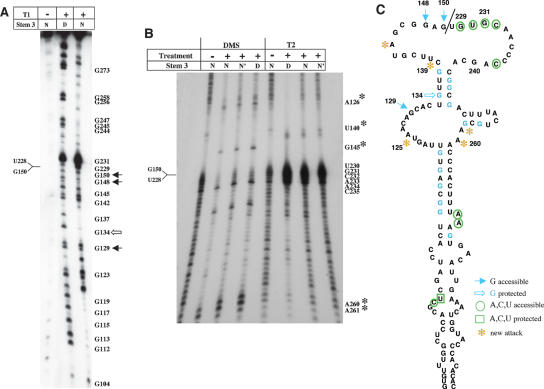
To ensure that RNAs with substitutions in motifs A and B induced a change in RNA structure, we performed enzymatic and chemical probing using RNase T2 and DMS (see Figs. 3 ▶, 4 ▶). As shown in Figure3B ▶, new RT stops were detected in motif A mutant in residues A181, A202–A199, C207, C220–221, and C245–G244. These changes indicated that the region around the C-rich bulge was significantly more open than the wild-type RNA, albeit C238 was protected from DMS attack (Fig. 4B ▶; cf. to 4A).
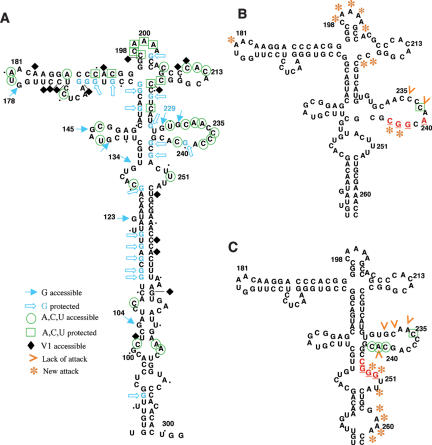
FIGURE 4.
Reorganization of domain 3 RNA structure in substitution mutants of motifs A and B. (A) Secondary structure probing of the wild-type IRES using DMS and RNase A, T1, T2, and V1. (B) RNase T2 and DMS probing of motif A mutant. (C) RNase T2 and DMS probing of motif B mutant. Substituted residues are highlighted in red. For simplicity, only main changes relative to the wild type are depicted in the structure of mutants in motif A and B.
The strong RT stop in residue A181 demonstrated that the distant GNRA loop was more accessible to RNAse T2 attack than the wild-type sequence (Fig. 3B ▶). It is noteworthy that adenines 201–199 in the RAAA motif were also more accessible to DMS attack in this mutant (Fig. 4B ▶).
Regarding the motif B mutant, results in Figure3B ▶ showed that it was more accessible to RNase T2 around the mutated region, nt 249–251, but no reciprocal change in A180 was observed. New strong RT stops were detected between positions C262 and A259 (Fig. 3B ▶), and the C-rich bulge seemed less accessible than in motif A mutant RNA (Fig. 4C ▶; cf. to 4B).
In summary, the mutant in motif A showed reciprocal changes affecting the GNRA tetraloop and the region around residue 240, strongly suggesting that the nucleotide region substituted behaves as a functional GUAA receptor.
The RAAA motif also contributes to the interaction between the apical and proximal region
The stem–loop around the second purine-rich conserved motif, A199AAA202, located in the apical part of the central domain was reorganized in the GUAG mutant (Fernández-Miragall and Martínez-Salas 2003), as well as in motif A mutant (see Fig. 4A,B ▶), suggesting that organization of this short stem–loop was dependent on both the GNRA tetra-loop and motif A sequences.
We have shown previously that a short synthetic hairpin carrying a GUAA motif specifically interacts with the central domain of the FMDV IRES. In addition, GUAG or UCCG replacements in the GUAA motif significantly reduced its binding capacity to the central domain (Fernández-Miragall and Martínez-Salas 2003). To test potential interactions mediated by the apical region of the central domain, we performed gel shift analysis using as probe transcripts of increasing size encompassing different motifs of the central domain (Fig. 5A ▶). RNA–RNA interaction dependent on the synthetic RAAA oligoribonucleotide, spanning nt G195–C219, was weak but specific for domain 3, its basal region, and the full-length IRES (Fig. 5B ▶). None of the other IRES domains or control RNAs (tRNA and antisense transcripts) used as ligands led to the formation of retarded complexes with this probe. RNA interaction between the RAAA oligoribonucleotide and D3 transcript relied on the presence of Mg2+ ions in the binding buffer and was dosage dependent. RNA–RNA binding was more efficient using as probe the transcript D3160–196 (Fig. 5C ▶), which encompasses the long GNRA bulged hairpin (Fig. 5A ▶). This interaction was specific for the central domain of the FMDV IRES (Fig. 5C ▶) and dosage dependent (Fig. 5D ▶), with a Kd ~100 nM. The specificity of these interactions was further assessed using labeled anti-sense domain 1–2-as probe (Fig. 5E ▶). No interaction was detected with any of the transcripts tested even in long exposures, with the exception of the entire IRES, as expected from their complementary sequence.
