Abstract
Cellular RNA chaperones are crucial for the genesis of correctly folded functional RNAs. Using several complementary in vitro assays we find that the bunyavirus nucleocapsid protein (N) is an RNA chaperone. In the Bunyaviridae genomic RNA is in stable “panhandle” formation that arises through the hydrogen bonding of the terminal nucleotides of the RNA. The RNA chaperone function of N facilitates panhandle formation even though the termini are separated by >2 kb. RNA panhandle formation is likely driven by the exceptionally high base-pairing specificity of the terminal nucleotides as evidenced by P-num analysis. N protein can nonspecifically dissociate RNA duplexes. In addition, following panhandle formation, the RNA chaperone activity of N also appears to be involved in dissociation of the RNA panhandle and remains in association with the 5′ terminus of the viral RNA following dissociation. Thus, N likely functions in the initiation of genome replication to allow efficient initiation of RNA synthesis by the viral polymerase. The RNA chaperone activity of N may be facilitated by an intrinsically disordered domain that catalyzes RNA unfolding driven by reciprocal entropy transfer. These observations highlight the essential features that are probably common to all RNA chaperones in which the role of the chaperone is to nonspecifically dissociate higher order structure and formation of functional higher order structure may often be predicted by RNA P-num value. The data also highlight features of N that are probably specifically important during replication of bunyavirus RNA.
Keywords: Bunyaviradae, replication, RNA structure, RNA chaperon, panhandle, nucleocapsid protein
INTRODUCTION
Hantaviruses comprise a genus in the Bunyaviridae family, whose genome is composed of three minus-strand viral RNA (vRNA) segments. The vast majority of these genome RNAs, both intracellularly and in virions, are found as RNPs (or nucleocapsids) complexed with the viral N protein, which form a sharp band on CsCl density gradient centrifugation at a density (1.3 g/mL) indicative of a mostly protein nature. Early electron microscopic analysis indicated that the termini of the Uukuniemi virus genome RNAs interact to form circular structures that dissociate under denaturing conditions (Hewlett et al. 1977). Furthermore, the genome RNPs of other Bunyaviruses have also been observed to be circular, indicating that intramolecular association of the vRNA termini form in vivo, and that the terminal panhandle ostensibly forms in the context of bound N protein (Pettersson and von Bonsdorff 1975; Obijeski et al. 1976; Raju and Kolakofsky 1989). All members of the Bunyaviridae family contain terminal nucleotide sequences that could facilitate the association of termini through the formation of short panhandles, and for LaCrosse virus, these termini have been shown to be base-paired within the nucleocapsids, via psoralen cross-linking in situ. As with all minus-strand RNA viruses, these genome RNPs or nucleocapsids serve as templates for both mRNA and anti-genome synthesis. Recent reverse genetic studies using Bunyamwera virus replicons have revealed that the ability of the genomic RNA termini to base-pair is an important feature of bunyavirus replication (Barr and Wertz 2005). However, how these termini base-pair in the context of the nucleocapsid and precisely what role they play in viral RNA synthesis are unclear.
RNA and protein chaperones facilitate the correct folding of RNA and protein substrates into correct functional higher order structures. Because RNAs readily form mutually exclusive alternative higher order structures, they easily enter “kinetic traps” as they fold into their lowest energy structures (Treiber and Williamson 2001; Cristofari and Darlix 2002). These kinetic traps consist of local stable higher order structures that would require energy to dissociate incorrectly hydrogen-bonded nucleotides to permit the formation of globally stable RNA structures containing biologically functional secondary and tertiary signals. Even relatively short RNA molecules such as tRNA are subject to significant kinetic traps during folding process and this problem becomes more complicated for longer RNA molecules (Herschlag 1995). Chaperones appear to function by repeatedly binding to a wide variety of substrate molecules, unfolding higher order structures and allowing the formation of thermodynamically stable and functional states (Herschlag et al. 1994; Todd et al. 1996). In contrast to this nonspecific activity of RNA chaperones, structure-specific RNA binding proteins can stabilize correctly formed structure through the recognition and specific binding (Weeks 1997).
Examples of cellular RNA chaperones include hnRNP A1, which can facilitate both the disassembly and the annealing of complementary nucleic acid strands (Pontius and Berg 1992), cold shock protein CspA, host factor 1, protein Hfq, yeast LA protein, and the tumor suppressor protein p53 (Tsuchihashi et al. 1993; Jiang et al. 1997; Pannone et al. 1998; Negroni and Buc 1999). As might be expected, some RNA viruses also appear to require RNA chaperone activity and encode proteins with this function. An RNA chaperone activity associated with the nucleocapsid protein of HIV-1 appears to be required for the formation of several RNA duplexes that function in nucleic acid replication (for review, see Rein et al. 1998). More recently, a similar general activity has been attributed to the core protein of hepatitis C virus and to hepatitis delta antigen (Wang et al. 2003; Cristofari et al. 2004).
In vitro, the viral nucleocapsid protein (N) interacts in a specific manner with vRNA (Gott et al. 1993; Richmond et al. 1998; Severson et al. 1999, 2001; Osborne and Elliott 2000) owing, at least in part, to a crucial central RNA binding domain of N (Xu et al. 2002; Severson et al. 2005). N exhibits high-affinity binding to the vRNA panhandle in vitro and has therefore been proposed to recognize the panhandle structure during the encapsidation process (Mir and Panganiban 2004). N also recognizes the cRNA panhandle but with lower affinity (Mir and Panganiban 2005).
Using Sin Nombre hantavirus, we describe in vitro experiments indicating that the bunyavirus N protein is an RNA chaperone capable of promoting correct annealing of the panhandle nucleotides. N protein contains an extensive disordered amino acid sequence that likely functions in RNA chaperone activity. We propose a model in which the chaperone activity of N facilitates formation of the RNA panhandle in the nucleoprotein complex in conjunction with the remarkably low P-num values of the terminal nucleotides. Following high-affinity binding to the panhandle we observe that N dissociates the panhandle and remains associated with nucleotides near the 5′ end of the viral RNA. Dissociation of the panhandle is probably related to the RNA chaperone activity of N and is likely to function during initiation of genome replication.
