Abstract
Domain 5 (D5) is absolutely required for all catalytic functions of group II introns. Here we describe the solution NMR structure, electrostatic calculations, and detailed magnesium ion-binding surface of D5 RNA from the Pylaiella littoralis large ribosomal RNA intron (D5-PL). The overall structure consists of a hairpin capped by a GNRA tetraloop. The stem is divided into lower and upper helices of 8 and 5 bp, respectively, separated by an internal bulge. The D5-PL internal bulge nucleotides stack into the helical junction, resulting in a coupling between the bulge A25 and the closing base pair (G8–C27) of the lower helix. Comparison of the D5-PL structure to previously reported related structures indicates that our structure is most similar, in the helical regions, to the crystal structure of D5 from yeast Ai5γ (D5-Ai5γ) and the NMR structure of the U6 snRNA stem–loop region. Our structure differs in many respects from both the NMR and X-ray structures of D5-Ai5γ in the bulge region. Electrostatic calculations and NMR chemical shift perturbation analyses reveal magnesium ion-binding sites in the tetraloop, internal bulge, and the AGC triad in the lower stem. Our results suggest that the structure, electrostatic environment, and the magnesium ion-binding sites within the tetraloop, bulge, and triad regions are conserved features of the splicing machinery of both the group II introns and the spliceosome that are likely key for catalytic function.
Keywords: Adaptive Poisson-Boltzmann electrostatics, magnesium ion binding, NMR structure, P. littoralis ribosomal RNA intron, RNA catalysis, residual dipolar coupling
INTRODUCTION
Self-splicing group II introns and the megadalton spliceosome share many strikingly conserved and functionally important structural features that suggest a linked evolutionary history (Sharp 1985; Cech 1986; Jacquier 1990; Villa et al. 2002). In particular, the highly conserved and catalytically important group II intron domain 5 (D5) element and the conserved intramolecular stem–loop (ISL) found in the spliceosomal U6/U6atac small nuclear RNAs (snRNAs) have similar structural features. Beyond the structural similarities, D5 can also replace U6/U6atac in an in vivo functional assay (Shukla and Padgett 2002).
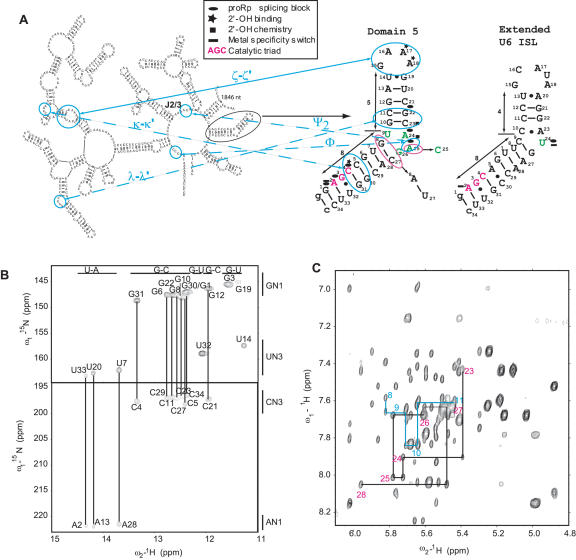
D5 is a key component of the catalytic core of the self-splicing group II introns, and it interacts with domains 1, 2, and 3 (D123) to activate both chemical steps of splicing. Extensive biochemical studies have shown that D5 is a centerpiece for organizing the catalytic core of this group II intron ribozyme (Michel et al. 1989; Koch et al. 1992). Through an extensive network of tertiary contacts, it stabilizes the rest of the group II intron active folded core involving D123 (Fig. 1 ▶; Costa and Michel 1999; Boudvillain et al. 2000). Previous biochemical studies have shown that the 2′-OH groups of the last two adenines in the GAAA tetraloop (A17 and A18) and the highly conserved C–G base pairs (C5–G30 and C4–G31) in the lower helix tether D5 onto the tetraloop receptor in D1 via the ζ–ζ′ and κ–κ′ interactions, respectively (Fig. 1A ▶; Abramovitz et al. 1996; Boudvillain and Pyle 1998; Boudvillain et al. 2000). Base pairs in the upper helix of D5 (G12–C21, C11–G22) are also expected to form a network of base triple λ–λ ′ interactions with domain 1 (D1) (Fig. 1A ▶; Boudvillain et al. 2000). Finally, the D5 bulge nucleotide C25 (A25) forms a Φ interaction with intron nucleotide 5 in D1 and a ψ2 interaction with one of the highly conserved nucleotides in the J2/3 linker region (Fig. 1A ▶; Podar and Perlman 1999; de Lencastre et al. 2005). D5 is therefore at the heart of the catalytic machinery of this class of ribozymes.
FIGURE 1.
Sequence and secondary structure of the Pyllaiela littoralis (PL) large ribosomal subunit group II intron and the D5 RNA 2D structure as deduced from the NMR data. (A) D5 (circled) is shown in the context of the rest of the “Six-wheel” domain architecture of the PL group II intron. The D5-PL sequence highlighting sites for metal ion binding, intronic component binding, and chemistry are shown together with the extended U6 intramolecular stem–loop structure. The sequences for D5-PL and yeast D5-Ai5γ differ in only three positions indicated by magenta circles. The corresponding Ai5γ sequence of U27–A8 and C25 are indicated with arrows pointing from the D5-PL to D5-Ai5γ sequence. The AGC triad nucleotides are highlighted in magenta for both D5 and U6 ISL. The various tertiary interactions between D5 and D123 are also indicated. (B) NMR evidence for Watson-Crick base pairs in D5-PL shown in A using direct detection of Watson-Crick G–C and U–A base pairs at pH 6.5 and 298 K. Except for U9 and G26, all the imino correlations are observed. (C) Sequential base-ribose walk for the bulge region in D5-PL. A 500 MHz 1H–1H NOESY spectrum of D5-PL was acquired at 45°C in D2O on an ~1 mM RNA sample in 10 mM phosphate buffer (pH 6.5) and 100 mM KCl. The NOESY mixing time was 250 msec. The sequential walk is continuous from G8 to C11 and C23 to A28 and depicted, for clarity, only for the bulge region.
Recent solution NMR and X-ray crystal structures of D5 from yeast Ai5γ group II intron (D5-Ai5γ) and an NMR structure of an extended U6 ISL revealed some similarities and differences in their conformational fold (Zhang and Doudna 2002; Sashital et al. 2004; Sigel et al. 2004). The base-pairing pattern within the important D5 internal bulge region remains controversial, however. In the D5 structure, an initially proposed C25–G26 bulge was revised based on chemical probing and sequence covariation analysis to a U9○G26 wobble pair and unpaired A24 and C25 (see Fig. 1A ▶; Michel et al. 1989; Costa et al. 1998; Konforti et al. 1998). The crystal structure confirmed the U9○G26 wobble configuration and showed the bulge nucleotides A24 and C25 flipped out from the helix (Zhang and Doudna 2002). The NMR structure, in contrast, indicated that the bulge nucleotides A24 and C25 are stacked into the helical structure, while the U9○G26 wobble was not observed. Instead, G26 had a syn conformation and was flipped down into the major groove of the lower helix in close proximity to the AGC triad (Sigel et al. 2004).
A striking similarity between group II intron D5 and the spliceosomal U6/U6atac snRNAs is the presence of an invariant RGC (R = A or G) triad within their respective conserved lower helical RNA elements (Fig. 1A ▶). In a metal-specificity switch experiment, the replacement with sulfur of the nonbridging oxygen 5′ of A2 of the AGC triad of Ai5γ inhibited activity in the presence of Mg2+. Activity was restored by addition of Cd2+, suggesting that this oxygen coordinates an essential metal ion (Gordon and Piccirilli 2001). The inference of coordinated metal ion within this RGC triad, however, has not been demonstrated conclusively for yeast U6 within the spliceosome (Fabrizio and Abelson 1992; Yu et al. 1995). What has been demonstrated for U6 is that a phosphorothioate adjacent to the highly conserved bulge nucleotide U80 undergoes a similar metal rescue (Yean et al. 2000). The equivalent position in D5, A24, is a divalent ion-binding site that does not undergo metal rescue (Chanfreau and Jacquier 1994; Sigel et al. 2000; Gordon and Piccirilli 2001; Zhang and Doudna 2002).