FIGURE 5.
RNA–RNA interactions dependent on the apical region of the central domain. (A) Schematic representation of domain 3 with indication of the regions encoded in transcripts used as probes used in gel shift assays. (B) 5′-end-labeled RAAA synthetic oligoribonucleotide (50 nM) was incubated in binding buffer with the indicated sense RNAs (400 nM), antisense (as), or tRNA; the last lane was incubated without Mg+2. Transcript 3ABC corresponds to the apical region of domain 3, nt 151–227. Residues encompassing domains 1–2 and 4–5 are indicated in Figure1 ▶. RNA complexes were separated in native 6% acrylamide gels in TBM buffer. (C) Interaction of the FMDV GNRA hairpin is specific for the central domain. Gel shift analysis carried out with D3160–196 transcript (50 nM) as probe and the different unlabeled FMDV IRES domains (400 nM). (D) Titration curve of transcript D3160–196 interaction with the central domain. (E) Specificity of the interactions studied in gel shift assays. The antisense sequence of domain 1–2 (50 nM) was used as probe to interact with each of the indicated IRES sequences (400 nM) as indicated in panel B. The FMDV IRES was included as positive control. (F) Efficiency of RNA–RNA interaction within the central domain is increased by residues in the apical region. Gel shift performed with the enlarged version of the apical region D3121–261 (50 nM) and the indicated transcripts (400 nM). The control antisense (c-as) RNA is described in Materials and Methods.
The use of a larger transcript termed D3121–261, encoding the entire apical region as probe in the gel shift analysis, indicated a significant increase in the efficiency of formation of the retarded complex (Fig. 5F ▶). Comparison of the efficiency of retarded complex formation with unlabeled D3 transcript indicated an increase from 40% in the case of labeled D3160–196 GNRA hairpin to 68% in the case of labeled D3121–261. Besides the GNRA motif-dependent interactions, this increment was consistent with the presence of additional contacts in the apical region, including the RAAA motif, contributing to distant contacts. As the size of the central domain probe increased, a new contact with domain 1–2 was detected, in agreement with previous data using the entire domain 3 as probe (Ramos and Martínez-Salas 1999). Conversely, the intensity of interaction with the entire IRES decreased (Fig. 5F ▶), likely because of a reduced accessibility imposed by a folded IRES structure.
Mutational analysis of the RAAA motif showed that nucleotide substitution A199AAAG203 to CGCCC reduced IRES activity ~90-fold (López de Quinto and Martínez-Salas 1997). In contrast, other substitutions predicted to preserve the RNA structure of this region were active. No change in the pattern of RNA–protein binding was detected in UV-cross-link assays carried out with the modified IRES (data not shown) as was demonstrated previously for the GNRA mutants (Fernández-Miragall and Martínez-Salas 2003). Therefore, to determine the functional relevance of the RAAA motif, we carried out structural probing of the substitution mutant CGCCC. Partial RNase T1 digestion revealed the presence of three new G residues (196, 199, and 208) accessible to T1, irrespectively of the analysis being carried out by primer extension or direct cleavage (Fig. 6A,B ▶). With the exception of protection of T1 in native RNA at position G134, the pattern of T1 cleavage in the basal part of domain 3 was similar to that obtained in wild-type RNA. Partial RNase A digestion showed that residues C211 and C204 became accessible, indicating that they were unpaired (Fig. 6C ▶). Results of DMS accessibility were consistent with enzymatic probing. The substitution in the RAAA loop to GCUGC had a similar DMS-induced RT stop pattern (data not shown). Thus, the CGCCC mutant offered an RNA conformation locally more open than the wild-type RNA structure (Fig. 6C ▶; cf. to Fig. 4A ▶). However, organization of the adjacent stem–loops was unaffected. This is in contrast to the long-distance effect originated by mutations affecting the GNRA motif.
FIGURE 6.
