Abstract
The 18S rRNA of the small eukaryotic ribosomal subunit contains several expansion segments. Electron microscopy data indicate that two of the largest expansion segments are juxtaposed in intact 40S subunits, and data from phylogenetic sequence comparisons indicate that these two expansion segments contain complementary sequences that could form a direct tertiary interaction on the ribosome. We have investigated the secondary structure of the two expansion segments in the region around the putative tertiary interaction. Ribosomes from yeast, wheat, and mouse—three organisms representing separate eukaryotic kingdoms—were isolated, and the structure of ES3 and part of the ES6 region were analyzed using the single-strand-specific chemical reagents CMCT and DMS and the double-strand-specific ribonuclease V1. The modification patterns were analyzed by primer extension and gel electrophoresis on an ABI 377 automated DNA sequencer. The investigated sequences were relatively exposed to chemical and enzymatic modification. This is in line with their indicated location on the surface at the solvent side of the subunit. The complementary ES3 and ES6 sequences were clearly inaccessible to single-strand modification, but available for cleavage by double-strand-specific RNase V1. The results are compatible with a direct helical interaction between bases in ES3 and ES6. Almost identical results were obtained with ribosomes from the three organisms investigated.
Keywords: eukaryotes, expansion segment, ribosomes, rRNA, structure
INTRODUCTION
Eukaryotic ribosomal RNA contains more than 50 so-called expansion segments (ES), that is, additional nucleotide clusters inserted at specific positions in the common conserved rRNA core (Gerbi 1996). The role of these extra sequence elements has been widely debated (Houge et al. 1995; Houge and Doskeland 1996; Van Nues et al. 1997; Morgan et al. 2000). Even though it seems improbable that these extra sequences have persisted in the eukaryotic genome without contributing to the function of the ribosomal RNA, experimental confirmation of their functional role is largely lacking. Experiments have shown that complete removal of ES27 in the eukaryotic 23S-like rRNA affects processing and stability of the rRNA (Sweeney et al. 1994). Replacement with ES27 sequences from other organisms restores the function. Similarly, insertions of short sequence into ES3 in 18S rRNA interferes with ribosome assembly (Musters et al. 1990), whereas changes in others, for example, ES19 in the eukaryotic 23S-like rRNA, do not affect ribosomal function (Musters et al. 1991).
Twelve of the expansion segments, also referred to as variable regions (Gorski et al. 1987; Neefs and De Wachter 1990), are found in the 16S-like rRNA. The size of these expansion segments varies considerably between species (Gonzalez et al. 1985, 1988; Hancock and Dover 1988). ES3 and ES6 are two of the largest expansion segments found in 18S rRNA. Although ES3 varies in size in different organisms, the secondary structure models indicate a similar basic structure in all organisms (Fig. 1 ▶; Wuyts et al. 2002; R.R. Gutell, S. Subashchandran, M. Schnare, Y. Du, N. Lin, L. Madabusi, K. Muller, N. Pande, N. Yu, Z. Shang, et al., in prep.). In the secondary structure of 18S rRNA, ES3 is depicted as a helix extending from the 5′-domain of the conserved core structure (helix H10_1 in Fig. 1 ▶).
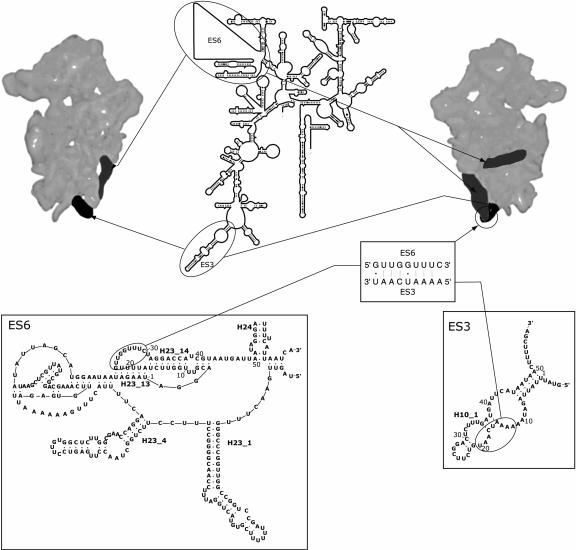
FIGURE 1.
