Abstract
The Noc1–4p proteins were previously reported to be involved in intranuclear and nucleocytoplasmic transport of pre-ribosomes. Using fold recognition and structural modeling, we show that Noc1–4p are largely comprised of α-helical repeats similar to HEAT repeats. Because other HEAT-repeat proteins play key roles in transport processes, this finding provides a plausible mechanistic explanation for the function of the Noc proteins.
Keywords: rRNA, ribosome formation, ribonucleoproteins, molecular models, yeast
Eukaryotic ribosomes are assembled in the nucleolus in a series of highly coordinated events (for review, see Fatica and Tollervey 2002; Tschochner and Hurt 2003). In recent years, several reports have described around 140 nonribosomal factors involved in this multistep process (Harnpicharnchai et al. 2001; Dragon et al. 2002; Fatica et al. 2002; Grandi et al. 2002; Nissan et al. 2002). An important aspect of ribosome subunit synthesis is their transport from the nucleolus to nucleoplasm and then to the cytoplasm. Two recent reports identified a family of pre-ribosome-associated transport factors termed Noc proteins (Milkereit et al. 2001, 2003). These proteins are involved in intranuclear transport and export of the pre-60S subunit (Noc1/2/3p) and nuclear export of the pre-40S subunit (Noc4p). Moreover, Noc3p also plays a key role in the initiation of DNA replication (Zhang et al. 2002). The biochemical and genetic characterization of the Noc proteins (Milkereit et al. 2001, 2003) did not, however, reveal the mechanism(s) by which they mediate ribosomal subunit transport.
Limited sequence similarity between the Noc1/3/4p proteins over a short region of ~45 residues has been noted previously (Milkereit et al. 2001, 2003). Using profile consistency analysis (Pei et al. 2003), we have extended this alignment into a larger Noc domain (Fig. 1 ▶). This extended similarity provides further evidence that these proteins have related functions despite their nonredundancy. Orthologs of Noc proteins are present in all higher eukaryotes and were used as starting queries for PSI-BLAST searches (Altschul et al. 1997). We could not identify convincing sequence similarity to proteins of known structure or function. The search of the hidden Markov model (HMM) database of protein families (Bateman et al. 2000) resulted in Noc1p and Noc3p matching CBF/Mak21 domain (PF03914). The annotation for this domain, however, contains no additional information beyond what is already known about Noc proteins. Multiple alignments of Noc proteins were then used to train HMMs and scan the protein database, but again, no informative matches were found. We therefore resorted to fold recognition using the 3D-PSSM server (http://www.sbg.bio.ic.ac.uk/~3dpssm/; Kelley et al. 2000). With confidence in the range 70%–90%, the server returned predictions that all Noc proteins share structural similarity with proteins containing HEAT/Armadillo repeats (Andrade and Bork 1995; Andrade et al. 2001). This prediction was confirmed using the consensus of multiple fold recognition methods at the 3D-Jury metaserver (Ginalski et al. 2003). In a follow-up to this prediction, we compared Noc proteins to HMMs trained on all major classes of HEAT-repeat proteins (Andrade et al. 2001). Noc2p and Noc4p had no sequence similarity to known HEAT repeats with E < 10, while Noc1p and Noc3p had five and two HEAT repeats, respectively, with statistically insignificant E-values (E = 6.9 for Noc1p and E = 1 for Noc3p). We conclude that at the sequence level Noc proteins do not have convincing sequence similarity to any of the major classes of HEAT repeats previously described (Andrade et al. 2001).
FIGURE 1.
Multiple sequence alignment of Noc1/3/4p proteins. Numbers flanking the alignment correspond to parts of proteins that were aligned. Numbers in parentheses indicate lengths of sequences that were omitted due to insertions or deletions. Protein names are separated by an underscore from species abbreviations: YEAST, Saccharomyces cerevisiae; SCHPO, Schizosaccharmyces pombe; HUMAN, Homo sapiens; ARATH, Arabidopsis thaliana. Letters on the consensus line are: s, small residues; t, tiny; b, big; h, hydrophobic; a, aromatic; l, aliphatic; p, polar; c, charged; −, negatively charged; +, positively charged. Individual residues with more than 80% identity in the entire alignment are colored yellow and shown as capital letters on the consensus line.