Two well-studied group II introns are from yeast Ai5γ and Pylaiella littoralis (PL). The mitochondrial genes encoding the large ribosomal RNA (LSU) are interrupted by four group II introns in PL, of which the second intron of the LSU not only self-splices but also does so under most of the conditions tested (Fontaine et al. 1995, 1997; Costa et al. 1997). These self-spicing reactions are critical for the expression of the mitochondrial genes. Even though Ai5γ and PL share significant similarities in their domain organization at the secondary structure level, at least four important differences exist between them. First, PL has an overall higher GC content (40.9%) than Ai5γ (20.5%) (Costa et al. 1997). Second, the catalytically important D5 structural elements differ at one position in the bulge region (A25 in PL and C25 in Ai5γ) (Fig. 1A ▶) and two positions in the lower helix (G8–C27 in PL and A8–U27 in Ai5γ) (Fig. 1A ▶). Intriguingly, the 8–27 base pair and the nucleotide at position 25 covary in a catalog of D5 sequences, suggesting a coupling between these positions (Shukla and Padgett 2002). Third, in vitro self-splicing of Ai5γ requires rather unphysiological salt conditions [100 mM Mg2+ and 500 mM (NH4)2SO4], whereas the PL intron retains the ability to self-splice at low magnesium ion concentrations (0.5–10 mM) and moderate monovalent salt concentrations [100 mM KCl or (NH4)2SO4] (Jarrell et al. 1988; Costa et al. 1997). Fourth, for PL, tight binding of the 5′-exon requires both intron domain I (D1) sequences and the lower helix of D5 (Costa and Michel 1999), whereas neither D5 nor domain III (D3) contributes any energy to D1 binding of a 5′-exon substrate in the yeast Ai5γ intron (Qin and Pyle 1997). These four differences suggest that either the structures of the two D5s are not the same, and/or they may engage the rest of the intronic elements differently for both steps of splicing. Either way, a solution structure of a distinct D5 is desirable both as an independent structure from a different group II intron ribozyme and as a molecular probe to further our understanding of the mechanism of splicing.
As a first step toward determining the conformation of the key active-site components of the PL group II intron, we have carried out solution NMR structural studies of an isolated D5-PL. The bulge region of D5-PL adopts a conformation different from the X-ray crystal structure of D5-Ai5γ, while both D5-PL and the X-ray D5-Ai5γ structures are significantly different in orientation relative to the lower helix of the NMR D5-Ai5γ structure. Moreover, electrostatic calculations and detailed NMR Mg2+ titration analyses indicate that magnesium ions bind specifically to the minor groove surfaces of the bulge nucleotide A24 and the tetraloop residue G19, and the major groove face of the AGC triad nucleotide A2 but not G3.
RESULTS
Spectral analysis of D5-PL
To understand the role of structure and electrostatics in D5 group II intron catalytic function, we determined the structure of D5-PL by NMR spectroscopy, used the coordinates to compute the electrostatic properties, and mapped the detailed magnesium ion-binding surface of D5. For this purpose, we used a 36-nt model RNA corresponding to nucleotides 2366 (designated nucleotide 1)–2399 (designated nucleotide 34) (Costa et al. 1998) with an extra GC pair appended to the end to improve transcription efficiency.
Assigning the D5 spectra proved particularly challenging, requiring a battery of homonuclear and multidimensional NMR experiments coupled with nucleotide specific labeling. These included through-bond experiments that directly link base–base, base–ribose, and ribose–ribose functional groups (Dayie 2005).
From the analyses of NMR spectra, we obtained the secondary structure of D5-PL shown in Figure1A ▶. All the imino resonances, except those of the bulge U9, were observed in the 1H–15N HSQC and could be assigned in a two-dimensional (2D) 1H–1H NOESY spectrum. Using a newly proposed experiment that links exchangeable imino protons with nonexchangeable aromatic protons, the U9 imino proton could be correlated with its aromatic C6 carbon and imino N3 nitrogen resonances (Dayie 2005). A Watson-Crick conformation for U9–A24 is ruled out by the imino proton chemical shift of U9 (11.2 ppm) and the absence of hydrogen-bond correlations between H2 of A24 and N3 of U9. The Watson-Crick pair G8–C27 that is adjacent to the bulge is readily seen in the JNN HNN-COSY experiment (Dingley and Grzesiek 1998): we observed a cross-peak between the G8 imino proton (H1) and nitrogen (N1) and the acceptor N3 resonance of C27 (Fig. 1B ▶). In the bulge region, characteristic sequential and stacking interactions are observed that indicate continuous nuclear Overhauser effect (NOE) contacts from G8 to G10 and C23 to C27. On the 3′-side of the bulge, we observe the following NOE: C23 H1′–A24 H8, A24 H1′–A25 H8, A25 H1′–G26 H8, G26 H1′–C27 H6, C27 H1′–A28 H8. On the 5′-side, we detect G8 H1′–U9 H6, and U9 H1′–G10 H8 (Fig. 1C ▶). Unlike what was reported for D5-Ai5γ (Sigel et al. 2004), we do not observe NOE contact between H8 of G26 and H1′ of A28. A24 H2 forms an interstrand NOE with G10 H1′, makes a sequential contact with A25 H1′, and stacks with the H2 proton on the base of A25. A25 H2 forms NOE contacts with G26 H1′ and H8 but not with U9 H1′ or C27 H6/H5. The 1H–1H TOCSY experiment to determine the ribose pucker and the 1H–1H NOESY experiment to determine the glycosidic torsion indicate all but A16, A24, and A25 adopt 3′-endo sugar puckers and all nucleotides except G26 adopt an anti conformation. We observe very strong intranucleotide NOE between resonances of H8 and H1′ of G26 at long mixing times (250 msec) and medium to strong NOE from G26 H8 to H1′ at shorter mixing times (50 msec), suggesting a syn glycosidic geometry for G26.
In agreement with other studies, upfield-shifted imino resonances were observed for the G19 H1 (11.45 ppm) and U14 H3 (11.35 ppm) protons of the G○U wobble, and their imino protons had strong NOE contacts with each other. As expected for a stacked G○U base pair, the U14 H3 imino proton also has NOE contacts to those of G15 and U20. The GAAA tetraloop exhibits NOE cross-peaks and chemical shifts consistent with earlier NMR structures (Jucker et al. 1996). For example, the H1′ proton of the loop-closing nucleotide G19 is unusually upfield-shifted to 3.88 ppm as expected for a classical GNRA fold (Jucker et al. 1996). Similarly, the H1 imino proton of G15, which is protected from solvent, is upfield shifted (10.51 ppm), and has NOE contacts with the H1 imino proton of U14 and the H8 aromatic proton of A18 as expected for a G–A mismatch and formation of a GNRA tetraloop.
Structure of D5-PL
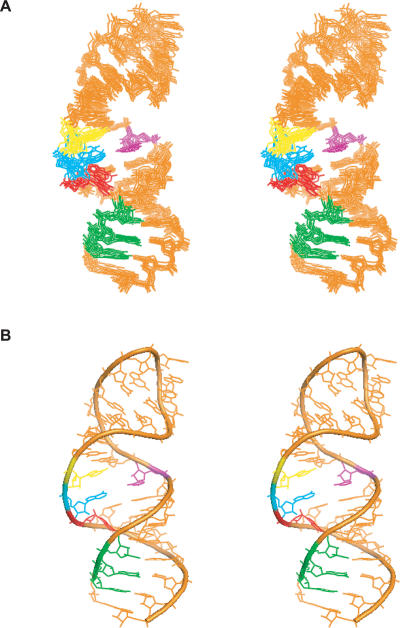
The structure of D5-PL was calculated from 549 NOE distances, 304 dihedral torsion angles, and 37 residual dipolar coupling (RDC) restraints. Measurement of intrabase 2JNN couplings across hydrogen bonds enabled the unambiguous identification of the base-pairing scheme within D5 and provided restraint for structure calculations (Table 1 ▶). Only NMR-derived restraints were used in the structure calculations. Initial structure calculations used only NOE distance, hydrogen bonds, and dihedral angle restraints. This resulted in well-defined local structural elements and good overall global structure. The root mean square deviation (RMSD) of the 10 lowest energy structures over all heavy atoms from the mean structure for the lower helix is 0.72 Å, for the upper helix is 0.63 Å, and for all residues is 1.34 Å (Table 1 ▶).
TABLE 1.
