Abstract
Rrp5p is a trans-acting factor important for biogenesis of both the 40S and 60S subunit of the Saccharomyces cerevisiae ribosome. The protein contains 12 tandemly repeated S1 RNA binding motifs in its N-terminal region, suggesting the ability to interact directly with the pre-rRNA. In vitro binding studies, using immunopurified Rrp5p and in vitro transcribed, 32P-UTP-labeled RNA fragments, revealed that Rrp5p is a general RNA-binding protein with a strong preference for single-stranded sequences rich in uridines. Co-immunoprecipitation studies in yeast cells expressing ProtA-tagged Rrp5p showed that the protein is still associated with pre-ribosomal particles containing 27SA2 pre-rRNA but not with particles containing the 27SB precursor. Thus, Rrp5p appears to dissociate from the 66S pre-ribosome upon or immediately after further processing of 27SA2 pre-rRNA, suggesting the presence of (an) important binding site(s) within the 3′-terminal portion of ITS1. The location of these possible binding site(s) was further delimited using rrp2-1 mutant cells, which accumulate the 5′-extended 5.8S pre-rRNA species. The results indicate that association of Rrp5p with the pre-ribosome is abolished upon removal of a 30-nt region downstream from site A2, which contains two short, single-stranded U stretches. Sequence comparison shows that only the most 5′ of these two U-rich stretches is conserved among yeast species whose ITS1 can functionally replace the S. cerevisiae spacer. The implications for the role of Rrp5p in yeast ribosome biogenesis are discussed.
Keywords: yeast, ribosomes, trans-acting factor, RNA-binding, S1 motif
INTRODUCTION
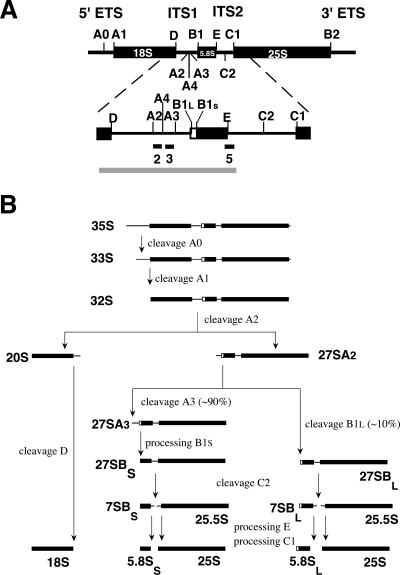
Eukaryotic 18S, 5.8S, and 25S–28S rRNA species are encoded by multiple, tandemly repeated transcription units, each of which is transcribed by RNA polymerase I into a single, large precursor rRNA molecule that contains an external transcribed spacer at either end (5′-ETS and 3′-ETS) as well as two internal transcribed spacers separating the 18S from the 5.8S (ITS1), and the 5.8S from the 25S–28S sequence (ITS2) (Fig. 1A ▶). This polycistronic pre-rRNA is first extensively modified by ribose methylation and pseudouridylation (for review, see Kiss 2001), followed by removal of the spacer regions via a complex series of endo- and exonucleolytic steps. During this processing the ribosomal proteins assemble with the intermediate pre-rRNA species in an ordered fashion.
FIGURE 1.
Pre-rRNA processing in Saccharomyces cerevisiae. (A) Structure of the 35S primary transcript. Thick bars represent the mature 18S, 5.8S, and 25S rRNA sequences; thin lines, the spacer regions. Cleavage sites are indicated as are the location of the probes used for Northern and primer extension analysis. The gray bar corresponds to the in vitro transcript used in the binding experiments. (B) The pre-rRNA processing pathway.
The whole process of ribosome maturation, which takes place largely in a specialized region of the nucleus called the nucleolus, is best characterized in the yeast Saccharomyces cerevisiae (for reviews, see Warner 2001; Fatica and Tollervey 2002; Fromont-Racine et al. 2003; Raué 2004), in which pre-rRNA processing starts with cotranscriptional removal of most of the 3′ ETS by the RNase III homolog Rnt1p (Abou-Elela et al. 1996; Kufel et al. 1999). The resulting 35S precursor, which is part of a large 80S/90S pre-ribosomal particle (Dragon et al. 2002; Grandi et al. 2002; Bernstein et al. 2004), then loses its 5′ ETS by successive cleavages at sites A0 and A1, to give rise to a 32S precursor (Fig. 1B ▶). Further cleavage of this precursor at site A2 separates the 90S particle into a 43S and 66S pre-ribosome, containing 20S and 27SA2 pre-rRNA, respectively. The 43S particle is transported to the cytoplasm where cleavage of the 20S precursor at site D produces the mature 18S rRNA (Stevens et al. 1991; Moy and Silver 1999; Vanrobays et al. 2003). The 66S pre-ribosome enters either of two pathways: In most cases the 27SA2 pre-rRNA is cleaved at site A3, followed by exonucleolytic digestion to site B1S, the 5′ end of the short form of 5.8S (5.8SS) rRNA. In roughly 10% of the 66S pre-ribosomes, however, 27SA2 pre-rRNA undergoes a single endonucleolytic cleavage at site B1L, located 6 nt upstream of B1S, which eventually leads to the minor 5.8SL rRNA species. An additional processing site, called A4, whose role is not yet fully understood, is located midway between A2 and A3 (Eppens et al. 2002; Faber et al. 2004). The resulting 27SBS and 27SBL pre-rRNA species are further matured identically by endonucleolytic cleavage at site C2 and subsequent exonucleolytic removal of the ITS2 sequences still present in the 7S and 25.5S precursors to give the mature 5.8S and 25S rRNAs. The whole ribosome maturation process critically depends on several ribonucleoprotein particles (snoRNPs) and a large number of nonribosomal proteins collectively known as trans-acting factors that constitute transient components of the different pre-ribosomal particles (Kressler et al. 1999; Venema and Tollervey 1999; Fromont-Racine et al. 2003; Raué 2004).
Almost all of these trans-acting factors are involved in the production of either the 40S or the 60S ribosomal subunit. A striking exception is Rrp5p, first identified in yeast as being essential for the production of both subunits (Venema and Tollervey 1996). Recently, another trans-acting factor with such a dual role, called Rrp12p (Oeffinger et al. 2004), was identified.