Schematic illustration of the positions of expansion segments ES3 (black) and ES6 (dark gray) on the yeast 40S ribosomal subunits (Spahn et al. 2001) and in the secondary structure model of 18S rRNA (Comparative RNA Web site; http://www.rna.icmb.utexas.edu/; Cannone et al. 2002). The 40S subunit is viewed from the interface side (left) and from the solvent side (right). (Insets) Secondary structure models for yeast ES3 and ES6 (The European ribosomal RNA database, http://oberon.rug.ac.be:8080/rRNA/; Wuyts et al. 2002). The nucleotides are numbered in accordance with Figure 2 ▶. The proposed tertiary interaction between sequences in yeast ES3 and ES6 is based on the consensus sequences (Alkemar and Nygard 2003).
Expansion segment ES6 is located in the central domain of 18S rRNA (Fig. 1 ▶). The average length of the expansion segment is 250 nucleotides (Neefs and De Wachter 1990), but in certain taxa such as Insects and Protists, this region can be up to 800 nt long (Hancock et al. 1988; Crease and Colbourne 1998; Choe et al. 1999). The variability in size is mainly caused by inserts in the 5′-part of ES6 (Wuyts et al. 2000), but inserts at the 3′-end, immediately preceding the structural core (helix H24 in Fig. 1 ▶), are also found in some Euglenozoans (Wuyts et al. 2000; Busse and Preisfeld 2002). In the latest phylogenetic secondary structure model of ES6 (Wuyts et al. 2000), the 5′-part of ES6 is modeled as two long hairpins (H23_1 and H23_4 in Fig. 1 ▶), whereas the 3′-part is suggested to form an intricate system of helices and pseudoknots (Fig. 1 ▶).
The position of ES3 and ES6 on the yeast 40S subunit has recently been determined using cryo-electron microscopy and 3D image reconstruction (Spahn et al. 2001). The morphologic feature referred to as the left foot, located at the lower part of the body of the 40S subunit, contains ES3 and part of ES6 (Fig. 1 ▶; Spahn et al. 2001). The part of ES6 that is juxtaposed to ES3 in the left foot seems to derive from the 3′-half of the expansion segment as the two 5′-helices H23_1 and H23_4 appear to hold a position on the yeast ribosome similar to that occupied by helix 21 in the prokaryotic 30S ribosomal subunit, that is, on the back of the subunit (Spahn et al. 2001).
We have recently suggested that the two juxtaposed expansion segments ES3 and ES6 may be in direct contact in the 40S subunit (Alkemar and Nygard 2003). This suggestion was based on a comparative sequence analysis of ES3 and ES6 sequences from six eukaryotic taxa, representing >2900 discrete species. The sequence analysis showed that ES3 and ES6 contained two complementary sequence motifs with the consensus sequences AAAACCAAU and GUUGGUUUU, respectively, that allowed formation of a helix containing seven to nine contiguous base pairs in ~95% of the species analyzed (Alkemar and Nygard 2003).
Here we have studied the suggested tertiary interaction by analyzing the structure of ES3 and ES6 in intact ribosomes from fungi, plants, and animals, organisms representing three different eukaryotic kingdoms. The analysis was performed using the secondary-structure-sensitive chemical (CMCT and DMS) and enzymatic (RNase V1) reagents in combination with primer extension and gel electrophoresis on an ABI 377 automated DNA sequencer. The structural data are compatible with an involvement of the two complementary sequences in a helical structure.
RESULTS
Expansion segments ES3 and ES6 found in eukaryotic 18S rRNA are juxtaposed in the left foot of the yeast 40S ribosomal subunit (Spahn et al. 2001). The two expansion segments contain a complementary sequence motif that allows formation of a 7–9-bp helix in most eukaryotes (Alkemar and Nygard 2003). Here we have analyzed the secondary structure of the two regions in ES3 and ES6 to determine the possibility of a tertiary interaction between these two eukaryote-specific sequences in intact ribosomes.