Structural statistics for D5-PL
| With RDC | Without RDC | |
| A. Restraints for structure calculations | ||
| No. of structures | ||
| Accepted | 10 | 30 |
| Calculated | 30 | 100 |
| No. of intraresidue distance restraints | 349 | 349 |
| No. of interresidue distance restraints | 200 | 200 |
| No. of dihedral restraints | 304 | 304 |
| No. of hydrogen-bond restraints | 60 | 60 |
| No. of dipolar coupling restraints | 37 | 0 |
| B. Average RMS deviation of experimental data | ||
| Average RMS deviation of distance (Å) | 0.019 | 0.018 |
| Average RMS deviation for dihedral angle (L) | 0.676 | 0.528 |
| Average RMS deviation for RDC (Hz) | 2.5 | 0 |
| C. Average RMS deviation from idealized geometry | ||
| Bonds (Å) | 0.017 | 0.018 |
| Angles (L) | 0.628 | 0.659 |
| Improper(L) | 0.344 | 0.330 |
| D. Violations | ||
| NOE violations >0.2 A ° | 0 | 0 |
| Dihedral violations >5L | 0 | 0 |
| E. Heavy atoms average RMS deviation from the mean structure and (RMS pairwise difference between structures) (Å) | ||
| Overall | 0.65 ± 0.13 (0.97 ± 0.20) | 1.34 ± 0.38 (1.99 ± 0.52) |
| Lower helix (residues 1–8, 27–34) | 0.43 ± 0.07 (0.64 ± 0.14) | 0.72 ± 0.13 (1.07 ± 0.27) |
| Upper helix (residues 10–14, 19–23) | 0.33 ± 0.07 (0.49 ± 0.11) | 0.63 ± 0.16 (0.93 ± 0.22) |
| Tetraloop (residues 15–18) | 0.34 ± 0.07 (0.51 ± 0.18) | 0.47 ± 0.08 (0.70 ± 0.16) |
| Bulge (residues 9, 24, 25, 26) | 1.35 ± 0.16 (2.00 ± 0.34) | 1.42 ± 0.42 (2.12 ± 0.66) |
| Bulge (residues 9, 24, 26) | 0.86 ± 0.07 (1.28 ± 0.34) | 1.42 ± 0.42 (2.12 ± 0.66) |
To further improve the accuracy and precision of the NMR structure, we derived long-range structural information from measured residual dipolar couplings (RDCs). Partial alignment of the 13C, 15N-labeled D5-PL sample was achieved using ~22 mg/mL Pf1 phage as cosolute. One-bond 1H–13C RDCs were measured for 34 aromatic and 18 ribose groups. Use of all 52 measured RDC restraints increased the precision of the structures to an overall RMSD (over all heavy atoms) of 1.08 ± 0.21 Å, and RMSD (over all heavy atoms) for the local lower and upper helical structural elements of 0.61 ± 0.23 Å and 0.60 ± 0.19 Å, respectively (data not shown). The RDCs did not, however, improve the precision of the bulge and tetraloop structures. Because this is likely caused by the flexibility of these two regions (K.T. Dayie and N.V. Eldho, unpubl.), we carried out a systematic refinement process that used relaxation as a filter. Omitting all the flexible tetraloop and bulge A25 nucleotides in the RDC refinement further improved the RMSD of both the global and local structures of D5-PL (Fig. 2 ▶; Supplemental Fig. 1 [see below for Supplemental Data information]). The RMSDs improve to 0.65 ± 0.13 Å for the overall structure, and the upper and lower helices improve to 0.33 ± 0.07 Å and 0.43 ± 0.07 Å, respectively (Table 1 ▶). The RMSD for superimposition of the tetraloop and bulge regions also improves to 0.34 ± 0.07 Å and 0.86 ± 0.07 Å, respectively (Table 1 ▶). Refinement with RDC for the entire or part of the tetraloop residues leads to increased heavy-atom RMSD values for the loop residues (0.5–0.6 Å). Similarly, inclusion of the RDC data for the bulge A25 nucleotide leads to an increased RMSD value for the entire bulge region, and the conformation of A25 remains ill-defined. Thus, use of 37 measured RDC values (that omitted those of the tetraloop and A25) led to a further improvement of the D5-PL structure. Back-calculations of RDCs were in agreement with the experimental values for the RDC refined structures (Supplemental Fig. 1E). Overall, these values indicate the structure of D5-PL is of high quality.
FIGURE 2.
Solution NMR structure of D5-PL. (A) A stereo representation of the superposition over the 10 lowest energy conformers of D5-PL. (B) A stereo representation of the D5-PL structure closest to the mean.
D5-PL adopts lower and upper helical stems of 8 and 5 bp, respectively, separated by an internal bulge. Nucleotides from G10 to U14 and G19 to C23 form the A-form upper stem, while those from G1 to G8 and C27 to C34 form an A-form lower stem. The four nucleotides U9, A24, A25, and G26 are unpaired and form the internal bulge (Fig. 3A ▶). The upper helical stem is terminated by a conserved GNRA-type (N is any nucleotide, R is a G or A) tetraloop.
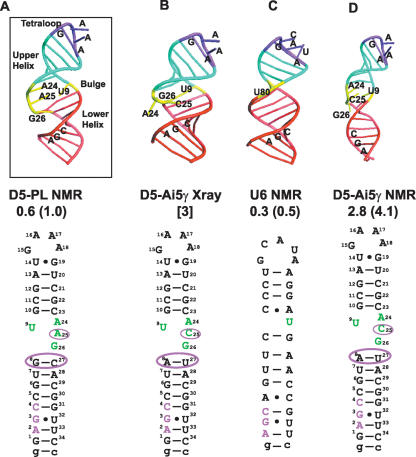
FIGURE 3.
Schematic of D5 and U6 structures highlighting differences and similarities. (A) Solution NMR structure of D5-PL depicting the three critical structural regions of the tetraloop, bulge, and triad AGC regions. (B) X-Ray crystal structure of D5-Ai5γ depicting extra helical conformation of the bulge A24 and C25 nucleotides. (C) Solution NMR structure of U6 ISL depicting the three critical structural regions of the tetraloop, bulge, and triad AGC regions. (D) Solution NMR structure of D5-Ai5γ. The atomic coordinates for the D5-Ai5γ and U6 structures are from PDB codes 1KXK, 1R2P, and 1XHP (6–8). The overall RMSD from the mean structure and the RMS pairwise difference between structures are shown below each structure based on the 10 lowest energy conformers for the NMR structures. For the X-ray structure, we show the resolution. For ease of comparison, we also depict the 2D sequences below each structure.
The overall D5-PL structure compares quite favorably with the high-resolution structure of U6 ISL and less well with the D5-Ai5γ NMR structure (Fig. 3 ▶; Sashital et al. 2004; Sigel et al. 2004). The pairwise RMSD, over the helical regions, of D5-PL NMR to U6 structure is 2.0 Å, to D5-Ai5γ X-ray is 3.9 Å, and to D5-Ai5γ NMR is 6.2 Å. The upper helical region is the most similar among the four structures with pairwise RMSDs of 0.5 Å (U6), 1.2 Å (D5-Ai5γ NMR), and 1.4 Å (D5-Ai5γ X-ray). Compared to the upper helix, the lower helical region has higher pairwise RMSDs of 1.2–2.5 Å between the four structures. On the other hand, the tetraloop region has the most similar structure among the three D5 structures (0.8–0.9 Å) and is slightly different from the U6 structure (1.6 Å). This is not surprising since U6 has an extra nucleotide in the loop.