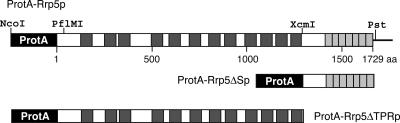
Rrp5p is a 1729 amino acid–long protein with a distinct two-domain structure (Torchet et al. 1998; Eppens et al. 1999; Fig. 2 ▶): The N-terminal two-thirds of the protein consists of 12 S1 RNA binding motifs, while the remaining C-terminal region contains seven tetratricopeptide repeats (TPR), generally thought to be involved in protein–protein interactions (Blatch and Lassle 1999). Mutational analysis has shown that several of the TPR motifs are required for cleavages at sites A0–A2, whereas deletions in the S1 domain result in inhibition of cleavage at either site A3 or site A2, depending on which particular motifs are deleted (Eppens et al. 1999; Vos et al. 2004b).
FIGURE 2.
Domain structure of Rrp5p. The N-terminally attached ProtA tag is shown in black. The 12 S1 RNA-binding motifs are hatched, and the seven tetratricopeptide repeats (TPR) are in gray. Numbering is based on the sequence of Rrp5p.
Large-scale immunopurification studies have shown Rrp5p to be already present in the early 80S/90S pre-ribosomal particle (Dragon et al. 2002; Grandi et al. 2002; Nissan et al. 2002; Schäfer et al. 2003). Although assembly of this particle requires the U3 snoRNP (Dragon et al. 2002), we have recently shown that genetic depletion of U3 snoRNA does not affect the association of Rrp5p with 35S pre-rRNA in vivo (Vos et al. 2004a), which, together with the presence of the multiple S1 motifs, suggests that this protein recognizes the precursor directly.
The experiments described in this article demonstrate that Rrp5p indeed is an RNA-binding protein and that this property is confined to the S1 domain. In vitro, the protein does not specifically recognize pre-rRNA but does display a distinct preference for single-stranded U-rich sequences. Analysis of pre-rRNA species that coprecipitate with epitope-tagged Rrp5p in vivo clearly suggested the presence of an Rrp5p binding site closely downstream from site A2 in ITS1.
RESULTS
Rrp5p is an RNA-binding protein
To determine whether Rrp5p is capable of binding pre-rRNA, we set up an in vitro RNA-binding assay based upon incubation of in vitro transcribed, radiolabeled (r)RNA with IgG-Sepharose beads to which ProtA-tagged protein had been attached. The beads were loaded with Rrp5p by mixing them with a whole-cell extract prepared from exponentially growing yeast cells that expressed ProtA-tagged Rrp5p from a plasmid-encoded gene (Eppens et al. 1999). In order to limit proteolysis, the low-protease strain HI227 (Abou-Elela et al. 1996) was used throughout these experiments. Extract from untransformed HI227 cells, which express only untagged Rrp5p from the genome, served as a control. Prior to incubation with the RNA, the beads were washed with high-salt buffer to remove contaminating proteins. SDS-PAGE analysis followed by silver staining of the protein retained on the beads indeed detected only Rrp5p and IgG (data not shown).
Rrp5p-loaded and control beads were incubated with an in vitro transcribed, α-32P-UTP-labeled pre-rRNA fragment spanning ITS1 and the adjacent mature 5.8S rRNA sequence (~500 nt) (Fig. 1A ▶). This region was chosen because the domain of Rrp5p containing the S1 motifs is known to be involved in cleavage at A2 and A3 within ITS1 (Eppens et al. 1999; Vos et al. 2004b), making this spacer a plausible candidate for the location of (a) site(s) recognized by the protein. The amount of bound RNA relative to the input was quantified by scintillation counting.
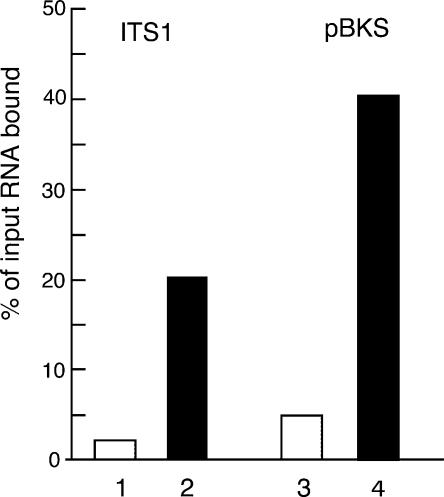
Figure 3 ▶ depicts a representative experiment showing that, when saturating amounts of labeled RNA were used, Rrp5p-loaded beads bound ~10 times the amount of the ITS1 fragment as did control beads. While this figure varied by no more than 10% between different experiments, the actual amount of the fragment retained by the beads, relative to the input, varied from 15% to 40%, depending on the cell extract used for the loading. We cannot exclude the possibility that differences in folding of the synthetic RNA used in different experiments also contribute to this variation. However, in view of the recognition of a broad range of RNAs by Rrp5p in vitro discussed below, this seems less likely.
FIGURE 3.
In vitro RNA binding by Rrp5p. 32P-UTP-labeled, in vitro transcribed RNA fragments were incubated with IgG Sepharose beads loaded with ProtA-Rrp5p. (Bars 1,2) ITS1 fragment (cf. Fig. 1A ▶); (bars 3,4) fragment transcribed from plasmid pBKS. (Gray bars) Control beads prepared with extract from cells expressing untagged Rrp5p; (black bars) beads prepared with extract from cells expressing tagged Rrp5p. Binding is expressed as the percentage of the input radioactivity retained by the beads.
To determine whether the retention of the ITS1 fragment by Rrp5p-loaded beads represents a (pre-)rRNA-specific interaction, we tested binding of an unrelated RNA (pBKS-RNA) obtained by in vitro transcription of ~1400 bp of the pBluescriptKS+ cloning vector (Stratagene). Figure 3 ▶ shows that the amount of pBKS transcript bound to ProtA-Rrp5p-loaded beads is also ~10-fold higher than the amount retained by control beads. Similar results were obtained for synthetic fragments corresponding to the 5′ ETS or ITS2 of yeast pre-rRNA (data not shown). These data indicate that Rrp5p is a general RNA-binding protein with, at least in vitro, affinity for sites of low complexity, a feature common to all proteins containing S1 RNA binding motifs studied so far (Subramanian 1983; Merendino et al. 2003; Schubert et al. 2004).
Rrp5p has a distinct preference for single-stranded U-rich sequences
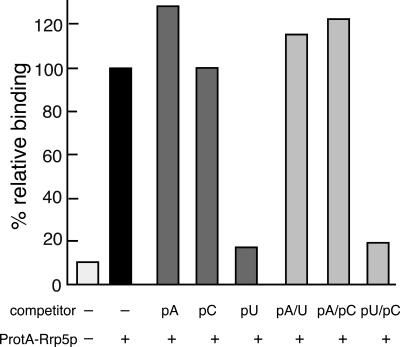
To investigate whether Rrp5p displays any sequence preference in its recognition of RNA, we performed in vitro competition binding experiments using ~1000-fold excess (1 μg) of unlabeled RNA homopolymers (poly[A], poly[C] or poly[U]) as competitor. Poly(G) was not used because of its known tendency to form a highly stable self-complementary structure (Zimmerman et al. 1975). Figure 4 ▶ shows that addition of neither poly(A) nor poly(C) significantly diminished binding of the labeled ITS1 fragment. Addition of poly(U), on the other hand, reduced binding to almost the level of the control, demonstrating a pronounced specificity of Rrp5p for homopolymeric stretches of U residues.