RESULTS
Hantavirus N nonspecifically alters global higher order RNA structure in vitro
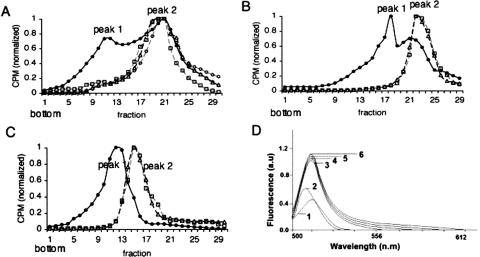
To determine whether hantavirus N protein can affect the overall structure of RNA, we expressed full-length, radioactively labeled, Sin Nombre hantavirus S segment vRNA by in vitro transcription. This RNA was then either incubated with Sin Nombre hantavirus N protein, or without N protein for 3 h. An additional RNA sample was heat denatured for 5 min and renatured for 3 h. The RNA in each sample was then purified and fractionated on 10%–60% sucrose gradients. The S-segment vRNA that had not been either treated with N or heat denatured was present in two major peaks (labeled “1” and “2” in Fig. 1A ▶). In contrast, vRNA that had been denatured and renatured resulted in the sedimentation of the RNA to a single position in the gradient corresponding to the more slowly migrating RNA peak 2 (Fig. 1A ▶). Similarly, treatment of S segment RNA with N protein for 2.5 h resulted in sedimentation to position 2 (Fig. 1A ▶). To examine the integrity of the vRNA following sedimentation, samples from peaks 1 and 2 from the untreated RNA, peak 2 of the denatured and renatured RNA, and peak 2 from N-treated RNA were analyzed on a 5% denaturing polyacrylamide gel. This revealed that the S segment vRNA from each sample was of the same length (data not shown). From these data, it appeared that S segment vRNA folds into at least two populations that are different in higher order structure. However, treatment of the RNA with either hantavirus N protein or heat converts the RNA to one or more forms that migrate to a single position (peak 2) in a gradient. The relatively slower sedimentation of the heat-denatured, or N-treated, RNA is consistent with a less compact or alternative higher order structure.
FIGURE 1.
Effect of N on global RNA structure. (A) A full-length, radioactively labeled S segment RNA was either untreated, treated with N, or HIV-1 Gag as described in the text, sedimented on 10%–60% sucrose gradients, and RNA was detected by scintillation counting. (•) Untreated RNA, (▵) heat treated RNA, (□) N-treated RNA, (○) RNA treated with HIV-1 Gag. Similar results were obtained in three independent experiments. (B) RNA containing a deletion of the 3′ nucleotides such that normal panhandle formation would be blocked. (C) Nonviral RNA. (D) The fluorescence spectra of 120 nM of acridine orange is shown: (1) in the absence of RNA, (2) the presence of 95 pM SNV S segment RNA from sucrose gradient peak 1, (3) peak 2, (4) from thermally denatured and renatured RNA, (5) from N treated RNA, or (6) from HIV-1 Gag-treated RNA sample.
Since N protein appeared to affect the apparent conformation of S segment vRNA in the same way as thermal denaturation, this was consistent with the activity of an RNA chaperone. Chaperones facilitate the general nonspecific denaturation of RNA such that the RNA can fold into an optimally low free-energy state (Herschlag et al. 1994; Todd et al. 1996). Thus, we compared the effect of N protein to that of retroviral nucleocapsid protein, a previously characterized viral RNA chaperone (for review, see Rein et al. 1998). Hantavirus S segment RNA was incubated with human immunodeficiency virus type-1 (HIV-1) Gag protein, the protein was removed, and the RNA was analyzed on sucrose gradients in parallel with heat-treated or hantavirus-N treated RNA (Fig. 1A ▶). The RNA that had been treated with Gag migrated to a fraction coincident with the RNA that had been treated with N protein or treated with thermal denaturation.
The termini of the Bunyaviridae vRNA are complementary and facilitate the generation of RNA panhandles. To examine the effect of N on a vRNA that is unable to form the terminal panhandle we synthesized both an RNA lacking the 3′-terminal 42 nt of the hantavirus S segment RNA and a nonviral RNA molecule of approximately the same length as the vRNA S segment. As shown in Figure 1, B and C ▶, these RNAs both sedimented to two primary positions in the gradient in a manner similar to that observed for full length S segment RNA, although the absolute position of the two peaks was different than that for the full length S segment. Moreover, thermal denaturation and renaturation or incubation with hantavirus N protein resulted in an RNA population of a single slower mobility on sucrose gradients (peak 2) (Fig. 1B,C ▶). To further evaluate the effect of N on general RNA structure we examined the interaction of S segment RNA with acridine orange, a molecule that intercalates into double-stranded nucleic acids, that binds with single-stranded nucleic acid through an uncharacterized interaction, and that typically exhibits a measurable increase in fluorescence quantum yield when associated with RNA. The fluorescence spectra of acridine orange from 500 to 650 nm, in the absence and the presence of treated and untreated RNA, is shown in Figure 1D ▶. Addition of untreated S segment RNA from peak 1 of the sucrose gradient resulted in a marginal increase in the fluorescence quantum yield and a detectible shift in the maximum peak emission wavelength. In contrast, addition of RNA from peak 2 of the sucrose gradient, and from RNA that was treated with hantavirus N, HIV-1 Gag, or thermal denaturation and renaturation resulted in a threefold increase in fluorescence quantum yield of acridine orange without alteration of other spectral features. From these various data it appears that hantaviral N protein and thermal denaturation affect general RNA conformation in a similar way. N protein appears to interact with RNA nonspecifically to generate RNA that is similar to RNA that has been heat denatured and renatured, or treated with an established RNA chaperone.
Hantavirus N protein facilitates RNA panhandle formation
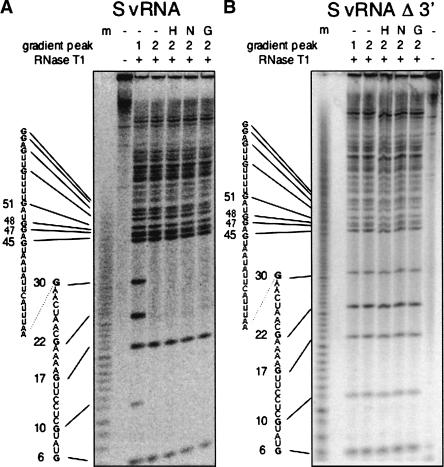
We next specifically analyzed the effect of N on nucleotides that comprise the terminal vRNA panhandle. Hantavirus S segment vRNA was 5′-end labeled. The RNA was then either denatured and renatured, treated with N, or treated with HIV-1 Gag. Subsequently, the RNA was fractionated on sucrose gradients as described previously and RNA from peaks 1 and 2 were recovered. RNA was then digested with RNase T1 and fractionated on a 20% denaturing gel (Fig. 2A ▶). RNase T1 preferentially cleaves single-stranded RNA at G residues. However, G residues that are situated in double-stranded regions are relatively resistant to RNase T1. Interestingly, all terminal G residues in vRNA from peak 1, which was not treated with N protein, were readily cleaved by RNase T1 suggesting that the 5′ terminus is in largely single-stranded conformation. In contrast, the terminal G residues at positions 10, 22, and 30 of RNA from peak 2 were markedly resistant to RNase T1 treatment consistent with their position in double-stranded RNA. Further, the RNase T1 digestion of RNA that had been thermally denatured and renatured, treated with hantavirus N, or treated with HIV-1 Gag also resulted in protection of some of these same terminal G residues. This indicates that the 5′ terminus of SNV S segment RNA in peak 1, which was initially relatively single-stranded in character, became more double-stranded following treatment with N protein.
FIGURE 2.