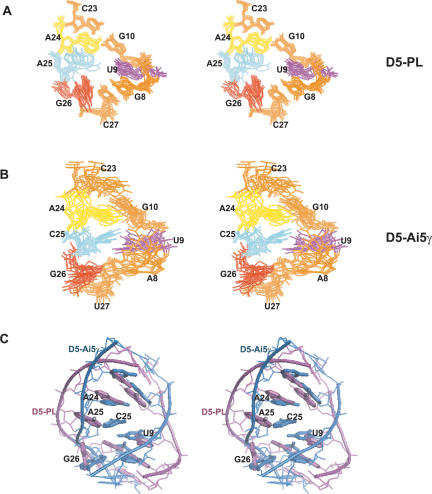
The conformation of the critical bulge region in D5-PL differs from previously determined X-ray and NMR structures of D5-Ai5γ. Similar to the NMR D5-Ai5γ structure, the bulge nucleotides in D5-PL also form a loose stacking interaction (Fig. 4 ▶). U9 stacks onto the preceding G8 and the succeeding G10. G8 base-pairs to C27 at the top of the lower helix, and G10 base-pairs to C23 at bottom of the upper helix. These two base pairs form the floor and roof of the bulge (Fig. 4A ▶). Unlike the arrangement of the analogous C25 in the NMR D5-Ai5γ structure, A25 adopts at least two conformations, one of which forms a continuous stacking interaction with A24; the other conformers extrude slightly into the minor groove of the bulge and pack at ~90° against A24, away from the Hoogsteen face of G8 (Fig. 4A ▶). This latter conformation is intermediate to the complete extrusion of C25 into the minor groove seen in the X-ray structure of D5-Ai5γ. These conformations come out of the structure calculation regardless of whether RDCs were included or omitted for this residue. G26 does not pack against the AGC triad as observed in the NMR D5-Ai5γ structure. Instead, it packs into the major groove against G8 to potentially shield the G8–C27 base pair from solvent exposure and allow the direct observation of the imino proton resonance of G8. The contortion of the lower and upper helices and the severe kink observed in the D5-Ai5γ NMR structure is not seen in D5-PL (Fig. 4C ▶). For instance, in D5-PL, the backbone phosphate–phosphate distances from U9 to A24, A25, and G26 are 22.1–23.4 Å, 21.3–22.7 Å, and 19.4–21.4 Å, respectively. In D5-Ai5γ NMR structures, the corresponding distances are 13.8–16.2 Å, 12.3–14.6 Å, and 13.5–16.8 Å, respectively. The width of the bulge in the D5-Ai5γ NMR structure is comparatively shorter than in D5-PL (Fig. 4C ▶). Interestingly, in the D5-Ai5γ X-ray structure, the equivalent distances for U9 to A24, C25, and G26 are 21.5, 21.96, and 17 Å, respectively. The U9 to A24 and C25 distances are much closer to what is observed in D5-PL. Not surprisingly, because of the U9○G26 wobble pair observed in the X-ray structure, the U9 to G26 distance is much shorter than that in D5-PL. It is harder to compare the U6 ISL bulge width because there is only one nucleotide within the bulge region. Nonetheless the backbone phosphate–phosphate distance from U80 (the equivalent of A24 in D5) to C66 and C67, the two flanking bases on the opposite strand, is 18.5 to 18.8 Å, respectively. One consequence of this bulge conformation is that the backbones of the two strands in D5-PL are held farther apart, compared to that of the D5-Ai5γ NMR structure (Fig. 4C ▶), to effectively create a hole in the center of the bulge region surrounded by the five nucleotides U9, G10, A24, A25, and C27. A similar hole is observed in the X-ray structure of D5-Ai5γ but not in the NMR structures of D5-Ai5γ and U6. Another striking consequence of this bulge arrangement within D5-PL is that the hydrogen-bonding groups of G26 point away from putative catalytic groups of the AGC triad, which likely rules out any joint participation of G26 and the AGC triad in catalysis. Indeed, as we show below, G3 is especially unresponsive to Mg2+ ions, which calls into question its presumed catalytic role.
FIGURE 4.
Comparison of D5-PL and D5-Ai5γ NMR structures. (A) Stereoview of the superposition over all heavy atoms of the 10 lowest energy structures of the bulge nucleotides (U9, A24, [excluding A25], G26) and surrounding base pairs (G8–C27 and G10–C23) within D5-PL. (B) Stereoview of the superposition over all heavy atoms of the 10 lowest energy structures of the bulge nucleotides (U9, A24, [excluding C25], and G26) and surrounding base pairs (A8–U27 and G10–C23) within D5-Ai5γ. (C) Stereoview of the superposition of representative (structure closest to the mean structure) D5-PL and D5-Ai5γ bulge structures (nucleotides 8–10, 23, 24, 26, 27).
Surface electrostatic potential distribution in D5 and U6
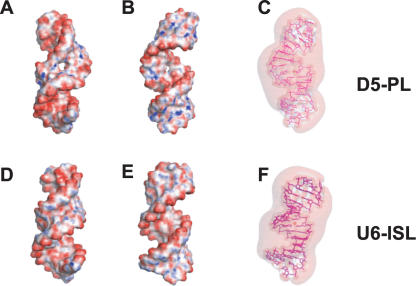
Electrostatic properties play an important role in RNA molecular interactions because of the high negatively charged backbone of nucleic acids (Honig and Nicholls 1995; Draper 2004). To visualize the charge distribution around D5, we calculated the surface electrostatic potential map using the nonlinear mode of the Adaptive Poisson Boltzmann Solver (APBS) (Holst et al. 2000; Baker et al. 2001) and visualized the results with Pymol (http://pymol.sourceforge.net/). Mapping the electrostatic potential to the molecular surface of D5-PL reveals a striking overall pattern of negative electrostatic potential of the D5 structure with smaller regions of positive potential. As expected for RNA structures (Honig and Nicholls 1995; Chin et al. 1999), regions of very high negative electrostatic potential cluster mostly in the major grooves of the tetraloop, AGC triad, and internal bulge regions (Fig. 5A,B ▶). The major groove of the triad region is substantially more electronegative than the minor groove (Fig. 5A,B ▶). Similarly, the tetraloop region is electronegative in both the major and minor grooves, but the minor groove is more electropositive than the major groove. Interestingly, the internal bulge has sites of intense negative potential in both grooves, especially around the cavity created by the bulge geometry in D5-PL (Fig. 5A,B ▶). The three-dimensional (3D) electrostatic potential contours at −1.5kT/e reveal two regions of isopotential cavities in the minor grooves of the tetraloop and AGC triad regions (Fig. 5C ▶). Both regions are likely sites of RNA–RNA interaction.
FIGURE 5.
Comparison of calculated molecular electrostatic surface potential and 3D isopotential contours of D5-PL and U6-ISL. Negative surfaces are in red (−25kT), neutral surfaces in white (0kT), and positive surfaces in blue (10kT) for D5 (A,B) and U6 (D,E). The 3D isopotential contours are displayed for D5 (C) and U6 (F) at −1.5kT/e. The major grooves of the tetraloop and the AGC triad regions and the minor groove of the internal bulge are shown for D5 (A) and U6 (D). The major groove of the bulge region and the minor grooves of the tetraloop and the AGC triad regions are shown for D5 (B) and U6 (E). The 3D isopotential contours are displayed from the major groove of the bulge region and the minor grooves of the tetraloop and the AGC triad regions for D5 (C) and U6 (F) at −1.5kT/e. Cavities appear over the minor groove faces of the tetraloop and bulge, and are indicative of likely sites of contact with other RNA elements.
Given the structural similarities between U6 ISL and D5, we tested whether they also share conserved electrostatic features. We calculated the electrostatic potential map and isopotential contours for D5-PL and U6 extended ISL. For both D5 (Fig. 5A–C ▶) and U6 (Fig. 5D–F ▶) structures, areas of large negative potential accumulate in the major grooves of the tetraloop, the internal bulge, and triad regions. The minor groove is not as negatively charged (Fig. 5A,B,D,E ▶). In D5-PL, a cavity exists in the internal bulge region (Fig. 5A,B ▶) that is absent in U6. These results suggest the bulge, tetraloop, and AGC regions are likely specific magnesium ion-binding sites in both U6 and D5 RNA substructures. The NMR titration experiments discussed next were used to test this hypothesis for D5-PL.
D5-PL has three Mg2+ ion-binding sites
Given that D5 is a potential metal-binding platform for the group II intron ribozyme (Steitz and Steitz 1993), we monitored directly the chemical shift changes of proton, carbon, and nitrogen sites for every nucleotide in D5 in the presence of increasing amounts of magnesium ions using resolution-optimized NMR spectroscopy experiments (Dayie 2005). Under identical experimental conditions, any observed differences in chemical shifts between unbound and magnesium-bound D5-PL would indicate a change in the local chemical environment of each nucleotide because of direct or indirect D5–metal ion interaction. Unlike a previous study limited to proton probes only (Sigel et al. 2004), this NMR approach provides exquisite detail about site-specific changes in the chemical environment of most atoms (proton, carbon, and nitrogen sites) in D5 (Figs. 6 ▶, 7 ▶; Supplemental Figs. 2–4), especially in the tetraloop (G15, A16, A17, A18, G19), the bulge (U9, A24, A25, G26) and the conserved (A2, G3, C4) triad regions.