FIGURE 4.
Competition binding experiments using homopolymers. ProtA-Rrp5p-loaded beads were incubated with 32P-UTP-labeled ITS1 fragment in the absence or the presence of excess homopolymer as indicated. Either single-stranded (dark gray bars) or (mock-) annealed (light gray bars) homopolymer was used as competitor. Binding is expressed relative to the sample containing no competitor. The white bar represents binding to control beads prepared with extract from cells expressing untagged Rrp5p.
Because pre-rRNA is highly structured, we also wanted to determine the effect of secondary structure on the recognition of RNA by Rrp5p. Addition of double-stranded poly(A:U), prepared by annealing equal amounts of the two homopolymers at 65°C, as competitor did not affect binding of the ITS1 fragment. Neither did mock-annealed poly(A) + poly(C), whereas addition of a mock-annealed mixture of poly(C) + poly(U) had the same strong competitive effect as addition of poly(U) by itself (Fig. 4 ▶). We conclude that the recognition of RNA by Rrp5p requires the presence of stretches of unpaired U-residues.
The S1 domain of Rrp5p is responsible for RNA binding in vitro
To test the RNA-binding properties of the individual domains of Rrp5p, we constructed two deletion mutants encoding either the S1 or the TPR domain by parting the RRP5 gene at the XcmI site (Fig. 2 ▶). Each of the two mutants was fitted with a sequence encoding the ProtA-tag at its 5′ terminus. When expressed together, the Rrp5ΔS1p and Rrp5ΔTPRp proteins restored normal growth of YPB203 (GAL::RRP5) cells on glucose-based medium (data not shown), demonstrating that the two fragments are properly folded and fully functional (data not shown), as are two slightly different contiguous fragments split between S1 motifs 9 and 10 (Eppens et al. 1999).
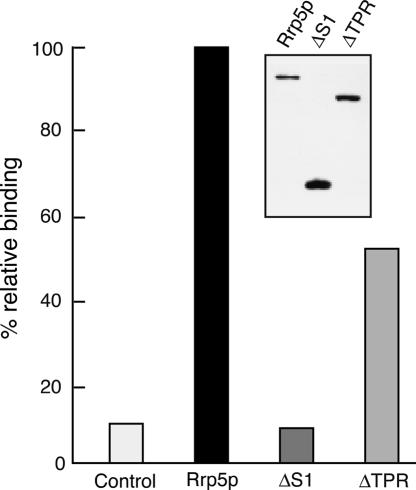
The two mutant proteins were tested for their ability to recognize the ITS1 fragment in vitro in the same manner as described above for the full-length protein, using extracts from HI227 cells that expressed either tagged fragment. As shown in the inset of Figure 5 ▶ the ProtA-Rrp5ΔTPRp-loaded beads carried about twice the amount of protein compared to beads loaded with intact ProtA-Rrp5p, while in the case of the ProtA-Rrp5ΔS1p protein the difference was even larger. These differences appear to be the result of variation in expression levels between the three types of protein (data not shown). The data presented in Figure 5 ▶ show that retention of the ITS1 fragment by beads loaded with Rrp5ΔS1p does not exceed the control value, even though these beads carry substantially more protein than those loaded with intact, tagged protein. Rrp5ΔTPRp-loaded beads, on the other hand, are still able to bind a significant amount of RNA, albeit less than the beads loaded with intact protein. We conclude that the RNA-binding ability of Rrp5p is confined to the S1 domain of the protein, but that the presence of the TPR domain may have a stimulatory effect, possibly by modulating folding of the S1 domain.
FIGURE 5.
RNA binding is confined to the S1 domain of Rrp5p. IgG Sepharose beads were incubated with extract obtained from cells expressing ProtA-tagged intact Rrp5p (black bar), the TPR domain (ΔS1), or the S1 domain (ΔTPR) and incubated with 32P-UTP-labeled ITS1 fragment. Binding is expressed relative to the sample containing intact Rrp5p. The white bar represents binding to beads prepared with extract from cells expressing untagged (intact) Rrp5p. (Inset) Western analysis of protein isolated from equal amounts of beads from the three samples.
In vivo pre-rRNA recognition by Rrp5p
The results of the in vitro binding experiments indicate that specific recognition of the pre-rRNA by Rrp5p in vivo cannot be based on sequence alone but must also depend on the architecture of the pre-ribosome that makes (a) particular site(s) accessible, while hiding others. Since in vitro reconstitution of pre-ribosomal particles is still beyond our capabilities, we have attempted to obtain more information on the nature of the in vivo binding site(s) within ITS1 by identification of the pre-rRNA species that are co-immunoprecipitated with tagged Rrp5p from yeast cell extracts. For this purpose, yeast strain YHV120 was constructed, which harbors an integrated ProtA::RRP5 locus, making this strain fully dependent on the tagged protein.
As shown in Figure 6A ▶, Northern analysis of coprecipitated RNA using probe 2, located shortly downstream from site A2 (Fig. 1A ▶), revealed the presence of a band corresponding to 27SA2 pre-rRNA (lane 2) that is absent from the control lane (lane 1). This confirms earlier reports that Rrp5p is still present in the 66S pre-ribosome, immediately after its separation from the 43S pre-ribosomal particle by the endonucleolytic cleavage of the 32S pre-rRNA at site A2 (Nissan et al. 2002). Primer extension analysis using probe 5, which hybridizes only to the 7S and 27S precursor species (Faber et al. 2004; Fig. 1A ▶) did not produce signals corresponding to sites B1L and B1S that exceed those seen in the control (Fig. 6B ▶), demonstrating that 27SBS and 27SBL pre-rRNA are not coprecipitated. Thus, further maturation of the 66S pre-ribosome containing 27SA2 pre-rRNA to the particle containing 27SB pre-rRNA is accompanied by the dissociation of Rrp5p. This suggests the presence of one or more functional binding sites for the protein in the 3′-terminal region of ITS1 between A2 and B1L and the absence of further such sites downstream from sites B1L/B1S. The latter is in accordance with the fact that Rrp5p does not have a role in the removal of ITS2 (Torchet et al. 1998; Eppens et al. 1999, 2002).