Complete ribosomes were isolated from yeast (Saccharomyces cerevisiae), wheat (Triticum aestivum), and mouse (Mus musculus), three organisms representing separate eukaryotic kingdoms. The secondary structure of ES3 and ES6 in the isolated ribosomes was analyzed using the single-strand-specific reagents DMS and CMCT and the double-strand-specific ribonuclease V1 (Favorova et al. 1981; Lowman and Draper 1986). The sites available for cleavage or modification were located by primer extension using fluorescent-labeled primers. The primer extension products were separated on an ABI 377 DNA sequencer (Fig. 2 ▶). Lanes containing 18S rRNA from control ribosomes incubated in the absence of modifying reagent were run in parallel to identify the reagent-independent termination products. These so-called natural stops were used as internal standards when evaluating the modification patterns generated by structure-sensitive reagents (Fig. 2 ▶). The investigated sequences showed relatively high availability for both chemical and enzymatic modification. This observation is in agreement with the suggested location of these expansion segments on the ribosomal surface (Spahn et al. 2001).
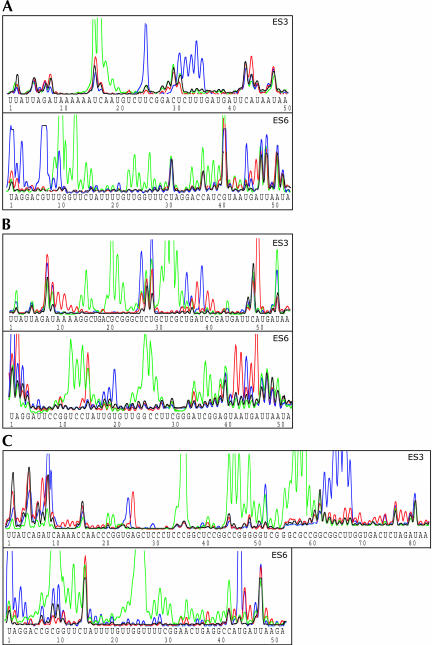
FIGURE 2.
Secondary structure data obtained for ES3 and ES6 in isolated yeast (A), wheat (B), and mouse (C) ribosomes. Native ribosomes isolated from yeast, wheat, and mouse were incubated in the presence of single-strand-specific reagents CMCT, final concentration 50 mM (blue), and DMS, final concentration 20 mM (red), or in the presence of the double-strand-specific ribonuclease V1, final concentration 1 unit/50 μL (green), as described in Materials and Methods. The generated modification patterns were analyzed using primer extension and gel electrophoresis in an ABI 377 automated DNA sequencer as described in Materials and Methods. Control samples (black) were incubated in the absence of modifying reagents but otherwise treated identically to the modified samples. The reagent-independent termination products seen in the control samples were used as internal standards for alignment and for adjusting the peak height for each of the individual overlaid lanes. Sequences are given in the 5′-to-3′ direction.
Yeast 18S rRNA has a short ES3 sequence containing only 50 nt. Bases available for single-strand-specific modification were found at positions U25 and A44 and in the sequence U31CUUU35 (Fig. 2A ▶). All bases in the latter cluster were modified by CMCT. We have previously noted that cytosines are occasionally available for modification by CMCT at the pH used in our modification experiments without being reactive to DMS (Holmberg et al. 1994).
Positions U31 and A44 also served as partial natural stops. However, the peak height at these sites was clearly increased upon addition of increasing concentrations of CMCT and DMS, indicating that these positions also were available for single-strand-specific modification. RNase V1 cleavage sites were found 3′ of the bases in the sequence U16CAA19 (Fig. 2A ▶), indicating that these bases were located in double-strand structures.