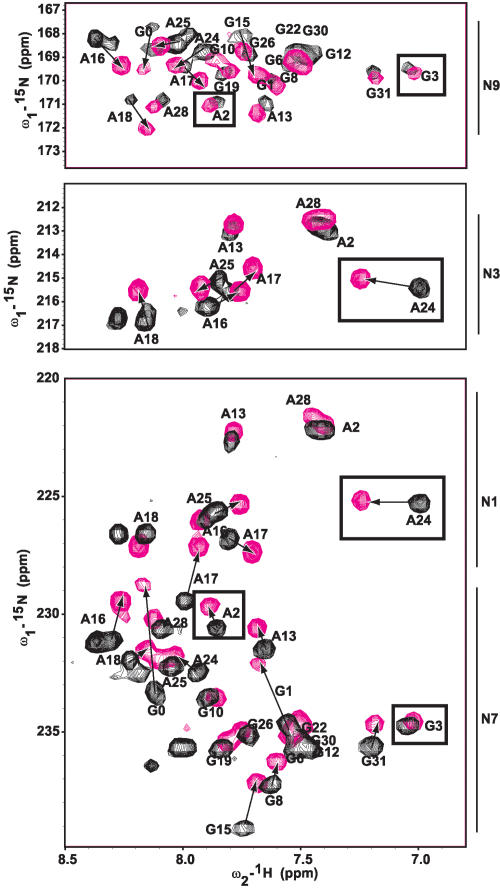
FIGURE 6.
NMR spectra showing the chemical shift perturbations of D5 nucleotides on binding MgCl2 in the N1, N3, N7, and N9 aromatic region. Arrows depict residues with the most chemical shift perturbation with increasing magnesium ion concentration (0 and 10 mM), indicative of a likely ion-binding site. Residues in the triad region (A2 and G3) and the bulge (A24) are boxed. Note how A2’s N7, but not N9 nor N3, is selectively perturbed and A24’s N3, but not N1, is selectively perturbed. G3 is not perturbed at the N7 or N9 positions.
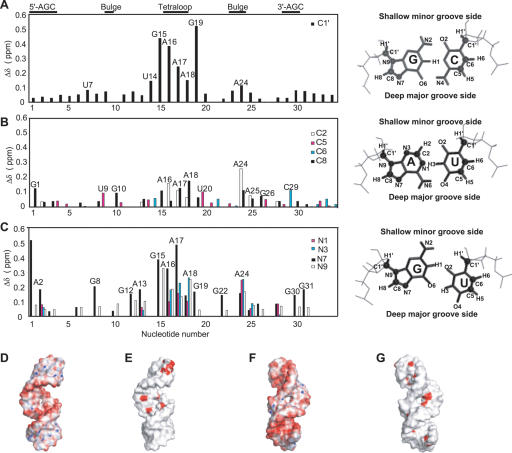
FIGURE 7.
Chemical shift perturbations from MgCl2 titration mapped onto the D5-PL solution NMR structure. Changes in chemical shift between free D5-PL and 10 mM bound D5-PL as a function of nucleotide sequence in the (A) ribose C1′, (B) aromatic carbon, and (C) aromatic nitrogen regions of D5-PL. Shown to the right of each panel are the configurations of the standard Watson-Crick base pairs for G–C and A–U as well as the wobble GU pair. The base and the ribose atoms probed in this study are highlighted as spheres, and the edges facing the deep major grooves and shallow minor grooves of RNA helices are indicated. Projection of the Mg2+ ion binding onto the D5 solution NMR structure indicates that metal ion binding occurs in both the major and minor grooves of the tetraloop and bulge regions (E,G), but only in the major groove of the triad (E,G). Shown also for comparison are the electrostatic calculations (D,F) taken from Figure5, A and D ▶.
Analyses of the ribose carbon (C1′) (Fig. 7A ▶; Supplemental Fig. 2), aromatic carbon (C2, C5, C6, C8) (Fig. 7B ▶; Supplemental Figs. 3, 4), and aromatic nitrogen (N1, N3, N7, N9) (Figs. 6 ▶, 7C ▶), groups reveal very strong magnesium ion binding primarily in the tetraloop and internal bulge regions for D5-PL (Figs. 6 ▶, 7 ▶). Mg2+ affects the tetraloop region more strongly than any other region, in contrast to what is observed for D5-Ai5γ, where the bulge region is the most affected (Sigel et al. 2004). For the ribose H1′–C1′ resonances, the tetraloop G15, A16, A17, A18, and G19 nucleotides undergo the greatest chemical shift changes (>0.1 ppm), with G19 having the largest shift of 0.52 ppm (Fig. 7A ▶). Other than G15, none of these resonances were significantly perturbed in the D5-Ai5γ NMR structure (Sigel et al. 2004). In the bulge region, A24 undergoes the largest chemical shift change (0.11 ppm), while U9, A25, and G26 are barely affected by Mg2+ (Fig. 7A ▶). This contrasts with the results for D5-Ai5γ, where C25 is the most perturbed bulge nucleotide in the H1′ region. For the aromatic carbons, a similar trend is observed (Fig. 7B ▶). The functional groups within the shallow minor groove (H2, C2, N1, N3) of the bulge A24 nucleotide undergo significant chemical shift change (>0.2 ppm), whereas those for U9, A25, and G26 are barely affected by Mg2+ (Fig. 7A,B ▶). However, the functional groups within the deep major groove (H5, C5, H8, C8, N7) of the surrounding bulge nucleotides are also perturbed (>0.1 ppm) (Fig. 7B,C ▶). Similar to D5-Ai5γ, H8–C8 of G10 is perturbed. Unlike D5-Ai5γ, N7 of G8 (H2 for A8 in D5-Ai5γ) and H5–C5 of U9 are also perturbed by Mg2+; yet, none of the functional groups probed for A25 in D5-PL are responsive to Mg2+ (Fig. 7A–C ▶). Some of these differences may be due to being able to probe more sites, with higher-resolution NMR experiments than has hitherto been done, and some could well be intrinsic differences between the two D5s.
Equally significant, none of the G3 functional groups probed (ribose C1′, base C8, aromatic N7 and N9) is responsive to Mg2+. Lack of responsiveness of G3’s N7 to Mg2+ (Figs. 6 ▶, 7C ▶) is particularly intriguing given a previous expectation that catalysis likely proceeds with N7 and O6 groups on G3 (Konforti et al. 1998). Similar to D5-Ai5γ, we observe only small to no changes for the aromatic and ribose carbons of the AGC catalytic triad region (Fig. 7A,B ▶); but unlike D5-Ai5γ, the deep major-groove nitrogen N7 positions of A2 and G31 are selectively responsive to Mg2+ ions (Figs. 6 ▶, 7C ▶). This is perhaps the single most significant difference between D5-Ai5γ and D5-PL. In the case of D5-Ai5γ, divalent ion binding was detected only with Mn2+ and only from the minor-groove face of the catalytic triad (Sigel et al. 2004). X-Ray analysis also failed to uncover any divalent ion binding within the AGC triad region (Zhang and Doudna 2002). It is likely that probing the N7 position in D5-Ai5γ by NMR could also uncover this Mg2+-binding site.
Projection of the NMR titration data onto the 3D D5-PL structure reveals an overlap with the regions predicted by the electrostatic calculations (Figs. 7D–G ▶). Consistent with the electrostatic calculations, all three critical regions important for binding and catalytic function are selectively sensitive to Mg2+ ions, indicative of likely metal ion-binding sites.
DISCUSSION
Because the highly conserved and catalytically important group II intron D5 element is absolutely critical for ribozyme function and shares similar structural features with the U6/U6atac ISL found in the spliceosomal snRNAs, much effort has gone into elucidating the X-ray and NMR solution structures of D5-Ai5γ and the NMR solution structure of the U6 ISL (Zhang and Doudna 2002; Sashital et al. 2004; Sigel et al. 2004). The analyses of this structure of D5-PL from a different group II intron ribozyme and identified Mg2+ ion-binding sites reveal striking similarities and differences with the previous structures. Our results lend further support, with some qualifications, to the resemblance of D5 to U6 within the catalytic core of each splicing machine.
The 3D structure of D5-PL has the same global architecture as the D5-Ai5γ and U6 ISL but with important local structural differences (Figs. 2 ▶–4 ▶ ▶). Both D5-Ai5γ and D5-PL structures adopt lower and upper helical stems of 8 and 5 bp, respectively, separated by an asymmetric internal bulge (Fig. 3A,B,D ▶). This contrasts with the U6 ISL that adopts a similar-sized lower helix but an upper helical stem of 4 nt (Fig. 3C ▶). The structural data, therefore, explain the observation that for D5 to function efficiently in a U12-dependent spliceosome in vivo, a single base pair from the upper helical stem of the D5 structure had to be removed (Shukla and Padgett 2002).