FIGURE 6.
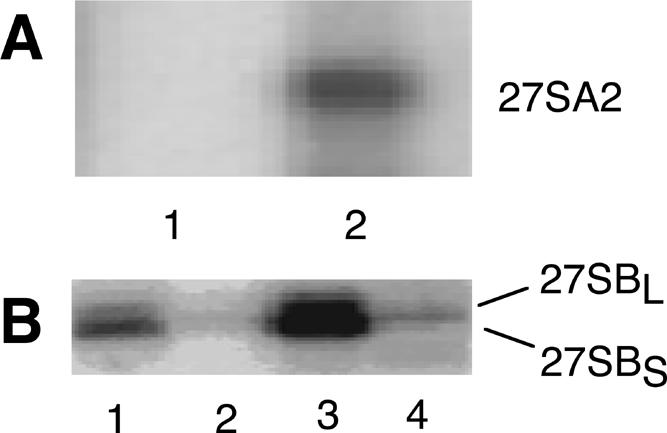
In vivo co-immunoprecipitation. (A) Extracts were prepared from cells expressing untagged Rrp5p (lane 1) or ProtA-Rrp5p (lane 2), treated with IgG Sepharose beads. Total RNA was isolated from the bead fraction and subjected to Northern analysis using probe 2, located closely downstream from site A2 (cf. Fig. 1 ▶). Only the region of the gel containing the 27S precursors is shown. (B) Total RNA was isolated from both the supernatant (lanes 1,3) and bead (lanes 2,4) fractions and subjected to primer extension analysis using probe 5, spanning the 3′ end of 5.8S rRNA (cf. Fig. 1 ▶). (Lanes 1,2) Samples prepared from cells expressing untagged Rrp5p; (lanes 3,4) samples prepared from cells expressing ProtA-Rrp5p. The stops corresponding to sites B1L and B1S are shown.
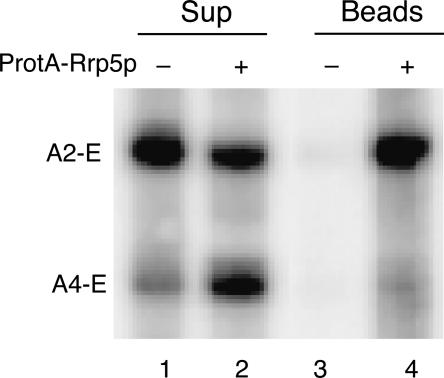
We have recently shown that the yeast rrp2-1 mutant, which carries a mutation in the RNA moiety of RNAse MRP, the snoRNP responsible for cleavage at site A3 (Shuai and Warner 1991; Schmitt and Clayton 1993; Chu et al. 1994), accumulates high levels of the 5′-extended pre-5.8S species A2-E and A4-E (Faber et al. 2004). This processing phenotype, therefore, allows us to investigate the possible role of the A2–A4 region within ITS1 in binding of Rrp5p. To that end, we transformed strain FVY05A (GAL::rrp5, rrp2-1) with a plasmid containing the ProtA: RRP5 gene. When grown on glucose as the carbon source, this strain becomes fully dependent on ProtA-Rrp5p, allowing ready identification of the pre-rRNA intermediates associated with the tagged protein. The results of the Northern analysis (Fig. 7 ▶) demonstrate efficient coprecipitation of the A2-E species, whereas the coprecipitate contains a negligible amount of the A4-E species (cf. lanes 4 and 2). This indicates that the region of ITS1 between sites A2 and A4 may contain sequences that interact directly with the protein.
FIGURE 7.
In vivo association of Rrp5p with 5′-extended pre-5.8S RNA species. Extracts were prepared from rrp2-1 cells expressing either untagged Rrp5p (lanes 1,3) or ProtA-Rrp5p (lanes 2,4) and treated with IgG Sepharose beads. Total RNA was isolated from the supernatant (lanes 1,2) and bead (lanes 3,4) fractions and subjected to Northern analysis using probe 3 (cf. Fig. 1 ▶) that detects the A2-E and A4-E, 5′-extended pre-5.8S species accumulating in rrp2-1 cells.
DISCUSSION
Although the presence of multiple copies of the S1 RNA binding motif in Rrp5p immediately suggested that the protein interacts directly with the pre-rRNA, the data presented above are the first experimental evidence that Rrp5p indeed is an RNA-binding protein.
RNA-binding properties of Rrp5p
Using an in vitro RNA-binding assay we have demonstrated that Rrp5p is able to recognize synthetic RNA transcripts corresponding to various regions of yeast pre-rRNA as well as sequences unrelated to pre-rRNA. Thus, at least in vitro, Rrp5p does not possess intrinsic specificity for pre-rRNA but is a general RNA-binding protein that appears to recognize low-complexity structural features.
Competition experiments using unlabeled homopolymers, however, established that Rrp5p does have a pronounced preference for single-stranded poly(U). In this respect it distinguishes itself from most other trans-acting factors that were either experimentally shown or are presumed to be (pre-)rRNA-binding proteins (Kressler et al. 1999; Dunbar et al. 2000; Valasek et al. 2001; Bjork et al. 2002; Jin et al. 2002; Wehner and Baserga 2002; Oeffinger and Tollervey 2003). In vitro studies on the RNA-binding properties of a number of these proteins, including several members of the Imp4p superfamily (Imp4p, Rpf1p, and Rpf2p) containing a σ70-like motif, as well as Rrp12p, carrying an RRM motif, demonstrated that they all interact with at least one, and in some cases two, other homopolymers besides poly(U) (Wehner and Baserga 2002; Oeffinger et al. 2004). Furthermore, Escherichia coli ribosomal protein S1 (rpS1p), the founding protein of the S1 motif, also recognizes a broad range of homopolymers, including poly(U), poly(A), and poly(C) (30), as does another E. coli protein containing S1 motifs, RNase E (Schubert et al. 2004).
Our data do not allow a firm conclusion regarding the minimum number of consecutive U residues required for interaction with Rrp5p. Inspection of the proposed secondary structure of the ITS1 fragment, as well as the 5′ ETS and ITS2 fragments bound by Rrp5p (Yeh and Lee 1990, 1992; Yeh et al. 1990; see also Fig. 8 ▶), however, indicates that this number is likely to be rather small, although occasional interruption with a different residue may not unduly disturb recognition of a U-rich sequence by the protein. It should be noted that E. coli rpS1p and RNase E are able to bind RNA molecules as short as 10 nt (Bisaglia et al. 2003; Schubert et al. 2004).