Enzymatic probing of RAAA substitution mutant. (A) RNase T1 probing analyzed by primer extension. RNA harboring the CGCCC substitution in the RAAA motif was incubated with RNase T1. A filled arrow denotes changes with the pattern obtained in the wild-type domain 3 (Fernández-Miragall and Martínez-Salas 2003). (B) Direct RNase T1 cleavage pattern of [γ-32P]ATP-5′-end-labeled CGCCC RNA, in comparison to the wild-type (AAAAG) RNA. Bands with the same electrophoretic mobility as cleavage fragments of the untreated transcript were not considered. Closed and empty arrows are used for new and lack of attack, respectively, relative to the wild-type sequence. (C) Reorganization of RNA structure in CGCCC mutant. Results from DMS attack have been represented on the secondary structure derived from RNase A, T1, and V1 digestion.
Analysis of the RNA structure of a truncated version of domain 3 reveals the absence of protections compatible with the lack of apical region interactions
The transcript termed stem 3 consists of a truncated version of domain 3 in which a deletion of nt 151–227 eliminates the apical region of the central domain. To identify a mark generated by the absence of the apical region, the structure of stem 3 was probed. Direct T1 accessibility to 5′-end-labeled RNA demonstrated that stem 3 differed from the entire domain 3 in the accessibility of four positions. G148 and G150, the residues that connect the left-hand arm of domain 3 with the right-hand arm in the truncated transcript, as well as G129 were accessible to T1, whereas G134 was protected (Fig. 7A ▶; cf. to wild-type, AAAAG, RNA in Fig. 6B ▶). Chemical probing followed of primer extension revealed significant changes, giving rise to RT stops at positions A261, A260, A145, and G126 (Fig. 7B ▶). These nucleotides were paired in the secondary structure of the entire domain 3 (cf. to Fig. 4A ▶). In agreement with these results, RNase T2 partial digestion of the truncated transcript originated three changes relative to the entire domain 3, U140, A260, and A261.
FIGURE 7.
Enzymatic and chemical probing of stem 3 RNA. (A) [γ-32P]ATP-5′-end-labeled RNA was incubated with RNase T1 and analyzed on denaturing gel electrophoresis. (B) DMS and T2 treatment followed by primer extension analysis. (C) Schematic representation of stem 3 secondary structure. A thin line between residues G150 and U228 depicts the junction of the left and right arms in the truncated version of the central domain, stem 3. Symbols used are as in Figures 2 ▶, 4 ▶, and 6 ▶.
Taken together, the evidence obtained from chemical and enzymatic RNA probing allowed us to define the secondary structure of stem 3 (Fig. 7C ▶). Reorganization of the central bulge was consistent with changes in A125, G129, and G134, together with A260 and A261. Changes in T1 protection led us to pair G256 and G258 with U251 and C249, respectively. Notably, G240 in motif A was no longer protected.
In summary, comparison of these results with the entire domain and that of mutants affected in conserved motifs described in previous sections (Fig. 4 ▶) led us to propose the G240CACG244 region as the residues where the GNRA motif is interacting. These data reinforce the role of the GNRA motif as a key structural element involved in the organization of the RNA structure of the central domain required for internal initiation translation.
DISCUSSION
The main goal of the present report was to study the biological significance of RNA structural motifs located in the central domain of the FMDV IRES element. To this end, first we examined in vitro the involvement of conserved motifs in RNA structure. Subsequently, we showed that reorganization of RNA structure had profound effects on IRES activity in vivo. Modification of the accessibility pattern of the different FMDV IRES mutants strongly suggests that the central domain acquires a folded structure that includes RNA tertiary interactions. Mapping of these contacts established a reciprocal link between the GNRA tetra-loop and the GCACG sequence, termed motif A. Fully consistent with our findings, both the GNRA and GCACG sequences are phylogenetically conserved, in spite of the high genetic variability of FMDV viral RNA (Martínez-Salas et al. 1993).
We have previously demonstrated by RNA probing techniques that the GUAA sequence of FMDV IRES acquires a tetraloop structure (Fernández-Miragall and Martínez-Salas 2003). GUAG and UNCG substitutions in this conserved motif led to reorganization of the apical region of the central domain. In full agreement with our results, recent NMR data confirm the presence of a GNRA tetraloop in the related EMCV IRES (Phelan et al. 2004) and poliovirus IRES (Du et al. 2004).
In this study RNase T2 accessibility indicated a change in RNA conformation in mutants affected in the GNRA motif when compared to wild-type GUAA RNA sequence. Together with an enhanced accessibility to RNases in the substituted motif, probing of these mutants identified two distant regions, encompassing nt 240 and 250 that became significantly more susceptible to RNase T2 attack. This finding was consistent with data indicating that a labeled GUAA hairpin specifically interacts with the central domain of the FMDV IRES transcript in gel shift assays. Additionally, and in support of distant interaction with residues within the central region of FMDV IRES, mutations in the GNRA motif significantly reduced their binding efficiency to IRES transcripts relative to the GUAA hairpin (Fernández-Miragall and Martínez-Salas 2003).