Effect of N on the 5′ end of vRNA. 5′ End-labeled S segment RNA or S segment RNA containing a 3′ deletion was treated with hantavirus N, HIV-1 Gag, or heat denatured and renatured and subsequently fractionated on sucrose gradients as in Figure 1 ▶. RNA from the gradient was then analyzed by RNase T1 digestion. A depicts RNase T1 digestion of S segment vRNA; B, RNase T1 digestion of S segment vRNA containing a 42-nt deletion at the 3′ terminus. (m) RNA sequence ladder generated by partial NaOH treatment, (−) no treatment of the RNA prior to sucrose gradient fractionation, (H) RNA that had been heat treated and renatured prior to sucrose gradient fractionation, (N) RNA treated with N before fractionation, (G) RNA treated with HIV-1 Gag prior to fractionation. RNA was derived from either sucrose gradient peak 1 or 2 (as in Fig. 1 ▶) as indicated. For both RNAs, thermal denaturation, treatment with N, or treatment with Gag yielded gradients with RNA present only in peak 2.
Although N rendered some of the nucleotides at the 5′ terminus resistant to RNase T1 digestion, it was not clear whether the 5′ end is in panhandle configuration. Thus, we carried out parallel analysis of S segment vRNA containing a deletion of the 42 nt at the 3′ end of the molecule. RNA from peak 1 was again readily accessible to RNase T1 (Fig. 2B ▶). However, following thermal denaturation and renaturation, treatment with SNV N, or treatment with HIV-1 Gag, the 5′-terminal nucleotides appeared to remain susceptible to RNase T1 digestion yielding a pattern indistinguishable from that of peak 1. This indicates that the RNase T1 resistance of the 5′-terminal G residues in full-length vRNA is dependent on the presence of the nucleotides of the 3′ terminus. The most direct interpretation of these data is that N protein facilitates the generation of the vRNA panhandle. Moreover, heat denaturation of the RNA likely creates an opportunity for the RNA to refold into a native, globally stable structure that includes a terminal panhandle. Similarly, N protein and HIV Gag enable RNA folding.
The RNase T1 digestion results with full-length S segment RNA are also noteworthy from the standpoint of characterization of higher-order panhandle structure. The secondary structure Bunyaviridae vRNA panhandles are usually envisioned in a simple manner featuring straightforward segments of paired and unpaired nucleotides. However, two G residues in the panhandle (at positions 6 and 17 from the 5′ end) are susceptible to digestion with RNase T1, which preferentially cleaves RNA at single-stranded G residues (Fig. 2 ▶). These nucleotides are predicted to be situated within double-stranded regions of the predicted panhandle. This suggests that the panhandle is likely to have a more complex secondary or tertiary structure than predicted by simple secondary structure modeling.
Hantavirus N protein has a general RNA helix destabilizing activity
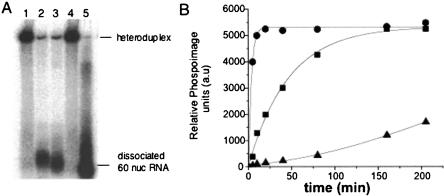
An RNA chaperone has the ability to destabilize and dissociate double-stranded RNA in a sequence independent manner. Thus, we carried out a standard helix destabilization experiment to determine whether N protein has such activity. A 60-nt-long, radioactively labeled RNA molecule was synthesized by in vitro transcription. The central 40 nt of this radioactively labeled RNA were annealed to a central region of an RNA ~2 kb in length. Ten bases on both the 5′ and 3′ sides would be noncomplementary and single stranded. Following hybridization to the 2-kb RNA the partially duplex RNA was purified and incubated with either hantavirus N protein, HIV-1 Gag protein, or BSA. As shown in Figure 3A ▶, both N protein and HIV-1 Gag efficiently released the radiolabeled RNA from the duplex. We also examined the kinetics of the RNA helix destabilizing activity by monitoring the amount of released RNA from the hybrid over time and determined the relative rate of helix dissociation with different amounts of N protein (Fig. 3B ▶). The rate of dissociation was similar for hantavirus N and for HIV-1 Gag. Similar results have been previously reported for known RNA helicases (Warrener and Collett 1995). Moreover, the rate of helix dissociation activity was dependent on N concentration. These data indicate that both proteins have an intrinsic helix destabilizing activity that can unwind double-stranded RNA. Similar RNA dissociation assays were also carried out using an alternative heteroduplex RNA containing a different sequence than that used in Figure 3 ▶. N and Gag both dissociated this alternative heteroduplex in a manner indistinguishable from that of the heteroduplex used in Figure 3 ▶ (data not shown).
FIGURE 3.
RNA helix destabilizing activity of hantavirus N and HIV-1 Gag. (A) A 60-nt-long oligoribonucleotide, labeled with α-32P-CTP was annealed to an RNA of ~2 kb. The resulting heteroduplex features a central base-paired region 40 nt in length and, for the short RNA, flanking nucleotides in single-stranded configuration. Dissociation of the two RNA would result in liberation of the 60-nt RNA from the heteroduplex. Samples were incubated as described in Materials and Methods for a total of 30 min. Lane 1, RNA heteroduplex without added protein; lane 2, RNA heteroduplex incubated with SNV N; lane 3, RNA heteroduplex incubated with HIV-1 Gag; lane 4, incubated with BSA; lane 5, the 60-nt-long oligoribonucleotide. Samples were incubated with 1:1 molar ration of Hetero duplex RNA:SNV N/HIV 1 Gag. Similar results were obtained in three or more separate experiments. (B) Kinetics of N-mediated heteroduplex dissociation using varying N:S segment RNA ratios. Samples were collected from a helix destabilization assay at the indicated times after initiation of the reaction and the dissociation of the heteroduplex was detected by gel analysis. (•) N:RNA = 6:1, (▪) N:RNA = 1:1, (▵) N:RNA = 1:6. The amount of released labeled RNA was quantified by phosphorimaging. Similar results were obtained from more than three separate experiments.
Some RNA chaperones, such as the DEAD-box RNA helicases, possess an ATPase activity that serves as a source of energy to unwind RNA (Mohr et al. 2002). However, there was no apparent difference in the helix destabilizing activity of either hantavirus N or HIV-1 Gag in the presence or absence of added ATP (data not shown). Thus, as discussed below, the energy required for N-mediated unfolding must arise from an alternative mechanism. This ATP-independent ability to dissociate RNA secondary structure is a general feature of RNA chaperones (Schroeder et al. 2004).
P-num analysis of hantavirus RNA
The formation of biologically functional higher order RNA structures is generally considered to be a thermodynamically favorable process that is driven by the RNA itself. The driving force behind the formation of biologically important RNA secondary structures could potentially be determined principally by average helix length; helix stability; number of helices/molecules; the size of hairpin, internal, and bulge loops; and overall stability. However, based on the examination of these RNA parameters using a large number of positive strand animal viruses, Palmenberg and Sgro (1997) found that none of these characteristics appeared to account for the generation of RNA secondary structures. Instead, the average “P-num” values for nucleotides is probably crucial for the formation of biologically meaningful RNA secondary structures (Palmenberg and Sgro 1997). Similarly, Jacobson and Zuker found that “well characterized” secondary structures in ribosomal and phage RNAs were composed of nucleotides of low P-num value (Jacobson and Zuker 1993; Zuker and Jacobson 1995). Thus, nucleotide P-num value is a predictive measure of pairing fidelity and can be used to identify RNA structures with the highest probability of assuming similar configurations among a series of energetically related structures (Jaeger et al. 1990).