FIGURE 8.
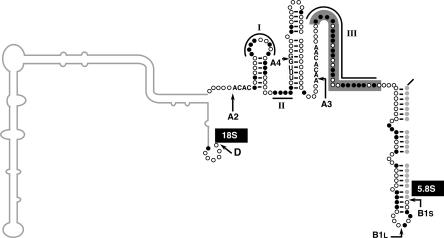
Schematic representation of the secondary structure of S. cerevisiae ITS1. Model according to Yeh et al. (1990) and Allmang et al. (1996). Cleavage sites A2, A4, A3, B1L, and B1S are indicated, including the conserved sequences known to be important for these cleavages. U residues in the remaining portion of the spacer downstream from A2 are shown as black circles; other residues, as open circles. The gray circles represent the nucleotides of the mature 5.8S rRNA sequence paired to ITS1. Potential, U-rich binding sites for Rrp5p, indicated as I–III, are over- or underlined. Shading indicates the extent of the previously tested deletion of site III (Van Nues et al. 1995), while the 5′ portion of the spacer known to be dispensable for processing (Henry et al. 1994; Van Nues et al. 1994) is represented by a gray line.
Target sites for Rrp5p in 35S pre-rRNA
The general RNA-binding capacity of Rrp5p implies that its specific interactions with pre-rRNA in vivo depend on the target site(s) being exposed in the architectural context of the pre-ribosome and possibly on its interaction with other trans-acting proteins. Given the present experimental possibilities, unequivocal identification of these target sites, therefore, would be a difficult task. To try to get an idea of their possible location we studied the pre-RNA species associated with Rrp5p in vivo, using immunoprecipitation. These studies confirmed that Rrp5p is associated with 35S and 27SA2 pre-rRNA but not anymore with the 27SB precursor species (Dragon et al. 2002; Nissan et al. 2002; Vos et al. 2004a). Thus it is still present in early 66S pre-ribosomes but dissociates on, or immediately after, removal of the remaining ITS1 sequences from the 27SA2 precursor, which suggests the presence of functional binding site(s) in the 3′-terminal half of the spacer, although it does not rule out the possibility of recognition sites located further downstream from which Rrp5p is detached upon processing of 27SA2 pre-rRNA.
According to the secondary structure model proposed for ITS1, the region downstream from site A2 contains three single-stranded, U-rich sequences, two short ones located between sites A2 and A4 (I and II in Fig. 8 ▶), and a longer one (III) downstream from site A3 (Yeh et al. 1990; Allmang et al. 1996). The results of the co-immunoprecipitation experiments using extracts from rrp2-1 cells that accumulate two 5′-extended pre-5.8S rRNA species (Faber et al. 2004) focused our attention on sequences I and II, which are still present in the A2-E pre-5.8S rRNA species, which remains associated with Rrp5p, but not in the shorter A4-E species, which fails to be coprecipitated with the tagged protein (Fig. 7 ▶). The fact that sequence I, but not sequence II, is conserved in the Kluyveromyces lactis ITS1, which can replace its S. cerevisiae counterpart without detectable consequences for processing (Van Nues et al. 1994), suggests that the former, but not the latter, is important for association of Rrp5p with the 66S pre-ribosome. Further experiments are required, however, to establish whether this sequence indeed interacts directly with Rrp5p or is involved in an indirect manner.
The failure of the A4-E pre-5.8S species to coprecipitate with ProtA-Rrp5p would rule out sequence III downstream from site A3 as a functional in vivo recognition site for Rrp5p. In vivo mutational analysis has shown that deletion of this region (cf. Fig. 8 ▶) does not affect pre-rRNA processing, including production of wild-type levels of 5.8SS and 5.8SL rRNA (Van Nues et al. 1994, 1995), which depends absolutely on Rrp5p (Eppens et al. 1999, 2002). Thus, sequence III is indeed dispensable for association of Rrp5p with the pre-ribosome and the subsequent role of this protein in processing at sites A0–A3.
The association of Rrp5p with the 35S precursor at a very early stage of ribosome biogenesis (Dragon et al. 2002; Vos et al. 2004a) indicates that direct recognition of the pre-RNA plays a major role in recruitment of the protein to the pre-ribosome. The presence of multiple TPR repeats (cf. Fig. 2 ▶), however, suggests that Rrp5p also interacts with other (pre-)ribosomal proteins. Such protein–protein interactions may well play a supporting role in tethering Rrp5p to the pre-ribosome and ensuring its timely release.
Implications for the role of Rrp5p in ribosome biogenesis
Although Rrp5p was considered to be a component of the SSU processome (Dragon et al. 2002), its association with 35S pre-rRNA does not depend on the U3 snoRNA (Vos et al. 2004a), which is essential for processome assembly. In this respect it resembles the recently identified t-Utp proteins, a subcomplex of seven processome proteins that associates with the rDNA to promote transcription and is then transferred to the nascent 35S precursor prior to association of the other components of the SSU processome (U3, Utps, and several SSU r-proteins) (Bernstein et al. 2004; Gallagher et al. 2004). However, whereas the t-Utps are essential for assembly of the SSU processome (Gallagher et al. 2004), Rrp5p is not, as association of U3 snoRNA with the 35S precursor does not require Rrp5p (Vos et al. 2004a). On the other hand, Rrp5p is critical for assembly of Rok1p into pre-ribosomes (Vos et al. 2004a), a putative RNA helicase essential for processing at sites A0–A2 (Venema et al. 1997) that associates subsequent to 80S processome formation (Dragon et al. 2002; Schäfer et al. 2003; Bernstein et al. 2004). We conclude that assembly of the complete early pre-rRNA processing machinery requires two platforms on the 35S pre-rRNA, one depending on the t-Utp complex, the other on Rrp5p. It should be noted that so far none of the t-Utps has been identified as an RNA-binding protein, either directly or by the presence of a known RNA-binding motif (Dragon et al. 2002; Gallagher et al. 2004).