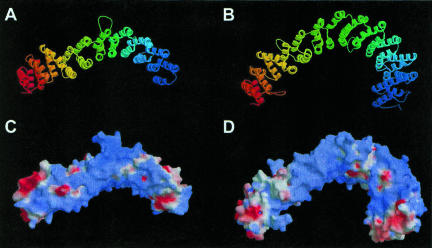
The plausibility of the fold recognition prediction and the degree of structural similarity with known HEAT-repeat proteins were further tested by building 3D models of all Noc proteins (Sali and Blundell 1993). Shown in Figure 2 ▶ are models for Noc1p (residues 349–771) and Noc2p (residues 146–699). The models were evaluated using quality criteria for comparative modeling (Sanchez and Sali 1998). A probability (pG) that estimates the reliability of the overall fold of protein models was calculated for each Noc protein. Models with pG > 0.7 are considered to have a correct overall fold (Sanchez and Sali 1998), although they will not be correct in all details. According to these criteria, models of Noc proteins are very reliable (pGNoc1p = 0.97; pGNoc2p = 0.96; pGNoc3p = 0.94; pGNoc4p = 0.78). This is particularly convincing because the Noc proteins show only 8–13% identity with the templates used for modeling, and in this range of sequence identity high pG values are unlikely unless the structural relationship between model and the template is genuine. The locations of the predicted HEAT repeat elements in the primary sequence of Noc1p are shown in Supplementary Figure S1 (http://www.homepage.montana.edu/~mdlakic/heat_Noc1p_suppl_FIG1.html).
FIGURE 2.
(A,B) 3D-models of Noc1p and Noc2p. Noc1p model (residues 349–771) and Noc2p model (residues 146–699) were built with MODELLER (Sali and Blundell 1993) using the PR65/A subunit of protein phosphatase 2A (Groves et al. 1999) as a template. Automatic alignments obtained by 3D-PSSM (Kelley et al. 2000) were corrected manually to minimize insertions and deletions within secondary structure elements of the template. Colors change from blue at the N-terminus to red at the C-terminus. (C,D) Electrostatic potential representations of Noc1p and Noc2p models. Electrostatic surface potential was calculated using GRASP (Nicholls et al. 1991). Blue and red colors correspond to positive and negative potential, respectively. Strong clusters of positive charge suggest that these proteins may interact with other negatively charged molecules, rRNA being an obvious candidate.
In addition to the Noc proteins, we have identified four other essential HEAT-repeat proteins that are associated with yeast pre-ribosomes: Rrp12p, Sda1p, Utp10p, and Utp20p (Oeffinger et al. 2004). It was recently estimated that at least 0.2% of eukaryotic proteins have HEAT or Armadillo repeats (Andrade et al. 2001). This abundance may reflect the functional versatility of proteins with HEAT repeats. The PR65/A subunit of protein phosphatase 2A (PP2A) functions as a scaffold for assembly of the catalytic and regulatory subunits (Groves et al. 1999), while the importin-β/karyopherin-β (imp-β/kap-β) family act as molecular transporters across the nuclear envelope (Gorlich et al. 1997; Malik et al. 1997; Chook and Blobel 1999; Cingolani et al. 1999; Kobe et al. 1999; Vetter et al. 1999). Many assembly and transport steps are critical for ribosome biogenesis, potentially involving multiple HEAT-repeat proteins.
Ribosome synthesis dominates nucleocytoplasmic transport in yeast, with each nuclear pore complex (NPC) importing ~1000 ribosomal proteins and exporting ~25 ribosomal subunits per minute (for review, see Jorgensen et al. 2004). Efficient import of ribosomal proteins relies on multiple, partially redundant members of the imp-β/kap-β family, the founding member of which has a HEAT-repeat structure (Chook and Blobel 1999; Cingolani et al. 1999; Vetter et al. 1999). Ribosome export is also known to require a member of the imp-β/kap-β family, Crm1p/Xpo1p (for review, see Johnson et al. 2002; Tschochner and Hurt 2003), but it is unlikely that single extrinsic factor mediates the export of the very large ribosomal subunits. We therefore predict that efficient subunit export will require multiple transport factors. At least one of the other HEAT-repeat proteins we have identified, Rrp12p, is required for ribosomal subunit export (Oeffinger et al. 2004).
Here we have reported that pre-ribosomes are associated with a family of divergent HEAT-repeat proteins, which are required for ribosomal subunit transport. The Noc proteins are structurally, and potentially functionally, related to general transport factors, despite lacking detectable sequence similarity to known HEAT-repeat proteins. Future studies will determine the relative contributions of these versatile proteins to ribosome assembly and subunit transport.
Acknowledgments
M.D. was supported in part by a Special Fellowship from the Leukemia & Lymphoma Society. D.T. is supported by the Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5184704.