Of great interest is the unusual configuration of bases in the internal bulge region, a region shown to be critical for metal ion binding and containing several catalytically important residues. D5 from Ai5γ and PL differ only at positions 8, 25, and 27 such that the 8–27 base pair is G–C or A–U in PL and Ai5γ, respectively, and the unpaired bulge nucleotide 25 is A or C in PL and Ai5γ, respectively. In a recent study, Shukla and Padgett evaluated a database of eukaryotic group II D5 elements, and they noted that when the 8–27 base pair is a G–C as in D5-PL, the 24 and 25 nucleotides are AA or AU 36% of the time each. Similarly, when the 8–27 base pair is A–U as in D5-Ai5γ, the 24 and 25 nucleotides are AC or AU 41% and 31% of the time, respectively. Only 1% of D5s have an Ai5γ-like A–U base pair and a PL-like AA bulge, and only 3% of D5s have a PL-like G–C base pair and an Ai5γ-like AC bulge. Intermediate sequences between D5-PL and D5-Ai5γ, therefore, appear highly disfavored and are inactive for splicing in vivo (Shukla and Padgett 2002).
The solution NMR structures provide a possible explanation for these observations. Both D5 NMR structures, but not the X-ray structure, indicate that the three unpaired bulge nucleotides (U9, A24, A25/C25) are stacked within the helix (see Fig. 4 ▶). The unpaired G26 adopts a syn conformation that enables it to pack against the major groove of the lower helix. The short backbone distances within the bulge of the D5-Ai5γ NMR structure allow U9 to come within hydrogen-bonding distance of C25, whereas the long backbone distances within the bulge region of the D5-PL structure preclude a U9–A25 hydrogen bond. In the D5-Ai5γ NMR structure, U9, C25, and U27 form tight packing interactions that likely create a restricted environment that may be obstructive for a purine ring at position 25. This contrasts with the D5-PL NMR structure. Herein, those three nucleotides are not as tightly packed, so as to contribute to the wider bulge width that can easily accommodate a purine at position 25. It is also instructive that A25 still does not have a well-defined conformation in the RDC-refined ensemble, ruling out a single rigid conformation in solution. A GC Watson-Crick base pair at positions 8 and 27 may, therefore, allow the unpaired residue 25 to be either a purine or a pyrimidine. An AU pair at positions 8 and 27 could likely restrict the unpaired residue 25 to be a pyrimidine. Given that D5 interacts with other intronic elements within the full intron, triple interactions are also likely. Nonetheless, a structural coupling probably exists between position 25 and the 8–27 base pair to modulate a conformation of the bulge important for catalysis in the group II intron.
In addition to coupling between structure and function, electrostatics is expected to modulate RNA catalytic function (Honig and Nicholls 1995; Draper 2004). Our electrostatic calculations and detailed NMR magnesium ion perturbation analyses reveal some similarities but striking and significant differences between D5-PL and D5-Ai5γ. Consistent with previous Tb3+ and Mg2+ metal ion cleavage experiments (Sigel et al. 2000) and X-ray analysis of D5-Ai5γ (Zhang and Doudna 2002), our current results implicate A24 in the bulge and G19 in the tetraloop nucleotides, but not the triad G3 nucleotide, in Mg2+ ion binding. This metal binding of the bulge A24 nucleotide within D5 parallels that of the U80 within the yeast spliceosomal U6 ISL implicated in binding and catalysis (Chanfreau and Jacquier 1994; Yean et al. 2000; Huppler et al. 2002). Our results, in agreement with all these previous studies, suggest that the bulge is likely one of the key metal-binding platforms in these ribozymes and the spliceosome. Surprisingly A25 (C25 in Ai5γ) is only slightly responsive to Mg2+ ions, in contrast to what is observed in D5-Ai5γ. Paradoxically, the triad G3, the single most important residue implicated in function, is also surprisingly unresponsive to magnesium ions in all the sites probed by NMR: ribose C1′, base C8, aromatic N7 and N9. This suggests that G3 N7 is unlikely to coordinate any metal ions as previously suggested (Konforti et al. 1998), at least in the ground state. However, unlike previous studies, we provide direct evidence for specific magnesium ion binding to the N7 position of A2 within the major groove face of the triad region (Figs. 6 ▶, 7C ▶). These results are consistent with a functional role for the major groove of the AGC triad in both the group II intron D5 and the spliceosomal U6 (Boulanger et al. 1995; Peebles et al. 1995; Gordon and Piccirilli 2001; Hilliker and Staley 2004). For instance, Gordon and Piccirilli (2001) showed that the Sp phosphorothioate diastereomer at position 2 and both phosphorothioate diastereomers at position 3 significantly reduced the efficiency of splice site hydrolysis. However, thiophilic metal ions could only rescue inhibition caused by the Sp diastereomer at position 2 but not those at position 3 (Gordon and Piccirilli 2001). These data and our current results suggest that both the pro-Sp oxygen and N7 at position 2 likely coordinate essential metal ions from the major groove of the AGC triad in D5, not from the minor groove as recently proposed (Sigel et al. 2004). This site was probably missed in the initial D5-Ai5γ studies because the N7 position was not probed. The similarity of the distribution of large negative electrostatic potentials in the AGC triad region of both D5 and U6 also suggests the N7 of A2 in U6 may be responsive to magnesium ions, and that both D5 and U6 are likely metal-binding substructures within their respective splicing machines. The hypothesis that the N7 of A2 in U6 (and D5-Ai5γ) is responsive to divalent ions is readily testable with the experiments described here, and could further strengthen the linkage between the group II intron ribozymes and the spliceosome.
In summary, the results presented here suggest that three critical structural motifs, namely, the tetraloop, the internal bulge, and the AGC triad within D5, are strategically placed to bind essential metal ions and to position functional groups essential for catalysis that have strong parallels in the spliceosome. Our results lend further support to similarities between D5 and U6 snRNA in their overall conformations, electrostatic environment, and the magnesium ion-binding sites of the tetraloop, bulge, and triad regions. These conserved features of the splicing machinery of both the group II introns and the spliceosome are likely key for their catalytic function. We hypothesize that strong negative electrostatic potential at the tetraloop, internal bulge, and triad regions are likely critical determinants for the catalytic function of this group of ribozymes and the corresponding U6/U6atac structures in the spliceosome.
MATERIALS AND METHODS
Sample preparation
The RNA samples were synthesized by in vitro transcription with T7 RNA polymerase and synthetic DNA templates (Operon Technologies, Inc.). Isotopically labeled NTPs (ISOTEC) were used to prepare the RNA using established protocols (Milligan and Uhlenbeck 1989; Batey et al. 1995) as described previously (Dayie 2005). After dissolving in NMR buffer (100 mM KCl, 10 mM potassium phosphate at pH 6.5, 8% D2O [or 100%], without or with magnesium [1–10 mM for the titration], and trace of sodium azide), the sample is heated to 363 K for 2 min and cooled rapidly on ice for 10 min. For residual dipolar coupling measurements, the Pf1 filamentous phage (ASLA Ltd.) was exchanged into the final NMR buffer by repeated 1-h ultracentrifugation at 95,000 rpm and dissolution of the bacteriophage pellet into the NMR buffer. 13C, 15N-labeled D5 RNA samples containing phage were prepared by dissolving the phage pellets into the NMR sample to a final phage concentration of ~22 mg/mL as checked by the quadrupolar splittings of the 2H NMR signal.
NMR spectroscopy
NMR experiments were run on four-channel UNITYINOVA 500 and 600 MHz spectrometers equipped with actively shielded z-axis gradient triple resonance probes. Details of the suite of NMR experiments used for both assignment and structure calculations are presented elsewhere (Dayie 2005). Hydrogen bonds were identified from a 2D 2JNN-COSY experiment (Dingley and Grzesiek 1998). One-bond residual dipolar couplings were measured from 2D 1H–13C TROSY (Meissner and Sorensen 1999) spectra on a fully labeled 13C, 15N D5 RNA sample, recorded with and without Pf1 phage. The experiments were measured using both constant time and nonconstant modes in the carbon dimension with separate experiments acquired for aliphatic and aromatic 1H–13C groups. One-bond 1H–13C couplings were measured for H1′–C1′, H2–C2, H5–C5, H6–C6, and H8–C8 sites within the RNA.
Magnesium ion titration
The D5-PL magnesium ion-binding surface was mapped by a chemical shift perturbation method that used resolution optimized 2D 1H–13C NMR (Dayie 2005) and two-bond 1H–15N HSQC experiments. Increasing amounts of MgCl2 solution (final total concentrations of 1, 4, and 10 mM) were titrated into ~0.28 mM 13C/15N-labeled D5-PL sample. Each complex was heated to 363 K for 1 min and rapidly cooled on ice for 10 min before taking the NMR spectra. Binding was monitored through changes in the peak positions of each nucleotide. Experiments were conducted in 10 mM potassium phosphate buffer, 100 mM KCl (pH 6.5) at 45°C. For each experiment, a sweep width of 16,000 Hz, a 1.3-sec recycle delay, and 256 scans were used. The combined ribose and base chemical shift perturbations were calculated as Δδ = [(ΔδH)2 + (ΔδX ·χ)2]1/2, where ΔδH and ΔδX are the chemical shift changes (for H = 1H and X = 13C or 15N, respectively) observed in D5 on adding Mg2+ ions, and the scaling factor χ reflects the relative chemical shift ranges for both protons, carbons, and nitrogens in each region (χ = 0.24, 0.18, and 0.20, respectively for the ribose carbon, aromatic carbon, and nitrogen regions) (Farmer et al. 1996).