Because the cleavages at A0 and A1 do not depend on any specific portion of the S1 domain (Eppens et al. 1999; Vos et al. 2004b), the requirement of Rrp5p for these processing events is most likely explained by its role as an assembly factor for part of the early processing machinery, including Rok1p and possibly also the snR10 snoRNP (Torchet and Hermann-Le Denmat 2002), which stimulates cleavage at A0–A2, though it is not essential (Tollervey 1987). The involvement of Rrp5p in A2 cleavage is more complex: It specifically requires S1 motifs 10–12 (Vos et al. 2004b), but this cleavage also critically depends on numerous other trans-acting factors, including Rok1p (Venema et al. 1997; Raué 2004). This suggests a parallel with the ability of E. coli rpS1p to enhance mRNA cleavage by the RegB endonuclease, which is thought to be caused by a favorable alteration in the structure of the RegB target site as a result of binding of the r-protein (Lebars et al. 2001). An attractive hypothesis, therefore, is that interaction of S1 motifs 10–12 with U-rich region I promotes recognition of site A2 by the (as yet to be identified) endonuclease by altering the local structure of ITS1. Interestingly, region I constitutes the loop of a small, evolutionarily conserved helix (Fig. 8 ▶) that has been implicated in correct and efficient processing at site A2 (Allmang et al. 1996). This Rrp5p-dependent restructuring of ITS1 must, however, be part of a larger series of events including the recruitment and/or correct positioning of the additional trans-acting factors (e.g., Rok1p) essential for processing at A2.
The molecular basis for the requirement of S1 motifs 1–8 of Rrp5p in cleavage at site A3 remains unresolved. Deletion of different nonoverlapping sets, and even individual members of motifs 1–8 (Eppens et al. 1999), results in identical phenotypes with regard to a block in A3 processing. We have already shown that these motifs do not constitute an essential scaffold for recruitment of RNAse MRP, the endonuclease responsible for the A3 cleavage, or helper proteins for this nuclease (Faber et al. 2004). On the other hand, mutant Rrp5p containing only S1 motifs 1–8 supports A3 cleavage in vivo (Vos et al. 2004b) and retains the ability to bind to RNA in vitro (H.R. Vos and P. de Boer, unpubl. results). Since deletion of the single-stranded U-rich region III from ITS1 does not affect A3-dependent, high-level production of 5.8SS rRNA (Van Nues et al. 1995), interaction between S1 motifs 1–8 and region III is not a prerequisite for A3 cleavage. Thus, the target site(s) for these motifs must be located elsewhere in the pre-rRNA. The redundancy in RNA-binding ability of the multiple S1 motifs and the lack of specificity in the interaction between Rrp5p and pre-rRNA in vitro, however, precludes unequivocal identification of these target site(s) by deletion analysis of either the protein or the pre-rRNA.
MATERIALS AND METHODS
Strains
E. coli strain MH1 (Hall et al. 1984) was used for cloning and propagation of plasmids. Yeast strains YPB01, YPB03, and YPB05 were obtained by transforming the protease-minus yeast strain HI227 (Abou-Elela et al. 1996) with plasmids pTRP1-ProtA-rrp5, pTRP1-ProtA-rrp5ΔS, and pTRP1-ProtA::rrp5ΔTPR, respectively, according to Gietz et al. (1992). YPB203 was obtained by cotransformation of YJV154 (Gal::rrp5) (Eppens et al. 1999) with plasmids pLEU-ProtA-rrp5ΔTPR and pTRP1-ProtA-rrp5ΔS. Transformants were selected on either SGlu or SGal plates using the marker gene(s) present on the respective plasmids. Yeast strain YHV120 was constructed by integration of the ProtA::rrp5 gene into the genome of YJV140 (Venema and Tollervey 1996). Yeast strain FVY05A (Gal::rrp5, rrp2-1) was obtained by crossing D308 (rrp2-1) with YJV154 (Gal::rrp5) (Faber et al. 2004).
Plasmids
A portion of the RRP5 gene was amplified by PCR using oligonucleotides rrp5ΔS (5′-CTCTTTTCCAAGGGGTGGAACCACTACT ACTGTGGATCAACTGG-3′) and rrp5ClaIrev (5′-GGATCGATA GAAATTGCTTCTTG-3′) to yield a fragment that contained the PflMI site fused to the XcmI site, thus creating a deletion of the sequence encoding the S1 motifs (cf. Fig. 2 ▶). After digestion of the PCR product with PflMI and XcmI, the mutant fragment was used to replace the original PflMI-XcmI fragment from pTRP1-ProtA-rrp5 (Eppens et al. 1999), resulting in plasmid pTRP1-ProtA-rrp5ΔS. Plasmid pTRP1-ProtA-rrp5ΔTPR was constructed by deleting the TPR motifs as an XcmI-SmaI fragment from pTRP1-ProtA-rrp5.
Plasmid pG3-SH22, which served as template for in vitro transcription of the ITS1 fragment used in the binding experiments, was constructed by cloning the 693-bp StuI-HpaI fragment of the rDNA unit from pJV12 (Venema et al. 1995), containing ITS1 and the 5.8S gene, flanked by small portions of the 18S gene and ITS2 (cf. Fig. 1 ▶), into the SmaI site of pGEM-3 (Promega).
In vitro transcription
α-32P-UTP-labeled RNA fragments for use as baits in the in vitro binding experiments were prepared by transcription of pG3-SH22 digested with HindIII or pBKS+ (Stratagene) digested with BsaI. In vitro transcription was performed using a large-scale in vitro transcription kit from Novagen according to the manual, with the exception that 2 μL of α-32P-UTP (10 mCi/mL) were used instead of α-35S-UTP. After the reaction, labeled RNA fragments were precipitated with NH4Ac according to the manual, and the pellet was redissolved in 50 μL double-distilled water (ddH2O) treated with diethylpyrocarbonate.
In vitro binding experiments
For in vitro binding experiments, 100 OD600 units of exponentially growing yeast cells, expressing plasmid-encoded, epitope-tagged, wild-type or mutant Rrp5p, were lysed by vortexing with 250 μL glass beads (diameter 0.8 mm) in 500 μL lysis buffer (50 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 150 mM NaCl, 0.1% Triton X-100, 1 mM DTT, 2 mM PMSF). For immunoprecipitation, 500 μL of IgG Sepharose beads (Amersham), prewashed 10 times with 1 mL portions of a high-salt binding buffer (50 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 500 mM NaCl, 0.1% Triton X-100, 1 mM DTT), were added to the cell extract. The mixture was incubated for 30 min with gentle agitation. After incubation, the beads were washed five times with 1-mL portions of high-salt binding buffer and resuspended in 500 μL of the same buffer. The whole procedure was performed at 4°C.
To 10 μL of the suspension containing the protein-loaded beads, 1 μL of the solution containing in vitro transcribed, radio-labeled RNA fragment was added, together with 1 μg of E. coli tRNA (Roche) as a nonspecific competitor. Care was taken to use a saturating amount of labeled RNA, which was determined in a separate dose-response experiment for each combination of extract and RNA sample. After adjusting the total volume to 100 μL with binding buffer, the reaction mixture was incubated for 30 min at 4°C with gentle agitation in a tabletop incubator. Unbound probe was removed by washing three times with 1-mL portions of cold binding buffer. The amount of bound radioactivity was measured by scintillation counting, and the intactness of the bound RNA was checked by gel electrophoresis.