The 53 nt at the 3′-end of yeast ES6 immediately preceding helix 24 in the conserved rRNA core (Wuyts et al. 2002) were available for single-strand-specific modification at positions U1, G3, G7, and U10 and in the sequence U45GA47 (Fig. 2A ▶). Interestingly, G46 was accessible for modification by both CMCT and DMS. As DMS usually modifies the N7 position in guanine, DMS modification of guanines should not be detectable by primer extension (Baudin et al. 1987). However, similar DMS-induced modifications of guanines were seen in the identical sequence context in ES6 from both wheat and mouse (Fig. 2B,C ▶).
RNase V1- induced cleavages were detected 3′ of the bases in the sequences U9UGGU13, C15U16, U23UG25, and C37AUC40 (Fig. 2A ▶). Additional cleavage sites were found following bases U27 and A35.
The results of the structural analysis of ES3 and ES6 in yeast 18S rRNA are summarized in Figure 3A ▶. No single-strand-specific modifications were seen in the sequences indicated to be involved in the interaction between ES3 and ES6. In contrast, both sequences contained sites available for RNase V1 cleavage, indicating that these sequences were engaged in helical structures (Fig. 3A ▶).
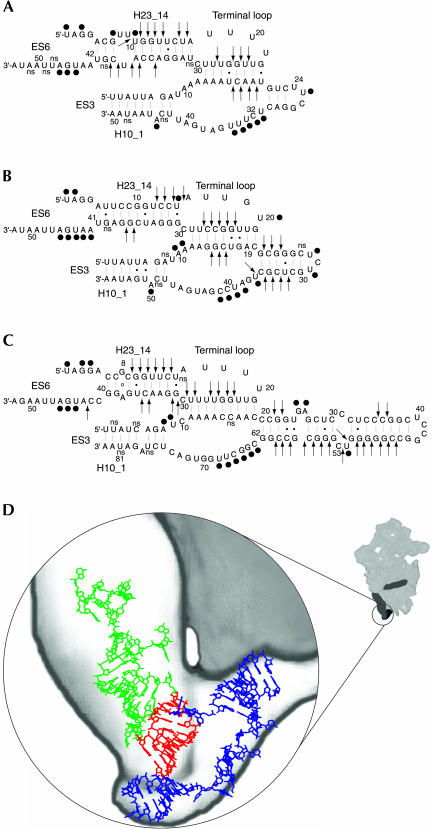
FIGURE 3.
Secondary structure models for ES3 and the 3′-part of ES6 in yeast (A), wheat (B), and mouse (C). The secondary structure models were generated using the Vienna RNA package (Hofacker et al. 1994). Sequence data were taken from GenBank accession nos. J01353, AY049040, and X00686 for Saccharomyces cerevisiae, Triticum aestivum, and Mus musculus, respectively. (•) Bases available for modification by single strand reagents; (arrows) phosphate bonds accessible to cleavage by the double-strand-specific RNase V1; (ns) reagent-independent stops. (D) Structural model of ES3 (blue), the ES6 structural elements helix H23_14, and its apical loop (green), with the bases proposed to participate in the tertiary interaction (red). The 3D model of ES3 and ES6 was constructed using ERNA–3D (Mueller et al. 1995).
The ES3 sequence in wheat 18S rRNA is slightly longer than the corresponding sequence in yeast ribosomes. Bases accessible for single-strand-specific modification were found at positions A10, A11, U27, U29, U36, and A50, and in the sequence A38UCC41 (Fig. 2B ▶). The single-strand-reactive clusters were interspaced by sequences susceptible to RNase V1 cleavage. Strong cleavages were seen 3′ of G15 and in the two sequences G21CG23 and C31UCG34 (Fig. 2B ▶). Less pronounced cleavages were also seen following bases G14, C16, and C35.
The 52-nt sequence at the 3′-end of wheat ES6 was available for strong single-strand-specific modifications at positions U1, A2, U15, and U20, and in the sequence A42AUGA46 (Fig. 2B ▶). Surprisingly, U15 was available for modification by DMS rather than by CMCT. Occasional DMS-induced modification of uracils has previously been reported (Moazed and Noller 1986). U15 was also part of a cluster of bases, U12CCU15, that were highly sensitive to cleavage by the double-strand-specific RNase V1 (Fig. 2B ▶). Additional strong V1 cleavages were seen following bases G25, C26, C36, and G37, with less intense cleavages found at U23, G24, and C27 (Fig. 2B ▶).