REFERENCES
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M.A. and Bork, P. 1995. HEAT repeats in the Huntington’s disease protein. Nat. Genet. 11: 115–116. [DOI] [PubMed] [Google Scholar]
- Andrade, M.A., Petosa, C., O’Donoghue, S.I., Muller, C.W., and Bork, P. 2001. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309: 1–18. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., and Sonnhammer, E.L. 2000. The Pfam protein families database. Nucleic Acids Res. 28: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook, Y.M. and Blobel, G. 1999. Structure of the nuclear transport complex karyopherin-β2-Ran•GppNHp. Nature 399: 230–237. [DOI] [PubMed] [Google Scholar]
- Cingolani, G., Petosa, C., Weis, K., and Muller, C.W. 1999. Structure of importin-β bound to the IBB domain of importin-α. Nature 399: 221–229. [DOI] [PubMed] [Google Scholar]
- Dragon, F., Gallagher, J.E., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A. and Tollervey, D. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14: 313–318. [DOI] [PubMed] [Google Scholar]
- Fatica, A., Cronshaw, A.D., Dlakic, M., and Tollervey, D. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9: 341–351. [DOI] [PubMed] [Google Scholar]
- Ginalski, K., Elofsson, A., Fischer, D., and Rychlewski, L. 2003. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics 19: 1015–1018. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., Dabrowski, M., Bischoff, F.R., Kutay, U., Bork, P., Hartmann, E., Prehn, S., and Izaurralde, E. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138: 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., Rybin, V., Bassler, J., Petfalski, E., Strauss, D., Marzioch, M., Schafer, T., Kuster, B., Tschochner, H., Tollervey, D., et al. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10: 105–115. [DOI] [PubMed] [Google Scholar]
- Groves, M.R., Hanlon, N., Turowski, P., Hemmings, B.A., and Barford, D. 1999. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96: 99–110. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai, P., Jakovljevic, J., Horsey, E., Miles, T., Roman, J., Rout, M., Meagher, D., Imai, B., Guo, Y., Brame, C.J., et al. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8: 505–515. [DOI] [PubMed] [Google Scholar]
- Johnson, A.W., Lund, E., and Dahlberg, J. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27: 580–585. [DOI] [PubMed] [Google Scholar]
- Jorgensen, P., Tyers, M., and Warner, J.R. 2004. Forging the factory: Ribosome synthesis and growth control in budding yeast. In: Cell growth: control of cell size (eds. M.N. Hall et al.). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (in press).
- Kelley, L.A., MacCallum, R.M., and Sternberg, M.J. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299: 499–520. [DOI] [PubMed] [Google Scholar]
- Kobe, B., Gleichmann, T., Horne, J., Jennings, I.G., Scotney, P.D., and Teh, T. 1999. Turn up the HEAT. Structure 7: R91–R97. [DOI] [PubMed] [Google Scholar]
- Malik, H.S., Eickbush, T.H., and Goldfarb, D.S. 1997. Evolutionary specialization of the nuclear targeting apparatus. Proc. Natl. Acad. Sci. 94: 13738–13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit, P., Gadal, O., Podtelejnikov, A., Trumtel, S., Gas, N., Petfalski, E., Tollervey, D., Mann, M., Hurt, E., and Tschochner, H. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105: 499–509. [DOI] [PubMed] [Google Scholar]
- Milkereit, P., Strauss, D., Bassler, J., Gadal, O., Kuhn, H., Schutz, S., Gas, N., Lechner, J., Hurt, E., and Tschochner, H. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278: 4072–4081. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296. [DOI] [PubMed] [Google Scholar]
- Nissan, T.A., Bassler, J., Petfalski, E., Tollervey, D., and Hurt, E. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21: 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger, M., Dlakic, M., and Tollervey, D. 2004. A pre-ribosome associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes & Dev. 18: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, J., Sadreyev, R., and Grishin, N.V. 2003. PCMA: Fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics 19: 427–428. [DOI] [PubMed] [Google Scholar]
- Sali, A. and Blundell, T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815. [DOI] [PubMed] [Google Scholar]
- Sanchez, R. and Sali, A. 1998. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl Acad. Sci. 95: 13597–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner, H. and Hurt, E. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13: 255–263. [DOI] [PubMed] [Google Scholar]
- Vetter, I.R., Arndt, A., Kutay, U., Gorlich, D., and Wittinghofer, A. 1999. Structural view of the Ran-Importin β interaction at 2.3 Å resolution. Cell 97: 635–646. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Yu, Z., Fu, X., and Liang, C. 2002. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell 109: 849–860. [DOI] [PubMed] [Google Scholar]