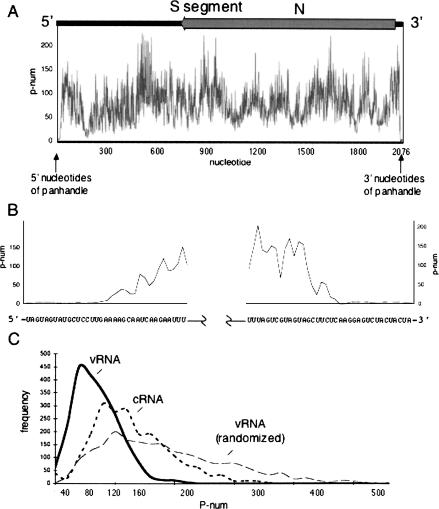
We carried out P-num analysis to examine the global topology of the hantavirus S segment vRNA. Figure 4A ▶ depicts the P-num values for the nucleotides of the hantavirus S-segment. The P-num values vary throughout the RNA and the average P-num of the S segment is 79 (i.e., on average, nucleotides of the S-segment could potentially form ~79 stable and reasonable interactions with complementary nucleotides elsewhere in the molecule). Thus a population of S segment vRNA molecules might be expected to contain RNAs of generally heterogeneous secondary structure. However, in contrast to the remainder of the genome, the termini of the S segment are composed of nucleotides with extraordinarily low P-num values. Moreover, the stable pairing partners of the 5′ terminus are those of the 3′ terminus such that the expected panhandle would be highly likely to form. Parallel analysis of the M and L segment RNAs yielded similar results (data not shown). In contrast to the native S segment sequence, a randomized S segment sequence contains significantly fewer nucleotides with low P-num values (Fig. 4C ▶). The strongly negative P-num skew of the S segment relative to its randomized counterpart is indicative of selective pressure for these preferred pairings. Moreover, the terminal nucleotides of the randomized S segment had high P-num values and would not be likely found in panhandle configuration (not shown). Comparison of the general P-num values of the minus strand vRNA with those of the plus stranded cRNA also indicate that vRNA is composed of nucleotides with lower P-num values than those in the cRNA (Fig. 4C ▶). This might reflect a greater requirement for overall order in the vRNA than in cRNA. However, this does not necessarily mean that the plus strand is devoid of functional higher order RNA structures.
FIGURE 4.
P-num analysis of the hantavirus S segment. The P-num values of the entire S segment along the length of the negative sense vRNA are shown in A. Plus stranded S segment cRNA contains the N gene. At the top of the plot is shown the location of the region complementary to the N gene in the vRNA S segment. An expanded view of the P-num values for the terminal nucleotides is shown in B. C compares the distribution of P-num values in S segment vRNA, S segment cRNA, and in a randomized vRNA sequence. The P-num values for the S segment vRNA are greater than those of the cRNA or the randomized vRNA sequence.
N protein unwinds the RNA panhandle
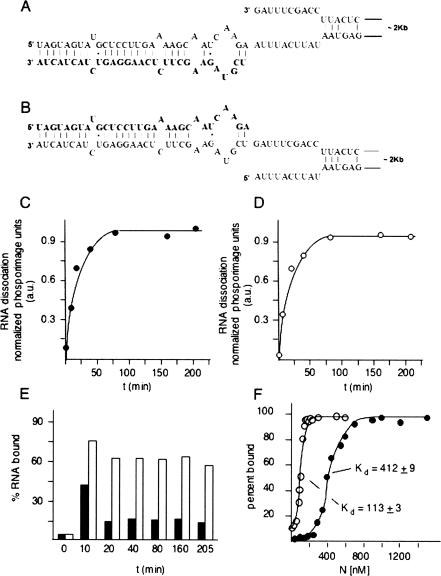
Trimeric N protein specifically interacts with the bunyavirus panhandle, and this specific interaction could have a role in either viral RNA encapsidation or initiation of genome replication or in both processes (Mir and Panganiban 2004). The latter process would be expected to require at least transient panhandle dissociation during replication initiation. Given the ability of N to nonspecifically dissociate RNA duplexes, N could hypothetically also play a role in dissociation of the panhandle. To test the idea that N can unwind the panhandle we generated RNA heteroduplexes that exactly correspond to the panhandle (Fig. 5A,B ▶). Previous analysis of N with a variety of vRNA panhandles indicate that a small panhandle composed of only the terminal 22 nt from the 5′ and 3′ termini is sufficient for high-affinity binding by N (Mir and Panganiban 2005). A radioactively labeled 32-nt RNA corresponding to the 3′ end of the viral genome was annealed to an S segment RNA containing a corresponding 32-nt deletion at its 3′ end (Fig. 5A ▶). We also generated a similar heteroduplex by annealing a short, labeled RNA corresponding to the 5′ end of the S segment with an S segment RNA containing a deletion of the 5′ end (Fig. 5B ▶). In a control experiment we found that heteroduplex formation required correct deletion of the appropriate terminus from the S segment; heteroduplexes did not form between the short 5′ and 3′ RNA with the full-length S segment (data not shown). Thus, both short, labeled RNAs almost certainly paired with the correct terminal partner on the long S segment derivative as shown in Figure 5A ▶. We carried out N-dependent dissociation assays with both heteroduplexes. The kinetics of dissociation for both terminal heteroduplexes are shown in Figure 5 ▶, C and D. N efficiently dissociated both panhandle heteroduplexes.
FIGURE 5.
Dissociation of the viral RNA panhandle by N. A depicts a heteroduplex between a 31-nt-long, radioactively labeled RNA (shown in bold lettering) corresponding to the nucleotides from the 3′ end of the viral S segment, and a nearly full length S segment containing a deletion at the 3′ end. Deletion of the 3′ terminal nucleotides was necessary for efficient heteroduplex formation. B shows a corresponding heteroduplex in which a short labeled RNA corresponding to the 5′ end of the genome is annealed to a nearly full-length S segment harboring a deletion of the 5′ terminal nucleotides. As with the duplex shown in A, deletion of the terminal nucleotides from the S segment is necessary for efficient formation of the heteroduplex. C and D show the kinetics of dissociation of the radioactively labeled, short, terminal RNAs from the heteroduplex for the duplex in A and in B, respectively. Helix dissociation experiments were carried out as in Figure 3 ▶ using a N:RNA ratio = 1:1. RNA filter binding data using the two heteroduplexes are shown in E. Each heteroduplex was incubated with N for the indicated period of time, and protein-dependent retention of the small, labeled RNA corresponding to the 3′ end of the panhandle (▪) or the 5′ end of the panhandle (□) was determined. Panel F shows the results of RNA filter binding with increasing concentrations of N and either the 3′ terminal RNA (•) or the 5′ terminal RNA (○). Measured dissociation constants for both RNAs are also indicated.