The distant regions that accumulated changes in chemical and enzymatic accessibility of UCCG and GUAG mutants were subjected to site-directed mutagenesis. Functional analysis indicated a relevant function of motif A, G240CACG244, as its substitution induced a 100-fold reduction in IRES activity, similar to that reported for GUAG mutant (López de Quinto and Martínez-Salas 1997). Conversely, chemical and enzymatic RNA probing showed that the motif A mutant contained an overall reorganization that affected not only the mutated region but also the distant A180 residue (Fig. 4B ▶). This result was not observed in mutant in motif B, albeit RNA probing also showed a change in RNA conformation around the substituted residues. On the basis of reciprocal changes presented in this study affecting both the GNRA tetraloop and the G240CACG244 motif, together with the decrease in IRES activity, we propose the GCACG region as the most probable candidate to function as a receptor of the GNRA motif.
Sequence variability of FMDV IRES from field isolates shows rare nucleotide substitutions in the GUAA sequence, GCAA or GCGA, always compatible with GNRA motifs. Phylogenetic conservation of the GCACG region was also noticed in a total of 36 different FMDV isolates (Fernández-Miragall and Martínez-Salas 2003). This result has been recently confirmed and extended to 103 different IRES sequences of FMDV (Carrillo et al. 2005). IRES residues involved in base pairing often show nucleotide covariation (Martínez-Salas and Fernández-Miragall 2004). However, it is remarkable that the GCACG region is invariant albeit potential Watson–Crick pairs are predicted with residues U228GUGC232 in computer folding programs (see Fig. 1 ▶). We interpreted the lack of covariation as a selective pressure to keep the primary sequence either, because of its involvement in distant contacts with other RNA motifs, as proposed here, or/and their recognition by auxiliary factors involved in translation initiation, not identified yet.
We have found a specific GNRA hairpin interaction with the central domain. This binding was consistent with previous data using a short synthetic GUAA oligoribonucleotide (Fernández-Miragall and Martínez-Salas 2003). In this case, substitution of GUAA to GUAG or UCCG impaired binding to the central domain. Here we have shown that the efficiency of retarded complex formation increased from ~1% in the short GUAA oligoribonucleotide to 40% in the case of the entire GNRA hairpin. This effect may be explained by an increase in binding efficiency generated by (1) a more stable hairpin or (2) the existence of additional contacts between transcript D3160–196 and the central domain. This second possibility is favored by gel shift assays in which substitution of the GUAA sequence to GUAG in transcript D3160–196 did not abolish binding to domain 3 (data not shown). Notably, a synthetic oligoribonucleotide encompassing the RAAA motif also weakly but specifically interacted with the central domain, including its basal region, stem 3. Taken together, these results may be interpreted as a cooperative binding exerted by multiple contacts between the apical region of the central domain. In support of this possibility, the intensity of the retarded complex formation was stronger with the transcript D3121–261 than with D3160–196.
Concerning the possibility of multiple RNA–RNA contacts within the central domain, it is of interest that a common structural modification was induced around residues 199–219 in both GUAG and motif A mutants (see Fig. 4A,B ▶; Fernández-Miragall and Martínez-Salas 2003), suggesting that RAAA stem–loop folding was dependent on the RNA structure determined by GNRA-dependent interactions. Remarkably, this reorganization is coincident to a large extent with that generated by a multiple substitution of the RAAA motif (see Fig. 6C ▶). The significant reduction of IRES activity in this mutant suggests impairment of an essential region. Neither RAAA nor GNRA motifs appear to be primarily involved in RNA–protein interactions, as revealed in UV-cross-linking assays performed with mutant IRES elements (Fernández-Miragall and Martínez-Salas 2003). The possibility of a second-site interaction of the RAAA loop with residues in the basal portion of domain 3, as deduced from gel shift assays, is likely; we hypothesize that this interaction may contribute to stabilize the GNRA–motif A interaction.
Probing of stem 3 RNA, encoding the proximal region of domain 3, strongly suggested the presence of a stable structure at its most basal region (Fig. 7C ▶). This region appears to fold independently of the surrounding IRES nucleotides, in agreement with previous mutational analysis that demonstrated the need of a base-paired structure involving residues G86UGU89 with A296CAC299 for IRES activity (Martínez-Salas et al. 1996). Thus, we propose that the basal portion of the central domain performs a stand function that contributes to IRES activity by holding the apical region in the appropriate conformation, compatible with its recognition by the translation machinery.