Structure calculation
All the NMR experiments described above were processed by NMRPipe (Delaglio et al. 1995) and analyzed by Sparky (T.D. Goddard and D.G. Kneller, SPARKY 3, University of California, San Francisco; http://www.cgl.ucsf.edu/home/sparky/). The structures were calculated based on a dynamical simulated annealing method implemented in the CNS program (Brunger et al. 1998). Planarity and hydrogen bonding for all base pairs indicated by exchangeable 2D-NOESY, 3D-15N-NOESY and trans-hydrogen bond experiments were enforced during the calculation. Weak planarity constraints (25 kcal/mol Å −2) were used for the bases involved in base-pairing as revealed by the trans-hydrogen bond experiments (Fig. 1B ▶). Consistent with these data, the backbone torsion angles for residues in the helical region (1–8, 10–14, 19–23, and 27–34) were constrained to A-form values (α = −62, β = 180, γ = 47, ɛ = −152, ζ = −73) with allowed deviations of ±15° (Sanger 1984). The bulge and tetraloop were left unrestrained. Table 1 ▶ is the summary of restraints used for the structure calculation. Nucleotides that showed strong H1′–H2′ and H1′–H3′ cross-peaks in a 50-msec 1H–1H TOCSY (A16, A24, A25) were restrained to a C2′-endo or S-type (145° ± 30°), and nucleotides that did not show H1′–H2′ peaks were restrained to C3′-endo or N-type (83° ± 15°). Analysis of H1′-aromatic NOEs from 50 msec and 250 msec 1H–1H 2D NOESY experiments indicated all nucleotides except G26 fell into the anti glycosidic torsion range. G26 was constrained to a χ angle of 70° ± 30°, and all the other nucleotides were constrained to χ angles of −160° ± 15°. Default values for NMR structure determination of nucleic acid using CNS were implemented with only minor changes to incorporate an RDC refinement protocol (Clore et al. 1998; Bax et al. 2001; Warren and Moore 2001).
Using CNS, a structure file and an initial extended structure were generated. From this extended structure, 100 structures were calculated with random initial velocities. An initial 40-psec (1-fsec time steps), high-temperature (20,000 K), torsion-angle-restrained molecular dynamics was followed by a 100-psec (2-fsec time steps) slow-cooling step (20,000 K to 1000 K) for the initial structures. At the final stage of the calculations, 35 psec (3.5-fsec time steps) of slow-cooling (1000 K to 300 K) restrained molecular dynamics was performed in Cartesian coordinate space. Finally the structures were subjected to 2000 steps of conjugate gradient minimization.
After this initial calculation, structures were evaluated for convergence, and a structure was accepted if it contained no violations of NOE restraints >0.2 Å and no dihedral angle violations >5°. Structures were also rejected if the RMSD of bonds from ideal values was >0.02 Å, or the RMSD of angles and impropers was >5°. Thirty structures that clustered in the lowest energy range were accepted and used to calculate the average structure. Structures were visualized using the software packages Insight II (MSI), Pymol (http://pymol.sourceforge.net/), and Nuccyl (http://www.biosci.ki.se/groups/ljo/software/nuccyl.html). These 30 structures were then used for the subsequent refinement by the measured residual dipolar coupling constants. Initial values of the magnitude of the axial (Da) and rhombic (R) components of the alignment tensor were estimated using both the histogram approach (Clore et al. 1998; Bax et al. 2001) and the best fit of the measured RDCs to the initial low-energy structures without violations using PALES (Zweckstetter et al. 2004). Values from these two methods were used to perform a 2D grid search. The minimum obtained was then used to refine the structures iteratively. The optimal values from the systematic grid search were Da = −34 and R = 0.2. Initially all 52 RDC values were used for the refinement. Because these did not improve the bulge and tetraloop structural regions, we performed a systematic refinement procedure. Using our unpublished observations that the tetraloop and bulge region of D5-PL were dynamic on the picoseconds to nanoseconds time scales (K.T. Dayie and N.V. Eldho, unpubl.), first we omitted all the RDCs belonging to the tetraloop nucleotides. Second, we omitted all the RDC values of the bulge nucleotides. Third, we replaced all or some of the RDC values of the tetraloop nucleotides and combined these with different permutations of all or part of the RDC values of A24, A25, and G26. For each of these bulge and/or tetraloop combinations, the structures were refined by cooling from 1000 K to 300 K in 50 cycles of restrained Cartesian coordinate space for a total of 40 psec. The force constant for the RDCs was increased from 0.0001 kcal mol−1 Hz−2 to 0.2 kcal mol−1 Hz−2. This was followed by a final round of 10 cycles of 500 steps of a Powell energy minimization. We used the Module program to visualize the results (Dosset et al. 2001).
Electrostatic calculations
The electrostatic potentials in and around D5-PL, D5-Ai5γ, and U6 extended ISL were calculated using the Adaptive Poisson-Boltzmann (APBS) software package, which incorporates the Manifold Code libraries to efficiently solve the Non-Linear Poisson-Boltzmann (NLPB) equation (Holst et al. 2000; Baker et al. 2001). The coordinates of U6 and D5-Ai5γ are taken from the published work (Zhang and Doudna 2002; Sashital et al. 2004; Sigel et al. 2004) deposited in the PDB (D5-Ai5γ NMR, 1R2P; D5-Ai5γ X-ray, 1KXK; U6, 1XHP). Hydrogens were added to the X-ray structure using Insight II (Version 2000; Molecular Simulations Inc., 2000). For all the structures, charges and atomic radii were assigned using the AMBER force field parameters (Cornell et al. 1995). The electrostatic potential was obtained by solution of the NLPB equation at 100 mM ionic strength with a solute dielectric of 2 and a solvent dielectric of 78.5. Calculations were repeated for other ionic strengths to match experimental conditions for each structure, where known. Because these had no effect on the overall electrostatic potential maps obtained, the results are presented for 100 mM.
[Note: Coordinates for the D5-PL structures calculated with RDCs have been deposited in the Protein Data Bank (http://www.rcsb.org), accession code 2F88.
SUPPLEMENTAL DATA
Supplemental materials are available from the authors (please send an e-mail message containing the keyword D5 to dayiek@ccf.org).
Acknowledgments
We thank LaPortia McElrath, Orlando Gumbs, Jun Qin, and Robert Silverman for many useful discussions and critical reading of the manuscript. NMR studies were carried out at the Cleveland Center for Structural Biology with support from the Cleveland Foundation, Hayes Fund, and additional equipment funding from the NIH Shared Instrumentation Program of the National Center for Research Resources. This investigation was supported by the Cleveland Clinic Foundation and US National Science Foundation (NSF) grant MCB-0316783 (K.T.D.) and a structural supplement to NIH grant GM55105 (R.A.P.).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2237806.