N binds stably and at high affinity to RNA containing the panhandle (Mir and Panganiban 2005). Since N also dissociates the panhandle it seemed likely that N remains stably associated with one terminus or both termini following dissociation. To examine maintenance of the interaction of N with viral RNA following panhandle dissociation, we incubated N using parallel reactions containing heteroduplexes with either the short, labeled 5′ RNA or 3′ RNA. At progressive time points, samples were then passed through nitrocellulose filters. Under the filter binding conditions that were used RNA alone does not bind to the filters but can be retained on filters by virtue of stable interaction with N. Retention of the labeled 5′ or 3′ terminus on filters is shown in Figure 5E ▶. At a relatively short time point (10 min) following incubation with N both the 5′ and 3′ termini are retained on filters. This corresponds to a time point where there is relatively little panhandle dissociation, as evidenced by the observed kinetics of heteroduplex dissociation (Fig. 5A,B ▶). However, over more protracted times of incubation there is marked preferential retention of the 5′ terminus on filters relative to the 3′ terminus indicative of stable longer term association of N with the 5′ terminus. Taken together, these helix dissociation and RNA filter-binding data indicate that high-affinity interaction of N with the panhandle leads to dissociation and that N remains bound to the 5′ end of the genome. This is also consistent with extensive data indicating that the preferred substrate for N is the panhandle. Such panhandle dissociation may play a role in the initiation of genome replication by the viral RNA-dependent RNA polymerase.
To further investigate the association of N with the termini, we carried out the filter-binding studies of 32 nt from both 5′ and 3′ ends of SNV S segment RNA with N protein. The corresponding binding profiles are shown in Figure 5F ▶. The dissociation constant (Kd) for the 5′ and 3′ ends indicate that the 5′ end of the viral RNA is recognized at higher affinity than is the 3′ end. These data are consistent with previous observations (Gott et al. 1993; Richmond et al. 1998; Severson et al. 1999, 2001; Osborne and Elliott 2000), and the notion that high-affinity recognition of the panhandle is followed by panhandle dissociation and maintenance of N with the 5′ end of the viral RNA.
DISCUSSION
Based on multiple in vitro criteria, hantavirus N protein is an RNA chaperone. Thus, N is potentially able to transiently reorder higher order RNA structure to allow escape from kinetic traps and to enable the formation of functional higher order structures such as the viral RNA panhandle. The general paradigm for in vivo replication of negative-strand RNA viruses includes the temporal regulation and synthesis of three types of viral RNAs. Following infection, the viral RNA-dependent RNA polymerase (RdRp), which was present in the infecting virion, transcribes the vRNA to produce mRNA. This results in the de novo expression of both additional RdRp and N (or its equivalent). Subsequent anti-genome RNA and vRNA synthesis takes place following sufficient accumulation of N. Both vRNA and anti-genome synthesis and their assembly with N appear to take place concurrently, and the templates for this synthesis are associated with significant amounts of N (Raju and Kolakofsky 1986, 1987).
How might the in vitro RNA chaperone activity of N that we have described function during virus replication in vivo? First, conventional RNA chaperone activity, which would help in escape from intramolecular kinetic traps, could be important for the correct handling and efficient translation of viral mRNA since, in contrast to vRNA, mRNA is not substantially found in complexes with N. Second, the RNA chaperone activity of N might facilitate correct in vivo panhandle formation of vRNA in the nucleoprotein complex, as reflected by the ability of N to facilitate panhandle formation in vitro (Fig. 2 ▶). Along these lines it is pertinent that the termini of intracellular vRNA in bunyavirus-infected cells are hydrogen-bonded and in panhandle configuration even in nucleoprotein complexes based on cross-linking studies with psoralen (Raju and Kolakofsky 1989). Though intracellular nucleocapsids harboring vRNA are stable, this protein–RNA complex also appears to be dynamic, as evidenced by the ability of specific DNA oligonucleotides to invade the viral nucleocapsid to generate an RNA–DNA substrate that can be cleaved with RNase H (Kolakofsky et al. 1987). Thus, transient reversible dissociation of N–RNA, and intermolecular N–N interaction within the nucleocapsid may be required for successful N-mediated panhandle formation. Third, the ability of N to dissociate RNA duplexes, including the panhandle (Fig. 5 ▶), is consistent with a subsequent role for N in initiation of RNA synthesis, as described in more detail below. Fourth, the chaperone activity of N could be required for local dissociation of higher order RNA structure on newly synthesized RNA prior to assembly into nucleocapsids. RNA dissociation may be intrinsically important for efficient in vivo loading of N onto vRNA and for correct intermolecular N–N interaction during replication. Finally, it is possible that the ability of N to nonspecifically dissociate RNA duplexes in vitro is related to a specific in vivo role, such as panhandle dissociation during genome replication, and that N does not function as a general non specific RNA chaperone in vivo. However, it is noteworthy that the general in vitro RNA chaperone activity of N is very similar to that of HIV-1 NC, an established RNA chaperone.
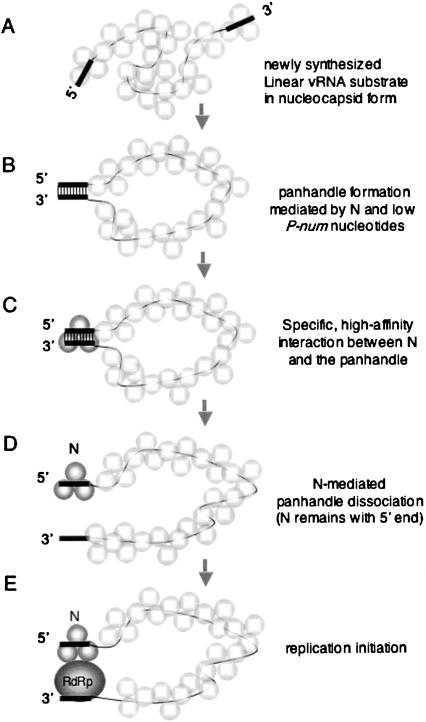
The in vitro interaction of hantavirus N protein with RNA provides a model that highlights key events that take place generally during RNA folding. The viral genome serves as an RNA substrate and features a functional biological structure (the panhandle), in this case composed of nucleotides separated by exceptionally long distance. Formation of the panhandle is driven by both N and the RNA sequence itself. In particular, those nucleotides that comprise the panhandle have very few possible pairing partners (i.e., nucleotides with low P-num values). In vitro formation of this functional structure is substantially hindered by alternative, locally stable, intramolecular interactions (kinetic traps). N protein (an RNA chaperone) then facilitates escape of the RNA substrate from kinetic traps by repetitive nonspecific helix destabilization to enable the opportunity for the panhandle to form. In vivo, the fact that vRNA is in the form of a nucleocapsid would likely preclude the formation of extensive kinetic traps. However, the chaperone function of N may nonetheless facilitate panhandle formation given the dynamic state of the nucleocapsid. Free N (not associated with the nucleocapsid) could function in panhandle formation. Alternatively, N molecules that are resident within the nucleocapsid could facilitate panhandle formation. In addition to its function as an RNA chaperone, N binds at high affinity and specifically with the RNA panhandle. The essential features of the chaperone activity of N, the specific recognition of the panhandle, and subsequent panhandle dissociation are presented in a temporal model (Fig. 6 ▶).