Experimental evidence for tertiary structure of IRES elements suggests a dynamic structure indicative of translation efficiency regulation at the level of RNA structure (Jan and Sarnow 2002; Yaman et al. 2003). Recently, it has been demonstrated that the CrPV IRES directly interacts within the ribosomal subunit space in the absence of canonical initiation factors (Spahn et al. 2004). Although different IRES elements such as CrPV and HCV appear to have a different structural organization, and their binding sites in the ribosomal subunit are different (Spahn et al. 2001), the conformational changes that they induce in the 40S ribosomal subunit show remarkable similarities. These results open the possibility that IRES elements could share the property of having structural elements that mediate its interaction with the 40S subunit. No binary complexes were detected in the absence of initiation factors in reconstitution assays with the related EMCV IRES (Pestova et al. 1996). However, the possibility that the central domain of the FMDV IRES is involved in facilitating direct RNA–RNA interaction with the 40S subunit still needs further investigation.
The flexible nature of the FMDV IRES may be important in the functional interaction with the ribosome. In this context, the central domain could play a significant role dictating the formation of a stable tertiary structure acquiring the correct orientation to recruit the ribosome subunits to the initiation sites.
MATERIALS AND METHODS
Generation of FMDV IRES constructs
The IRES of FMDV C-S8 consists of 462 nt, distributed in domains (Fig. 1 ▶). cDNAs corresponding to the subdomains studied and their mutated versions were cloned in the pGEM3 vector (Promega). The central domain 3 (nt 84–297) (Ramos and Martínez-Salas 1999) was further deleted to generate the proximal region, stem 3 (nt 84–297Δ151–227) devoid of the apical stem–loops. For that purpose, primers 5′-GCTTCGTAGCGGAGTGT GCAACCCC-3′, of sense polarity, and 5′-CTCCGCTACGAAGCA ACAGTGC-3′, antisense, were used in a double PCR including as template pBIC plasmid (Martínez-Salas et al. 1993). The construct D3160–196 encompassing the GNRA stem–loop was generated with primers 5′-GGAGCATGAATTCCGTGGGAAC-3′, sense, and 5′-GAGGCCCCGGGCCCCGTG-3′, antisense. Similarly, for D3121–260 construct primers 5′-GGCGAATTCTAGTAACAGCACTG-3′, sense, and 5′-GTCACTTTAACCCGGGTTTCGCAG-3′, antisense, were used.
To generate constructs with nucleotide substitutions in motifs A and B, the antisense primers 5′-GTCGCGCCGTTGGGGTTCC-3′ and 5′-CGCAGTAACCCGGCCGTGCTG-3′ were used in a double PCR reaction with external primers (Fernández-Miragall and Martínez-Salas 2003). Single substitution G240C was introduced using the antisense oligo 5′-GTCGCCGTGGTGGGGTTGCAC-3′. In all cases, the nucleotide sequence of the entire length of each cDNA under study was determined, using automatic sequencing (ABI PRISM dye terminator cycle sequencing ready reaction kit, Perkin Elmer).
In vitro transcription
Prior to RNA synthesis, plasmids were linearized to generate transcripts encoding domain 3, stem 3, or the full-length IRES using restriction enzymes SmaI or XhoI, as needed. Transcription was performed for 1 h at 37°C using 50 U of T7 RNA polymerase (New England Biolabs) in the presence of 0.5–1 μg of linearized DNA template, 50 mM DTT, 0.5 mM rNTPs, and 20 U of RNasin (Promega) or using the Megashortscript kit (Ambion) as recommended by the manufacturer. When needed, RNA transcripts were uniformly labeled using (α32P)-CTP (400 Ci/mmol). Reactions were incubated for 15 min with 1 U of RQ1 DNase (Promega), and unincorporated 32P-CTP eliminated by exclusion chromatography in TE-equilibrated sephadex columms.