Figure 3B ▶ summarizes the experimental results obtained in isolated intact wheat ribosomes. As seen in the figure, the two sequences in ES3 and ES6 indicated to be in direct contact (Alkemar and Nygard 2003) were only accessible for double-strand-specific modification.
The ES3 region in mouse 18S rRNA consists of 83 nt and is thus considerably longer than the homologous structural element in yeast and wheat. The ES3 region was generally accessible for modification and cleavage, but there was, however, longer stretches of bases within this region that were accessible neither to modification nor to cleavage by RNase V1. The ES3 sequence was accessible for modification by single-strand-specific reagents at 11 positions, A8, U9, G25, A26, U52, and C63GGCUU68. The latter six CMCT-reactive bases formed a highly reactive cluster (Fig. 2C ▶).
U52 was also part of an RNase V1-sensitive group spanning the sequence from C45 to C60 (Fig. 2C ▶). Additional RNase V1- accessible sites were found following bases C21, G22, C35, and C36. The first three bases showed low accessibility, whereas the phosphate bond following C36 was extremely sensitive to cleavage (Fig. 2C ▶).
The 3′-part of the mouse ES6 sequence was mainly accessible to cleavage by RNase V1 (Fig. 2C ▶). Sensitive sequences were mainly found in two large clusters containing the sequences C9GGUUC14 and U23GGUUUUCG31. The latter cluster contained one inaccessible phosphate bond 3′ of U27 (Fig. 2C ▶). In addition to the clustered RNase V1- sensitive sites, single cleavage sites were found following bases A34, C35, and C42 (Fig. 2C ▶). Bases accessible to single-strand-specific modification were found in two groups located at the beginning and at the end of the investigated ES6 sequence. The first group contained bases U1, G3, and G4, whereas the second group consisted of bases U44, G45, and A46 (Fig. 2C ▶).
The results are summarized in Figure 3C ▶. No single-strand cleavages were detected in the ES3 and ES6 sequences proposed to be in contact through base-pairing. Instead, the sequence element from ES6 was sensitive to RNAse V1 cleavage. Unlike the situation in yeast and wheat, no sites available for RNase V1 cleavage were found in the suggested interacting sequence in ES3 (Fig. 3C ▶).
DISCUSSION
Recent progress in cryo-electron microscopy and 3D image reconstruction has made it possible to compare the structure of prokaryotic and eukaryotic ribosomes and to locate specific features of the eukaryotic ribosome not present in the prokaryotic ribosome (Dube et al. 1998; Spahn et al. 2001). Such features are probably related to the additional ribosomal proteins and nucleotide sequences, expansion segments, that are only found in the eukaryotic ribosome. Thus, the positions of the two eukaryote specific expansion segments ES3 and ES6 on the yeast 40S subunit were recently determined (Spahn et al. 2001). ES3 emerges from the lower part of the body of the 40S subunit and protrudes into the morphological feature referred to as the left foot (Fig. 1 ▶).
ES6 has been associated with two structures located at the solvent side of the 40S subunit (Spahn et al. 2001). One of the structures forms a horizontal ridge in the center of the 40S body. This ridge seems to have a homologous position to helix 21 in the prokaryotic 30S subunits, indicating that the region around the ridge contains the two 5′-helical elements of ES6 (H23_1 and H23_4 in Fig. 1 ▶; Spahn et al. 2001). The second ES6-related morphological structure forms an arm that reaches and touches the left foot (Fig. 1 ▶; Spahn et al. 2001). Thus, sequences in ES3 and ES6 seem to be juxtaposed on the 40S subunit.