FIGURE 6.
N- and RNA-facilitated panhandle formation, specific recognition of the vRNA panhandle by N, and N-mediated panhandle dissociation. (A) vRNA synthesis results in an initial RNA with termini that are not base-paired and with associated N in the form of a nucleocapsid. Within the nucleocapsid there is both N–RNA interaction as well as intermolecular association between individual N trimers. (B) The RNA chaperone activity of N, in conjunction with the low P-num nucleotides that comprise the termini, facilitates panhandle formation. N that is intrinsically associated with the nucleocapsid or, alternatively, free N might mediate panhandle formation. (C) Following panhandle formation, N protein binds in a structure-specific manner, with high affinity, to the panhandle. In the figure the trimeric N that is stably bound to the panhandle is depicted in darker shading than the other associated N molecules simply for emphasis. (D) N protein mediates the unwinding of the panhandle and remains associated with the nucleotides at the 5′ end of the molecule. (E) Dissociation of the panhandle likely enables initiation of replication mediated by the viral RNA dependent RNA polymerase (RdRp). In the figure, N, which is bound to the 5′ end of the panhandle, is shown in association with the RdRp. This association is hypothetical.
Hantavirus N protein contains a prototypical “disordered” domain that likely functions during RNA chaperone activity. Recently, Tompa and Csermely (2004) made the intriguing observation that the theoretical disorder among the known RNA chaperones far surpasses that of any other class of proteins. Disordered regions in polypeptides are unlikely to assume uniform configuration in a population of protein molecules. This disorder could serve two important functions: First, lack of consistent structure would enable chaperones to interact with a variety of substrate molecules, at relatively low affinity, consistent with their ability to unfold many substrates. Second, binding of the disordered chaperone region to the misfolded RNA would result in transient ordering of the disordered region and concomitant unfolding of the bound RNA in a process of “reciprocal entropy transfer.” Thus, simultaneous ordering of the chaperone would provide the thermodynamic cost for unfolding the misfolded RNA.
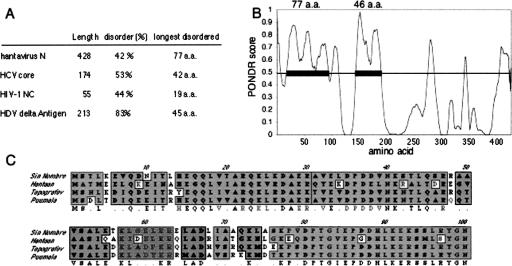
We used a “predictor of natural disordered regions” (PONDR) (Li et al. 1999) to characterize the theoretical disorder of the N protein and to compare N to the three other known viral RNA chaperones: HIV-1 nucleocapsid protein, hepatitis C virus core protein, and hepatitis delta antigen. All four proteins are highly disordered. Delta antigen features the highest overall percent composition of disordered residues. Hantavirus N contains the longest contiguous stretch of disordered 77 amino acid residues near the N terminus as illustrated in the PONDR plot (Fig. 7A,B ▶). Moreover, similar plots for diverse hantavirus species indicate that all have similar disordered regions in the N-terminal 94 amino acids (Fig. 7C ▶). This region of the hantavirus N protein also exhibits relatively high sequence conservation. This likely reflects a requirement for maintenance of this disordered region.
FIGURE 7.
Disorder in hantavirus N protein. In A the relative disorder of hantavirus N protein is compared to that of the three other known RNA chaperone proteins. The “PONDR” plot for the N sequence is shown in panel B. Amino acid sequences that score >0.5 are considered likely to be disordered. The SNV N protein has two putative disordered domains that are 77 and 46 amino acids in length. A ClustalW alignment of the putative disordered region of four diverse hantavirus species is shown in panel C. Shaded amino acids are conserved, while unshaded amino acids are divergent.
N was able to dissociate RNA duplexes even at a low chaperone:RNA ratio, although at a relatively slow rate compared with higher chaperone:RNA ratios (Fig. 3 ▶). It has been observed that a significantly higher chaperone:RNA ratio is required for the generation of annealed RNA products in assays to examine HIV-1 nucleocapsid or hepatitis C virus core function (Williams et al. 2002; Cristofari et al. 2004; Hargittai et al. 2004). These latter observations are consistent with a requirement for interaction between multiple HIV NC or HCV core molecules in the formation of annealed products.
It is likely that the majority, or perhaps all, of the RNA viruses make use of similar viral or cellular RNA chaperones during their replication. Based on the precedent with bunyaviruses, with the retroviruses, and with hepatitis C virus, it would seem likely that there is selective advantage to incorporating an RNA dissociation activity within the principle nucleocapsid peptide. For these viruses the nucleocapsid peptide necessarily interacts with the RNA genome with specificity during encapsidation and during replication. Given the close association between the nucleocapsid protein and the viral RNA, it would be efficient for this protein to harbor an intrinsic RNA chaperone activity to manage the architecture of the viral RNA genome.
MATERIALS AND METHODS
Hantavirus N protein and S segment RNA
The viral N protein and S segment RNA were derived from Sin Nombre hantavirus strain 77734 (Botten et al. 2000). N protein was expressed in E. coli, purified and prepared as described previously (Mir and Panganiban 2004). For all of the experiments presented we used this N-terminal GST-N fusion protein. However, we also used a bacterially expressed N protein containing a histidine 6-tag at the C terminus of N to verify the presented experiments. Both N proteins all yielded similar data. Based on previous analysis of hantavirus N protein, trimeric N exhibits specificity for the preformed viral RNA panhandle. However, the RNA chaperone activity of N was observed both with purified trimeric protein as well as with N preparations containing monomeric as well as trimeric molecules. The data presented here are derived from experiments containing a mixture of monomers, dimers, and trimers.
DNA containing the full length S segment RNA molecule with a proximally located T7 promoter was generated by PCR using an “upstream” primer containing a T7 promoter sequence, a “downstream” primer that overlaps the terminal sequences of the S segment transcription unit, and pAD-S as template as described previously (Mir and Panganiban 2004). The resulting DNA containing the entire S segment transcription unit was gel-purified and used directly in transcription reactions with T7 RNA polymerase (MBI Fermentase) in the presence of α-[32P]CTP. For the synthesis of an S segment RNA lacking the 3′ terminal nucleotides, we used a parallel strategy, such that the amplified PCR product contained a sequence lacking the 42 terminal nt at the 3′ end. Following transcription the DNA template was removed by digestion with DNase 1 and the RNA was purified using RNeasy kit (Invitrogen). Purified RNA was stored at −20°C in 25 μL aliquots for up to 2 wk.