REFERENCES
- Abramovitz, D.L., Friedman, R.A., and Pyle, A.M. 1996. Catalytic role of 2 prime-hydroxyl groups within a group II intron active site. Science 271: 1410–1413. [DOI] [PubMed] [Google Scholar]
- Baker, N.A., Sept, D., Joseph, S., Holst, M.J., and McCammon, J.A. 2001. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 98: 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey, R.T., Battiste, J.L., and Williamson, J.R. 1995. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 261: 300–322. [DOI] [PubMed] [Google Scholar]
- Bax, A., Kontaxis, G., and Tjandra, N. 2001. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 339: 127–174. [DOI] [PubMed] [Google Scholar]
- Boudvillain, M. and Pyle, A.M. 1998. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: A NAIM analysis. EMBO J. 17: 7091–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudvillain, M., de Lencastre, A., and Pyle, A.M. 2000. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature 406: 315–318. [DOI] [PubMed] [Google Scholar]
- Boulanger, S.C., Belcher, S.M., Schmidt, U., Dib-Hajj, S.D., Schmidt, T., and Perlman, P.S. 1995. Studies of point mutants define three essential paired nucleotides in the domain 5 substructure of a group II intron. Mol. Cell. Biol. 15: 4479–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N.S., et al. 1998. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Cryst. D54: 905–921. [DOI] [PubMed] [Google Scholar]
- Cech, T.R. 1986. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell 44: 207–210. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G. and Jacquier, A. 1994. Catalytic site components common to both splicing steps of a group II intron. Science 266: 1383–1387. [DOI] [PubMed] [Google Scholar]
- Chin, K., Sharp, K.A., Honig, B., and Pyle, A.M. 1999. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 6: 1055–1061. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Gronenborn, A.M., and Bax, A. 1998. A robust method for determining the magnitude of the fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. J. Magn. Reson. 133: 216–221. [DOI] [PubMed] [Google Scholar]
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., and Kollman, P.A. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117: 5179–5197. [Google Scholar]
- Costa, M. and Michel, F. 1999. Tight binding of the 5′ exon to domain I of a group II self-splicing intron requires completion of the intron active site. EMBO J. 18: 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M., Fontaine, J.M., Loiseaux-de Goer, S., and Michel, F. 1997. A group II self-splicing intron from the brown alga Pylaiella littoralis is active at unusually low magnesium concentrations and forms populations of molecules with a uniform conformation. J. Mol. Biol. 274: 353–364. [DOI] [PubMed] [Google Scholar]
- Costa, M., Christian, E.L., and Michel, F. 1998. Differential chemical probing of a group II self-splicing intron identifies bases involved in tertiary interactions and supports an alternative secondary structure model of domain V. RNA 4: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayie, K.T. 2005. Resolution enhanced homonuclear carbon decoupled triple resonance experiments for unambiguous RNA structural characterization. J. Biomol. NMR 32: 129–139. [DOI] [PubMed] [Google Scholar]
- de Lencastre, A., Hamill, S., and Pyle, A.M. 2005. A single active-site region for a group II intron. Nat. Struct. Mol. Biol. 12: 626–627. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G., Zhu, G., Pfeiffer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Dingley, A.J. and Grzesiek, S. 1998. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 120: 8293–8297. [Google Scholar]
- Dosset, P., Hus, J.C., Marion, D., and Blackledge, M. 2001. A novel interactive tool for rigid-body modeling of multi-domain macromolecules using residual dipolar couplings. J. Biomol. NMR 20: 223–231. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. 2004. A guide to ions and RNA structure. RNA 10: 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P. and Abelson, J. 1992. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res. 20: 3659–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer II, B.T., Constantine, K.L., Goldfarb, V., Friedrichs, M.S., Wittekind, M., Yanchunas Jr., J., Robertson, J.G., and Mueller, L. 1996. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat. Struct. Biol. 3: 995–997. [DOI] [PubMed] [Google Scholar]
- Fontaine, J.M., Rousvoal, S., Leblanc, C., Kloareg, B., and Loiseaux-de Goer, S. 1995. The mitochondrial LSU rDNA of the brown alga Pylaiella littoralis reveals α-proteobacterial features and is split by four group IIB introns with an atypical phylogeny. J. Mol. Biol. 251: 378–389. [DOI] [PubMed] [Google Scholar]
- Fontaine, J.M., Goux, D., Kloareg, B., and Loiseaux-de Goer, S. 1997. The reverse-transcriptase-like proteins encoded by group II introns in the mitochondrial genome of the brown alga Pylaiella littoralis belong to two different lineages which apparently coevolved with the group II ribosyme lineages. J. Mol. Evol. 44: 33–42. [DOI] [PubMed] [Google Scholar]
- Gordon, P.M. and Piccirilli, J.A. 2001. Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat. Struct. Biol. 8: 893–898. [DOI] [PubMed] [Google Scholar]
- Hilliker, A.K. and Staley, J.P. 2004. Multiple functions for the invariant AGC triad of U6 snRNA. RNA 10: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst, M., Baker, N.A., and Wang, F. 2000. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation I. Algorithms and examples. J. Comput. Chem. 21: 1319–1342. [Google Scholar]
- Honig, B. and Nicholls, A. 1995. Classical electrostatics in biology and chemistry. Science 268: 1144–1149. [DOI] [PubMed] [Google Scholar]
- Huppler, A., Nikstad, L.J., Allmann, A.M., Brow, D.A., and Butcher, S.E. 2002. Metal binding and base ionization in the U6 RNA intramolecular stem–loop structure. Nat. Struct. Biol. 9: 431–435. [DOI] [PubMed] [Google Scholar]
- Jacquier, A. 1990. Self-splicing group II and nuclear pre-mRNA introns: How similar are they? Trends Biochem. Sci. 15: 351–354. [DOI] [PubMed] [Google Scholar]
- Jarrell, K.A., Dietrich, R.C., and Perlman, P.S. 1988. Group II intron domain 5 facilitates a trans-splicing reaction. Mol. Cell. Biol. 8: 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker, F.M., Heus, H.A., Yip, P.F., Moors, E.H., and Pardi, A. 1996. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 264: 968–980. [DOI] [PubMed] [Google Scholar]
- Koch, J.L., Boulanger, S.C., Dib-Hajj, S.D., Hebbar, S.K., and Perlman, P.S. 1992. Group II introns deleted for multiple substructures retain self-splicing activity. Mol. Cell. Biol. 12: 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti, B.B., Abramovitz, D.L., Duarte, C.M., Karpeisky, A., Beigelman, L., and Pyle, A.M. 1998. Ribozyme catalysis from the major groove of group II intron domain 5. Mol. Cell 1: 433–441. [DOI] [PubMed] [Google Scholar]
- Meissner, A. and Sorensen, O.W. 1999. The role of coherence transfer efficiency in design of TROSY-type multidimensional NMR experiments. J. Magn. Reson. 139: 439–442. [DOI] [PubMed] [Google Scholar]
- Michel, F., Umesono, K., and Ozeki, H. 1989. Comparative and functional anatomy of group II catalytic introns—A review. Gene 82: 5–30. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F. and Uhlenbeck, O.C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Peebles, C.L., Zhang, M., Perlman, P.S., and Franzen, J.S. 1995. Catalytically critical nucleotide in domain 5 of a group II intron. Proc. Natl. Acad. Sci. 92: 4422–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar, M. and Perlman, P.S. 1999. Photocrosslinking of 4-thio uracil-containing RNAs supports a side-by-side arrangement of domains 5 and 6 of a group II intron. RNA 5: 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, P.Z. and Pyle, A.M. 1997. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry 36: 4718–4730. [DOI] [PubMed] [Google Scholar]
- Sanger, W. 1984. Principles of nucleic acid structure. Springer-Verlag, New York.
- Sashital, D.G., Cornilescu, G., McManus, C.J., Brow, D., and Butcher, S. 2004. U2–U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat. Struct. Mol. Biol. 11: 1237–1242. [DOI] [PubMed] [Google Scholar]
- Shukla, G.C. and Padgett, R.A. 2002. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol. Cell 9: 1145–1150. [DOI] [PubMed] [Google Scholar]
- Sigel, R.K., Vaidya, A., and Pyle, A.M. 2000. Metal ion binding sites in a group II intron core. Nat. Struct. Biol. 7: 1111–1116. [DOI] [PubMed] [Google Scholar]
- Sigel, R.K., Sashital, D.G., Abramovitz, D.L., Palmer, A.G., Butcher, S.E., and Pyle, A.M. 2004. Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif. Nat. Struct. Mol. Biol. 11: 187–192. [DOI] [PubMed] [Google Scholar]
- Steitz, T.A. and Steitz, J.A. 1993. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. 90: 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa, T., Pleiss, J.A., and Guthrie, C. 2002. Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell 109: 149–152. [DOI] [PubMed] [Google Scholar]
- Warren, J.J. and Moore, P.B. 2001. A maximum likelihood method for determining D(a)(PQ) and R for sets of dipolar coupling data. J. Magn. Reson. 149: 271–275. [DOI] [PubMed] [Google Scholar]
- Yean, S.L., Wuenschell, G., Termini, J., and Lin, R.J. 2000. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T., Maroney, P.A., Darzynkiwicz, E., and Nilsen, T.W. 1995. U6 snRNA function in nuclear pre-mRNA splicing: A phosphorothioate interference analysis of the U6 phosphate backbone. RNA 1: 46–54. [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. and Doudna, J.A. 2002. Structural insights into group II intron catalysis and branch-site selection. Science 295: 2084–2088. [DOI] [PubMed] [Google Scholar]
- Zweckstetter, M., Hummer, G., and Bax, A. 2004. A prediction of charge-induced molecular alignment of biomolecules dissolved in dilute liquid-crystalline phases. Biophys. J. 86: 3444–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]