Competition binding experiments were performed by adding 1 μL of a solution containing single-stranded, double-stranded, or mock-annealed homopolymer (1 mg/mL) to the incubation mixture, together with the radiolabeled RNA fragment. Single-stranded poly(A), poly(C), and poly(U) were obtained from Amersham Biosciences. Double-stranded poly(A:U) was prepared by mixing equal amounts of the two single-stranded homopolymers and heating the mixture to 65°C, followed by slow cooling to room temperature. Control mixtures of poly(A) + poly(C) or poly(U) + poly(C) were treated in the same manner.
Co-immunoprecipitation
Identification of pre-rRNA species that could be co-immunoprecipitated with Rrp5p in vivo was carried out by preparing an extract from 100 OD600 units of exponentially growing yeast cells expressing the tagged protein in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, 1 mM DTT, and 2 mM PMSF in a similar manner as described above. The cell extract was then mixed with 50 μL of IgG Sepharose beads, prewashed in a low-salt binding buffer lacking MgCl2 (50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 0.1% Triton X-100, 1 mM DTT). This mixture was further treated in the same manner as for the in vitro binding experiments, with the exception that the low-salt buffer was used for the washing steps. After washing, the beads were transferred onto a BIO-RAD Micro Bio-Spin Chromatography column (catalog no. 732-6204). Bound proteins and RNA were eluted with 500 μL 0.5 × GTC mix (4 M guanidinium thiocyanate, 50 mM Tris-HCl at pH 8.3, 10 mM EDTA, 2% N-lauryl sarcosyl, 1% β-mercaptoethanol, diluted with an equal volume of binding buffer). To separate proteins and RNA, 2 μL 1 M glycogen, 2 μL 10 mM E. coli tRNA (Sigma Aldrich), 0.1 volume 3 M NaAc (pH 5.2), and 1 volume phenol-chloroform-isoamylalcohol (25:24:1) were added to the eluate, followed by vigorous vortexing. The water and the phenol phases (the latter including the interphase) were separated into two different tubes and 2.5 volumes of ethanol were added to each tube to precipitate RNA and proteins, respectively.
Western analysis
Immunoprecipitations were verified by Western analysis of ProtA-tagged proteins using peroxidase anti-peroxidase (PAP) (Sigma) and an ECL detection kit from Amersham.
RNA analysis
Northern and primer extension analysis were carried out as described previously (Vos et al. 2004b) using oligonucleotides 2 (5′-GATCACCTAGCGACTCTCTCCACC-3′), 3 (5′-CCAGTTACGAAAATTCTTGTTTTTGAC-3′), and 5 (5′-GAATGTTTGAGAA GGAAATGACGCTC-3′) as probes. The positions of these probes on the 35S pre-rRNA are shown in Figure 1A ▶.
Acknowledgments
P.d.B. was supported by a grant from the Council for Chemical Sciences (CW) with financial aid from the Netherlands Foundation for Scientific Research (NWO).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2257606.
REFERENCES
- Abou-Elela, S., Igel, H., and Ares Jr., M. 1996. RNAase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124. [DOI] [PubMed] [Google Scholar]
- Allmang, C., Henry, Y., Wood, H., Morrissey, J.P., Petfalski, E., and Tollervey, D. 1996. Recognition of cleavage site A2 in the yeast pre-rRNA. RNA 2: 51–62. [PMC free article] [PubMed] [Google Scholar]
- Bernstein, K.A., Gallagher, J.E., Mitchell, B.M., Granneman, S., and Baserga, S.J. 2004. The SSU processome is a ribosome assembly intermediate. Eukaryot. Cell 3: 1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaglia, M., Laalami, S., Uzan, M., and Bontems, F. 2003. Activation of the RegB endoribonuclease by the S1 ribosomal protein is due to cooperation between the S1 four C-terminal modules in a substrate- dependent manner. J. Biol. Chem. 278: 15261–15271. [DOI] [PubMed] [Google Scholar]
- Bjork, P., Bauren, G., Jin, S., Tong, Y.G., Burglin, T.R., Hellman, U., and Wieslander, L. 2002. A novel conserved RNA-binding domain protein, RBD-1, is essential for ribosome biogenesis. Mol. Biol. Cell 13: 3683–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch, G.L. and Lassle, M. 1999. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. Bioessays 21: 932–939. [DOI] [PubMed] [Google Scholar]
- Chu, S., Archer, R.H., Zengel, J.M., and Lindahl, L. 1994. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl. Acad. Sci. 91: 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon, F., Gallagher, J.E.G., Compagnone-Post, P.A., Mitchell, B.A., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar, D.A., Dragon, F., Lee, S.J., and Baserga, S.J. 2000. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. 97: 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppens, E.A., Rensen, S., Granneman, S., Raué, H.A., and Venema, J. 1999. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA 5: 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppens, E.A., Faber, A.W., Rondaij, M., Jahangir, R.S., van Hemert, S., Vos, J.C., Venema, J., and Raué, H.A. 2002. Deletions in the S1 domain of Rrp5p cause processing at a novel site in ITS1 of yeast pre-rRNA that depends on Rex4p. Nucleic Acids Res. 30: 4222–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber, A.W., Vos, J.C., Vos, H.R., Ghazal, G., Abou Elela, S., and Raué, H.A. 2004. The RNA catabolic enzymes Rex4p, Rnt1p and Dbr1p show genetic interaction with trans-acting factors involved in the processing of ITS1 in Saccharomyces cerevisiae pre-rRNA. RNA 10: 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A. and Tollervey, D. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14: 313–318. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Senger, B., Saveanu, C., and Fasiolo, F. 2003. Ribosome assembly in eukaryotes. Gene 313: 17–42. [DOI] [PubMed] [Google Scholar]
- Gallagher, J.E.G., Dunbar, D.A., Granneman, S., Mitchell, B.M., Osheim, Y., Beyer, A.L., and Baserga, S.J. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes & Dev. 18: 2506–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., Jean, A.S., Woods, R.A., and Schiestl, R.H. 1992. Improved methods for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., Rybin, V., Baßler, J., Petfalski, E., Strauss, D., Marzioch, M., Schäfer, T., Kuster, B., Tschochner, H., Tollervey, D., et al. 2002. 90S Pre-ribosomes include the 35S pre-rRNA, the U3 sno RNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10: 105–115. [DOI] [PubMed] [Google Scholar]
- Hall, M.N., Hereford, L., and Herskowitz, I. 1984. Targeting of E. coli β-galactosidase to the nucleus in yeast. Cell 36: 1057–1065. [DOI] [PubMed] [Google Scholar]