To end-label RNA for analysis by RNase T1 digestion, a DNA template was used where the initiating nucleotide used in transcription was a G, followed by the SNV nucleotide sequence. End-labeling was performed using transcription reactions in the presence of γ-P32 GTP.
Sucrose density gradient centrifugation of RNA
Prior to sedimentation, RNA was heat denatured and renatured or treated with N protein. Twenty μL samples of labeled RNA in buffer A (40 mM HEPES at pH 7.4, 80 mM NaCl, 20 mM KCl, and 1 mM DTT) were prepared. The final RNA concentration was 3.6 nM. For heat denaturation the RNA was heat treated at 95°C for 2 min and kept at room temperature for 2.5 h to allow the renaturation. For treatment with N protein RNA that had not been heat treated was incubated with 450 nM N protein in buffer A for 2.5 h at room temp. Control samples lacking N protein were incubated in parallel with the N-treated samples. Following incubation RNA was purified using RNeasy and layered onto 10%–60% (w/v) linear sucrose gradients containing buffer A. RNA was centrifuged at 30,000 rpm at 4°C for 22 h in an SW40 rotor. Fractions (0.5 mL) were collected from the gradient. To monitor the presence of S segment RNA in different fractions, 20 μL aliquots from each fraction were monitored using a Scintillation counter. To examine the integrity and length of the RNA, an aliquot of RNA from each peak fraction was run on 5% acrylamide, 8 M urea denaturing gel to estimate the size of RNA.
Spectroscopic measurements
RNA from peaks 1 and 2 was characterized spectroscopically using the fluorescent dye acridine orange. This dye is extensively used for analytical purposes in nucleic acids (Porumb and Petrescu 1986). Acridine orange was dissolved in double distilled water and its concentration was determined by molar absorption coefficient of E470 = 4.33 × 104M−1cm−1. The fluorescence spectra (500–600 nm, λext = 474 nm) of 120 nM acridine orange in buffer A was determined in the presence and the absence of 95 pM S segment RNA from either peak 1 or peak 2. Acridine orange was incubated with RNA for 15 min before fluorescence spectra were recorded.
RNase T1 digestion analysis
End-labeled RNA from different peak fractions of the gradient was purified by RNeasy kit (Qiagen). End-labeled RNA was digested with one unit of RNase T1 (Ambion) in 100 mM Tris (pH 7), 1 M KCl, 100 mM MgCl2 at 37°C for 5 min. The reaction was terminated by the addition of 20 μL of inactivation/precipitation buffer (Ambion) per 10 μL reaction. The mixture was incubated at −20°C for 15 min and centrifuged at 13,200 rpm for 15 min. The pellet was washed with 70% ethanol, air-dried, and dissolved in 7 μL of gel loading buffer. The mixture was heated at 95°C for 5 min and fractionated on 20% acrylamide/8 M urea sequencing type gels. The gels were exposed to PhosphorImager screens for visualization.
RNA helicase assay
Standard helicase reactions (20 μL) were carried out as follows: We synthesized a small RNA molecule, 60 nt in length by in vitro transcription reaction with T7 polymerase, radiolabeled with α-P32-CTP. The middle 40 bases of this short RNA were complementary to SNV S segment RNA corresponding to the nucleotides 1440–1480 of the RNA. The radiolabeled short RNA and unlabeled long RNA mixed in a 10:1 ratio in buffer A, heated at 95°C for 3 min and incubated at RT for 3 h. Unhybridized radiolabeled RNA was removed from the reaction mixture by using RNeasy. The hybridized RNA was gel-purified and sued as substrates in the helix destabilization reactions. Twenty microliter reactions containing 10 nM N protein/HIV1 Gag protein, 10 nM RNA substrate in binding buffer containing 1 mM Mg2+ in the presence or the absence of ATP were incubated at 37°C for different time intervals and terminated by addition of 4 μL of RNA sample buffer (100 mM Tris HCl at pH 7.4, 50 mM EDTA, 0.1% triton X-100, 0.5% SDS, 50% glycerol, and 0.1% bromophenol blue). The products were fractionated on 12% SDS PAGE. Gels were exposed to PhosphorImager screens for ~3 h and analyzed. Helix destabilization activity was quantified by monitoring the intensity of bands corresponding to released short RNA and helicase substrate at different time points.
RNA filter binding assay
Thirty-two-nucleotide-long RNAs corresponding to either 5′ or 3′ terminus of SNV S segment RNA were synthesized by in vitro T7 transcription and radiolabeled with [α-32P]CTP as described above. All binding reactions were carried out in binding buffer (40 mM HEPES at pH 7.4, 80 mM NaCl, 20 mM KCl, 1 mM dithiothreitol) at a constant concentration of RNA with increasing concentration of N protein. Reaction mixtures were incubated at room temperature for 45 min and filtered through nitrocellulose filters under vacuum. Filters were washed with 10 mL of binding buffer, dried, and retained radioactivity monitored by scintillation counting. Nonspecific retention of RNA was monitored by filtering the complete reaction mixture in the absence of protein. Dissociation constants were calculated by fitting the experimental data points into either hyperbolic or sigmoidal curve using Origin 6 program (Microcal). The apparent dissociation constant (Kd) corresponds to the concentration of N protein required to obtain the half saturation, assuming the complex formation obeys a simple bimolecular equilibrium. We assumed that plateau in the binding profile represents complete binding of RNA, to allow the calculations at half saturation.
RNA secondary structure and P-num analysis
mFOLD analysis of Sin Nombre hantavirus strain 77734 used the energy parameters of Turner (Jaeger et al. 1989; Zuker 1989) at 37°C. A randomized version of this sequence (identical base composition) was generated after five iterations of SHUFFLESEQ (EMBOSS), then computed by mFOLD using the same energy parameters as the genomic RNA. The P-num value for each base in each fold was returned from the REGION file for a +12 Kcal window.
Acknowledgments
We thank Dave Bear, Brian Hjelle, Dave Peabody, and Jesse Summers for helpful discussion and for critical evaluation of the manuscript. This work was supported by the University of New Mexico School of Medicine Research Allocation Committee and research grant (R21AI059330) from the NIH.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2101906.