Gel shift assays
For RNA–RNA interactions, [γ-32P]-ATP-labeled oligoribonucleotide RAAA (5′-GGCCAAAAGCCACGCCCACACGGGC-3′) or the indicated uniformly [α-32P]-CTP-labeled IRES transcripts were incubated independently at 80°C for 1 min, chilled on ice, and then mixed with the indicated unlabeled RNAs in 50 mM sodium cacodylate (pH 7.5), 300 mM KCl, 10 mM MgCl2 (Ferrandon et al. 1997). For negative controls, either the antisense version of domain 1–2 (D1–2as) or a fragment of the 18S RNA derived from pT7-RNA-18S control template (Ambion) (c-as) was synthesized using SP6 RNA polymerase. RNA–RNA complexes were allowed to form for 90 min at 20°C and inmediately analyzed by electrophoresis in native acrylamide gels supplemented with 2.5 mM MgCl2 (Fedor and Uhlenbeck 1990; Paillart et al. 1996). Gels were run at 4°C for 33 min at 180 V in TBM buffer (45 mM Tris at pH 8.3, 43 mM boric acid, 0.1 mM MgCl2). A titration curve performed using a concentration range (25–2000 nM) of unlabeled D3 RNA and 50 nM of probe D3160–196 was carried out to estimate the Kd of RNA–RNA interactions (Setzer 1999). In all cases, dried gels were exposed for autoradiography, as well as to a phosphorimager plate, to quantify the intensity of the retarded bands as described (Ramos and Martínez-Salas 1999).
RNA structure probing
For chemical probing, RNA (~1.5 μg) was treated with DMS (Fluka) (1 μL of DMS freshly diluted 1:5 in ethanol; 15 min at 20°C) using native (buffer N, 50 mM sodium cacodylate at pH 7.5, 300 mM KCl, 10 mM MgCl2 or buffer N′, 20 mM HEPES at pH 7.5, 2 mM AcMg, 100 mM ClK, 0.1 mM EDTA), and denaturing conditions (buffer D, 10 mM Tris-HCl at pH 8, 1 mM EDTA) (Brunel and Romby 2000). For enzymatic probing, ribonuclease A (5 × 10−6 mg/mL, 30 min, 37°C) (Aldaz-Carrol et al. 2002), T1 (0.03 U, 30 min, 37°C) (Branch et al. 1989) (Boehringer Manheim), T2 (0.05 U, 20 min at 37°C) (Invitrogen) (Irie and Ohgi 2001), and V1 (0.2 U, 10 min, 37°C) (Ambion) (Kolupaeva et al. 2000) partial digestion was carried out using 1.5 μg of RNA, and then analyzed by primer extension (Brunel and Romby 2000).
For T1 direct cleavage, RNA was 5′-end labeled and the full-length product purified from 6% acrylamide, 7 M urea gels. Full-length transcript present in a thin gel slice was eluted overnight in 200 μL of 10 mM Tris (pH 8), 1 mM EDTA, 0.1% SDS, at 37°C with shaking. Following ethanol precipitation in the presence of oyster glycogen, the eluted RNAs were resuspended in water to ~50,000 cpm/μL and digested as described above.
Primer extension analysis
Antisense oligonucleotides, complementary to positions 296–287 (5′-CCCGGGTGTGGGTACC-3′) or 185–165 (5′-CTTGTCGCCA AGGAGGAGTTC-3′) were 5′-end labeled using [γ-32P]-ATP and T4 polynucleotide kinase (Boehringer Mannheim). Then, template RNA (1.5 μg) was denatured 3 min at 95°C and cooled down on ice. Annealing and extension of the labeled antisense primer was carried out in 15 μL of reverse transcriptase (RT) buffer (20 mM Tris-HCl at pH 7.5, 15 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.01% (v/v) NP-40, 50% (v/v) glycerol) in the presence of 100 U of SuperScript II RT (Life Technologies) and 1 mM each dNTPs during 1 h at 45°C. The RNA template was then hydrolyzed in 20 μL of 50 mM Tris-HCl (pH 7.5), 7.5 mM EDTA, 0.5% SDS in the presence of 3.5 μL 3 M KOH at 95°C for 3 min followed of incubation at 37°C during 1 h. Hydrolysis was stopped by adding 6 μL of 3 M acetic acid. RT extension products were then fractionated in denaturing 6% acrylamide, 7 M urea gels as described (Fernández-Miragall and Martínez-Salas 2003). To prepare the sequence ladder, the Thermo sequenase cycle sequencing kit (Amersham) was used with the same 5′-labeled antisense oligonucleotide used for primer extension and plasmid pGEM-IRES as template.
RNA–protein interaction assays
S10 extracts from BHK-21 cells were prepared as described (López de Quinto and Martínez-Salas 2000). UV-cross-linking assays were performed using 40 μg of native proteins present in S10 extracts of BHK-21 cells and 0.02 pmol of the specific 32P-labeled RNA. The RNA–protein cross-linked lysate was digested with an excess of RNase A during 30 min at 37°C, followed by addition of SDS loading buffer, heating 2 min at 95°C, and electrophoresis through SDS-polyacrylamide gels (López de Quinto et al. 2001). Then, dried gels were used to visualize the 32P-labeled proteins by autoradiography.