- Henry, Y., Wood, H., Morrissey, J.P., Petfalski, E., Kearsey, S., and Tollervey, D. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13: 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S.-B., Zhao, J., Björk, P., Schmekel, K., Ljungdahl, P.O., and Wieslander, L. 2002. Mrd1p is required for processing of pre-rRNA and for maintenance of steady-state levels of 40S ribosomal subunits in yeast. J. Biol. Chem. 277: 18431–18439. [DOI] [PubMed] [Google Scholar]
- Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20: 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler, D., Linder, P., and De La Cruz, J. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J., Dichtl, B., and Tollervey, D. 1999. Yeast Rnt1p is required for cleavage of the preribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebars, I., Hu, R.M., Lallemand, J.Y., Uzan, M., and Bontems, F. 2001. Role of the substrate conformation and of the S1 protein in the cleavage efficiency of the T4 endoribonuclease RegB. J. Biol. Chem. 276: 13264–13267. [DOI] [PubMed] [Google Scholar]
- Merendino, L., Falciatore, A., and Rochaix, D. 2003. Expression and RNA binding properties of the chloroplast ribosomal protein S1 from Chlamydomonas reinhardtii. Plant Mol. Biol. 53: 371–382. [DOI] [PubMed] [Google Scholar]
- Moy, T.I. and Silver, P.A. 1999. Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes & Dev. 13: 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T.A., Baßler, J., Petfalski, E., Tollervey, D., and Hurt, E. 2002. 60S pre-ribosome formation viewed from assembly in the nucleus until export to the cytoplasm. EMBO J. 21: 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger, M. and Tollervey, D. 2003. Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. EMBO J. 22: 6573–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger, M., Dlakic, M., and Tollervey, D. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes & Dev. 18: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raué, H.A. 2004. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae. In The nucleolus (ed. M.O. Olson), pp. 199–222. Kluwer Academic/Plenum Publishers, New York.
- Schäfer, T., Strauss, D., Petfalski, E., Tollervey, D., and Hurt, E. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22: 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M.E. and Clayton, D.A. 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 7935–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, M., Edge, R.E., Lario, P., Coork, M.A., Strynadka, N.C., Mackie, G.A., and McIntosh, L.P. 2004. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J. Mol. Biol. 341: 37–54. [DOI] [PubMed] [Google Scholar]
- Shuai, K. and Warner, J.R. 1991. A temperature-sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 19: 5059–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, A., Hsu, C.L., Isham, K.R., and Larimer, F.W. 1991. Fragments of the Internal Transcribed Spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′ → 3′ exoribonuclease-1. J. Bacteriol. 173: 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A.R. 1983. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 28: 101–142. [DOI] [PubMed] [Google Scholar]
- Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of preribosomal RNA. EMBO J. 6: 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet, C. and Hermann-Le Denmat, S. 2002. High dosage of the small nucleolar RNA snR10 specifically suppresses defects of a yeast rrp5 mutant. Mol. Genet. Genomics 268: 70–80. [DOI] [PubMed] [Google Scholar]
- Torchet, C., Jacq, C., and Hermannle Denmat, S. 1998. Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA 4: 1636–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek, L., Hasek, J., Nielsen, K.H., and Hinnebusch, A.G. 2001. Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J. Biol. Chem. 276: 43351–43360. [DOI] [PubMed] [Google Scholar]
- Van Nues, R.W., Rientjes, J.M.J., Van der Sande, C.A.F.M., Zerp, S.F., Sluiter, C., Venema, J., Planta, R.J., and Raué, H.A. 1994. Separate structural elements within Internal Transcribed Spacer I of Saccharomyces cerevisiae precursor ribosomal RNA direct the formation of 17S and 26S rRNA. Nucleic Acids Res. 22: 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nues, R.W., Venema, J., Rientjes, J.M.J., Dirks-Mulder, A., and Raué, H.A. 1995. Processing of eukaryotic pre-rRNA: The role of the transcribed spacers. Biochem. Cell Biol. 73: 789–811. [DOI] [PubMed] [Google Scholar]
- Vanrobays, E., Gelugne, J.-P., Gleizes, P.-E., and Caizergues-Ferrer, M. 2003. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 2083–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15: 5701–5714. [PMC free article] [PubMed] [Google Scholar]
- ———. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 216–311. [DOI] [PubMed] [Google Scholar]
- Venema, J., Dirks-Mulder, A., Faber, A.W., and Raué, H.A. 1995. Development and application of an in vivo system to study yeast ribosomal RNA biogenesis and function. Yeast 11: 145–156. [DOI] [PubMed] [Google Scholar]
- Venema, J., Bousquet-Antonelli, C., Gelugne, J.-P., Caizergues-Ferrer, M., and Tollervey, D. 1997. Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol. 17: 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, H.R., Bax, R., Faber, A.W., Vos, J.C., and Raué, H.A. 2004a. U3 snoRNP and Rrp5p associate independently with Saccharomyces cerevisiae 35S pre-rRNA, but Rrp5p is essential for association of Rok1p. Nucleic Acids Res. 39: 5827–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, H.R., Faber, A.W., De Gier, M.D., Vos, J.C., and Raué, H.A. 2004b. Deletion of the three distal S1 motifs of yeast Rrp5p abolishes pre-rRNA processing at site A2 without reducing the production of functional 40S subunits. Eukaryot. Cell 3: 1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J.R. 2001. Nascent ribosomes. Cell 107: 133–136. [DOI] [PubMed] [Google Scholar]
- Wehner, K.A. and Baserga, S.J. 2002. The σ(70)-like motif: A eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9: 329–339. [DOI] [PubMed] [Google Scholar]
- Yeh, L.-C. and Lee, J.C. 1990. Structural analysis of the Internal Transcribed Spacer 2 of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 211: 699–712. [DOI] [PubMed] [Google Scholar]
- ———. 1992. Structure analysis of the 5′ external transcribed spacer of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 228: 827–839. [DOI] [PubMed] [Google Scholar]
- Yeh, L.-C., Thweatt, R., and Lee, J.C. 1990. Internal Transcribed Spacer 1 of the yeast precursor ribosomal RNA. Higher order structure and common structural motifs. Biochemistry 29: 5911–5918. [DOI] [PubMed] [Google Scholar]
- Zimmerman, S.B., Cohen, G.H., and Davies, D.R. 1975. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 92: 181–192. [DOI] [PubMed] [Google Scholar]