REFERENCES
- Barr, J.N. and Wertz, G.W. 2005. Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J. Virol. 79: 3586–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botten, J., Mirowsky, K., Kusewitt, D., Bharadwaj, M., Yee, J., Ricci, R., Feddersen, R.M., and Hjelle, B. 2000. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus). Proc. Natl. Acad. Sci. 97: 10578–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari, G. and Darlix, J.L. 2002. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 72: 223–268. [DOI] [PubMed] [Google Scholar]
- Cristofari, G., Ivanyi-Nagy, R., Gabus, C., Boulant, S., Lavergne, J.P., Penin, F., and Darlix, J.L. 2004. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 32: 2623–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott, P., Stohwasser, R., Schnitzler, P., Darai, G., and Bautz, E.K. 1993. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology 194: 332–337. [DOI] [PubMed] [Google Scholar]
- Hargittai, M.R., Gorelick, R.J., Rouzina, I., and Musier-Forsyth, K. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 337: 951–968. [DOI] [PubMed]
- Herschlag, D. 1995. RNA chaperones and the RNA folding problem. J. Biol. Chem. 270: 20871–20874. [DOI] [PubMed] [Google Scholar]
- Herschlag, D., Khosla, M., Tsuchihashi, Z., and Karpel, R.L. 1994. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 13: 2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewlett, M.J., Pettersson, R.F., and Baltimore, D. 1977. Circular forms of Uukuniemi virion RNA: An electron microscopic study. J. Virol. 21: 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, A.B. and Zuker, M. 1993. Structural analysis by energy dot plot of a large mRNA. J. Mol. Biol. 233: 261–269. [DOI] [PubMed] [Google Scholar]
- Jaeger, J.A., Turner, D.H., and Zuker, M. 1989. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. 86: 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1990. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 183: 281–306. [DOI] [PubMed] [Google Scholar]
- Jiang, W., Hou, Y., and Inouye, M. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272: 196–202. [DOI] [PubMed] [Google Scholar]
- Kolakofsky, D., Bellocq, C., and Raju, R. 1987. The translational requirement for La Crosse virus S-mRNA synthesis. Cold Spring Harb. Symp. Quant. Biol. 52: 373–379. [DOI] [PubMed] [Google Scholar]
- Li, X., Romero, P., Rani, M., Dunker, A.K., and Obradovic, Z. 1999. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 10: 30–40. [PubMed] [Google Scholar]
- Mir, M.A. and Panganiban, A.T. 2004. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 78: 8281–8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2005. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J. Virol. 79: 1824–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, S., Stryker, J.M., and Lambowitz, A.M. 2002. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 109: 769–779. [DOI] [PubMed] [Google Scholar]
- Negroni, M. and Buc, H. 1999. Recombination during reverse transcription: An evaluation of the role of the nucleocapsid protein. J. Mol. Biol. 286: 15–31. [DOI] [PubMed] [Google Scholar]
- Obijeski, J.F., Bishop, D.H., Murphy, F.A., and Palmer, E.L. 1976. Structural proteins of La Crosse virus. J. Virol. 19: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, J.C. and Elliott, R.M. 2000. RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J. Virol. 74: 9946–9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg, A.C. and Sgro, J.Y. 1997. Topological organization of picornaviral genomes: Statistical prediction of RNA structural signals. Semin. Virol. 8: 231–241. [Google Scholar]
- Pannone, B.K., Xue, D., and Wolin, S.L. 1998. A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 17: 7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson, R.F. and von Bonsdorff, C.H. 1975. Ribonucleoproteins of Uukuniemi virus are circular. J. Virol. 15: 386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius, B.W. and Berg, P. 1992. Rapid assembly and disassembly of complementary DNA strands through an equilibrium intermediate state mediated by A1 hnRNP protein. J. Biol. Chem. 267: 13815–13818. [PubMed] [Google Scholar]
- Porumb, H. and Petrescu, I. 1986. Interaction with mitochondria of the anthracycline cytostatics adriamycin and daunomycin. Prog. Biophys. Mol. Biol. 48: 103–125. [DOI] [PubMed] [Google Scholar]
- Raju, R. and Kolakofsky, D. 1986. Inhibitors of protein synthesis inhibit both La Crosse virus S-mRNA and S genome syntheses in vivo. Virus Res. 5: 1–9. [DOI] [PubMed] [Google Scholar]
- ———. 1987. Unusual transcripts in La Crosse virus-infected cells and the site for nucleocapsid assembly. J. Virol. 61: 667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1989. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J. Virol. 63: 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein, A., Henderson, L.E., and Levin, J.G. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem. Sci. 23: 297–301. [DOI] [PubMed] [Google Scholar]
- Richmond, K.E., Chenault, K., Sherwood, J.L., and German, T.L. 1998. Characterization of the nucleic acid binding properties of tomato spotted wilt virus nucleocapsid protein. Virology 248: 6–11. [DOI] [PubMed] [Google Scholar]
- Schroeder, R., Barta, A., and Semrad, K. 2004. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 5: 908–919. [DOI] [PubMed] [Google Scholar]
- Severson, W., Partin, L., Schmaljohn, C.S., and Jonsson, C.B. 1999. Characterization of the Hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 274: 33732–33739. [DOI] [PubMed] [Google Scholar]
- Severson, W.E., Xu, X., and Jonsson, C.B. 2001. cis-Acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75: 2646–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson, W., Xu, X., Kuhn, M., Senutovitch, N., Thokala, M., Ferron, F., Longhi, S., Canard, B., and Jonsson, C.B. 2005. Essential amino acids of the hantaan virus N protein in its interaction with RNA. J. Virol. 79: 10032–10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, M.J., Lorimer, G.H., and Thirumalai, D. 1996. Chaperonin-facilitated protein folding: Optimization of rate and yield by an iterative annealing mechanism. Proc. Natl. Acad. Sci. 93: 4030–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa, P. and Csermely, P. 2004. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 18: 1169–1175. [DOI] [PubMed] [Google Scholar]
- Treiber, D.K. and Williamson, J.R. 2001. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 11: 309–314. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi, Z., Khosla, M., and Herschlag, D. 1993. Protein enhancement of hammerhead ribozyme catalysis. Science 262: 99–102. [DOI] [PubMed] [Google Scholar]
- Wang, C.C., Chang, T.C., Lin, C.W., Tsui, H.L., Chu, P.B., Chen, B.S., Huang, Z.S., and Wu, H.N. 2003. Nucleic acid binding properties of the nucleic acid chaperone domain of hepatitis delta antigen. Nucleic Acids Res. 31: 6481–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrener, P. and Collett, M.S. 1995. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol. 69: 1720–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, K.M. 1997. Protein-facilitated RNA folding. Curr. Opin. Struct. Biol. 7: 336–342. [DOI] [PubMed] [Google Scholar]
- Williams, M.C., Gorelick, R.J., and Musier-Forsyth, K. 2002. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein’s nucleic acid chaperone function. Proc. Natl. Acad. Sci. 99: 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Severson, W., Villegas, N., Schmaljohn, C.S., and Jonsson, C.B. 2002. The RNA binding domain of the hantaan virus N protein maps to a central, conserved region. J. Virol. 76: 3301–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244: 48–52. [DOI] [PubMed] [Google Scholar]
- Zuker, M. and Jacobson, A.B. 1995. “Well-determined” regions in RNA secondary structure prediction: Analysis of small subunit ribosomal RNA. Nucleic Acids Res. 23: 2791–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]