In vivo IRES activity
Relative IRES activity was quantified as the expression of luciferase normalized to that of chloramphenicol acetyltransferase (CAT) from bicistronic mRNAs (López de Quinto and Martínez-Salas 1997). Transfection of 80–90% confluent BHK-21 monolayers was carried out using cationic liposomes, 1 h after infection with the vaccinia virus recombinant vT7F-3 expressing the T7 RNA polymerase (Fuerst et al. 1986). Extracts from 1 × 105 to 2 × 105 cells were prepared 20 h postransfection in 100 μL of 50 mM Tris-HCl (pH 7.8), 120 mM NaCl, 0.5% NP40. Experiments were performed on triplicate wells, and each experiment was repeated at least three times.
Acknowledgments
We thank S. López de Quinto for early contribution to this work and J. Gómez and C. Gutierrez for helpful suggestions on the manuscript. This work was supported by grants BMC-2002-0983 and GRSAL/0006/2004 and by an Institutional grant from Fundación Ramón Areces.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2153206.
REFERENCES
- Aldaz-Carroll, L., Tallet, B., Dausse, E., Yurchenko, L., and Toulmé, J.-J. 2002. Apical loop-internal loop interactions: A new RNA–RNA recognition motif identified through in vitro selection against RNA hairpins of the hepatitis C virus mRNA. Biochemistry 41: 5883–5893. [DOI] [PubMed] [Google Scholar]
- Bedard, K.M., Walter, B.L., and Semler, B.L. 2004. Multimerization of poly(rC) binding protein 2 is required for translation initiation mediated by a viral IRES. RNA 10: 1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham, G.J. and Jackson, R.J. 2000. Translation initiation on picornavirus RNA. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 869–900. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Branch, A.D., Benenfeld, B.J., and Robertson, H.D. 1989. RNA fingerprinting. Methods Enzymol. 180: 130–154. [DOI] [PubMed] [Google Scholar]
- Brunel, C. and Romby, P. 2000. Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol. 318: 3–21. [DOI] [PubMed] [Google Scholar]
- Carrillo, C., Tulman, E.R., Delhon, G., Lu, Z., Carreno, A., Vagnozzi, A., Kutish, G.F., and Rock, D.L. 2005. Comparative genomics of foot-and-mouth disease virus. J. Virol. 79: 6487–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A.T., Robertson, M.E., Conn, G.L., and Belsham, G.J. 2003. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J. Virol. 77: 12441–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll, C.C, and Swinger, K. 2003. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 A resolution. RNA 9: 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M. and Michel, F. 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 16: 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino, D. and Kieft, J.S. 2005. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA 11: 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, E.A., Batey, R.T., Masquida, B., and Doudna, J.A. 2001. A universal mode of helix packing in RNA. Nat. Struct. Biol. 8: 339–343. [DOI] [PubMed] [Google Scholar]
- Drew, J. and Belsham, G.J. 1994. Trans-complementation by RNA of defective foot-and-mouth disease virus internal ribosome entry site elements. J. Virol. 68: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z., Ulyanov, N.B., Yu, J., Andino, R., and James, T.L. 2004. NMR structures of loop B RNAs from the stem–loop IV domain of the enterovirus internal ribosome entry site: A single C to U substitution drastically changes the shape and flexibility of RNA. Biochemistry 43: 5757–5771. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. and Uhlenbeck, O.C. 1990. Substrate sequence effects on “hammerhead” RNA catalytic efficiency. Proc. Natl. Acad. Sci. 87: 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Miragall, O. and Martínez-Salas, E. 2003. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 9: 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon, D., Koch, I., Westhof, E., and Nusslein-Volhard, C. 1997. RNA–RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16: 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst, T.R., Niles, E.G., Studier, F.W., and Moss, B. 1986. Eukaryotic transsient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. 83: 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U. and Sarnow, P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Irie, M. and Ohgi, K. 2001. Ribonuclease T2. Methods Enzymol. 341: 42–55 [DOI] [PubMed] [Google Scholar]
- Jan, E. and Sarnow, P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Jubin, R., Vantuno, N.E., Kieft, J.S., Murray, M.G., Doudna, J.A., Lau, J.Y.N., and Baroudy, B.M. 2000. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 74: 10430–10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Jubin, R., Murray, M.G., Lau, J.Y., and Doudna, J.A. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292: 513–529. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Grech, A., Jubin, R., and Doudna, J.A. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9: 370–374. [DOI] [PubMed] [Google Scholar]
- Kolupaeva, V., Pestova, T.V., Hellen, C.U.T., and Shatsky, I.N. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273: 18599–18604. [DOI] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Pestova, T.V., and Hellen, C.U. 2000. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 74: 6242–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Quinto, S. and Martínez-Salas, E. 1997. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 71: 4171–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6: 1380–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Quinto, S., Lafuente, E., and Martínez-Salas, E. 2001. IRES interaction with translation initiation factors: Functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7: 1213–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky, P.J., Kim, I., Otto, G.A., and Puglisi, J.D. 2003. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 10: 1033–1038. [DOI] [PubMed] [Google Scholar]
- Lyons, A.J., Lytle, J.R., Gomez, J., and Robertson, H.D. 2001. Hepatitis C virus internal ribosome entry site RNA contains a tertiary structural element in a functional domain of stem–loop II. Nucleic Acids Res. 29: 2535–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas, E. and Fernández-Miragall, O. 2004. Picornavirus IRES: Structure function relationship. Curr. Pharm. Des. 10: 3757–3767. [DOI] [PubMed] [Google Scholar]
- Martínez-Salas, E., Sáiz, J.C., Dávila, M., Belsham, G.J., and Domingo, E. 1993. A single nucleotide substitution in the internal ribosome entry site of foot-and-mouth disease virus leads to enhanced cap-independent translation in vivo. J. Virol. 67: 3748–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas, E., Regalado, M.P., and Domingo, E. 1996. Identification of an essential region for internal initiation of translation in the aphthovirus internal ribosome entry site and implications for viral evolution. J. Virol. 70: 992–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas, E., Ramos, R., Lafuente, E., and López de Quinto, S. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82: 973–984. [DOI] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Coldwell, M.J., Jackson, R.J., and Willis, A.E. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11: 757–771. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B., and Steitz, T.A. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 98: 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillart, J.C., Skripkin, E., Ehresmann, B., Ehresmann, C., and Marquet, R. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. 93: 5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U., and Shatsky, I.N. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16: 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan, M., Banks, R.J., Conn, G., and Ramesh, V. 2004. NMR studies of the structure and Mg2+ binding properties of a conserved RNA motif of EMCV picornavirus IRES element. Nucleic Acids Res. 32: 4715–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Pestova, T.V., Kolupaeva, V.G., Khitrina, E.V., Poperechnaya, A.N., Agol, V.I., and Hellen, C.U.T. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes & Dev. 14: 2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Ramos, R. and Martínez-Salas, E. 1999. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES). RNA 5: 1374–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand, R., Thiviyanathan, V., Kaluarachchi, K., Lemon, S.M., and Gorenstein, D.G. 2004. Mutational and structural analysis of stem–loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J. Mol. Biol. 343: 805–817. [DOI] [PubMed] [Google Scholar]
- Robertson, M.E., Seamons, R.A., and Belsham, G.J. 1999. A selection system for functional internal ribosome entry site (IRES) elements: Analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA 5: 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setzer, D.R. 1999. Measuring equilibrium and kinetic constants using gel retardation. In Methods in molecular biology. Vol. 118. RNA-protein interaction protocols (ed. S. Haynes), pp. 115–128. Humana Press Inc., Totowa, NJ. [DOI] [PubMed]
- Spahn, C.M., Kieft, J.S., Grassucci, R.A., Penczek, P.A., Zhou, K., Doudna, J.A., and Frank, J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291: 1959–1962. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Jan, E., Mulder, A., Grassucci, R.A., Sarnow, P., and Frank, J. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell 118: 465–475. [DOI] [PubMed] [Google Scholar]
- Stoneley, M. and Willis, A.E. 2004. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene 23: 3200–3207. [DOI] [PubMed] [Google Scholar]
- Terenin, I.M., Dmitriev, S.E., Andreev, D.E., Royall, E., Belsham, G.J., Roberts, L.O., and Shatsky, I.N. 2005. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol. Cell. Biol. 25: 7879–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, S., Berrisford, D.J., and Avis, J.M. 2002. Effect of magnesium ions on the tertiary structure of the hepatitis C virus IRES and its affinity for the cyclic peptide antibiotic viomycin. Biochemistry 41: 5383–5396. [DOI] [PubMed] [Google Scholar]
- Walter, B.L., Nguyen, J.H., Ehrenfeld, E., and Semler, B.L. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5: 1570–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A.A., Koromilas, A.E., Zhou, L., Snider, M.D., Scheuner, D., Kaufman, R.J., et al. 2003. The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113: 519–531. [DOI] [PubMed] [Google Scholar]