Secondary structure of ES3 and the 3′-region of ES6
Several attempts have been made to construct a phylogenetic secondary structure model of ES6 (Neefs and De Wachter 1990; Nickrent and Sargent 1991; Hancock and Vogler 1998; Wuyts et al. 2000). Most of the proposed models contain a common hairpin (H23_14 in Figs. 1 ▶, 3 ▶) located at the 3′-end of ES6. This hairpin contains a large terminal loop that could be involved in the formation of a pseudoknot as suggested by Wuyts et al. (2000) (helix H23_13 in Fig. 1 ▶). The bases in the helical stem were almost completely inaccessible to single-strand-specific modification in all three species investigated. Exceptions, however, were found at helix termini or next to bulged nucleotides, that is, at positions previously shown to be susceptible to single strand modification, presumably because of increased dynamic instability of the helices at such positions (Stebbins-Boaz and Gerbi 1991). In contrast to the low reactivity to single-strand-specific reagents, the helical stem was accessible to cleavage by RNase V1 in both complementary sequences in all three organisms studied. Thus, the results are in agreement with the positioning of these sequences in helical structures.
The bases in the large terminal loop were inaccessible to single-strand-specific modification in yeast, wheat, and mouse with one exception, U20 in wheat ES6 (Fig. 3B ▶). Instead, the 3′-half of the loop was accessible for RNase V1 cleavage in the organisms studied (Fig. 3 ▶), indicating that the low availability of the loop for single-strand-specific modification was not caused by protein shielding or steric hindrance. The lack of single strand modifications in the 5′-half of the loop could indicate that this part of the loop was involved in the formation of a pseudoknot as suggested by Wuyts et al. (2000) even if there were no RNase V1 cleavages to directly support the participation in a double-strand structure.
In the phylogenetic secondary structure models, ES3 consists of a short initial helix, facing the common rRNA core, followed by a large internal bulge and a terminal hairpin of variable length (Cannone et al. 2002; Wuyts et al. 2002). The nucleotides in the initial helix were inaccessible for modification by both single- and double-strand-specific reagents. This could be because of a shielding effect of ribosomal proteins.
The accessibility of the sequence in the variable terminal hairpin to single-strand and double-strand-specific reagents essentially supported the phylogenetic secondary structure models (Wuyts et al. 2000) with the exception that G25 in mouse ES3 was available for CMCT modification, indicating that the base was unpaired. Thus, an alternate base-pairing involving the two sequences G27CUC30 and G54GGC57 was suggested (Fig. 3C ▶).
The 3′-side of the internal bulge was available for modification by single-strand-specific reagents. Modifications occurred at the homologous positions in yeast, wheat, and mouse ES3 (Fig. 3 ▶), indicating that these bases were not involved in any base-pairing. Additional single-strand-specific modifications were seen at two positions at the 5′-side of the internal loop in wheat and mouse ES3 (Fig. 3B,C ▶). This side of the loop was also sensitive to RNase V1 cleavage in yeast and wheat ES3 (Fig. 3A,B ▶).
A possible tertiary interaction between ES3 and ES6
We have recently suggested that sequences in ES3 and ES6 are in direct contact on the ribosome. The suggestion was based on a comparative analysis of ES3 and ES6 sequences from six eukaryotic taxa, representing ~2900 discrete species. ES3 and ES6 contain the two complementary consensus sequence motifs AAAACCAAU and GUUGGUUUU, respectively (Alkemar and Nygard 2003). The two complementary sequences are located in the apical loop of helix H23_14 (ES6) and in the internal bulge of helix H10_1 (ES3; Fig. 3 ▶). The two guanines G4 and G5 in the middle of the ES6 sequence are conserved in almost all species examined. The putative complementary positions in ES3 always contain pyrimidines, most often cytosines. In some organisms, one or more of the uracils in ES6 are replaced by cytosines. Such changes are compensated for by a change from adenine to a guanine at the putative complementary positions in ES3 (Alkemar and Nygard 2003). The least conserved base is found at position nine in ES6 (Fig. 1 ▶). Base variations at this position are not always compensated for by changes in the suggested complementary sequence. The base complementarities between the sequences in ES3 and ES6 allow formation of a helix containing seven to nine consecutive base pairs in >95% of all eukaryotes investigated (Alkemar and Nygard 2003).
The experimental analysis of the secondary structure of the two putative complementary regions in intact ribosomes isolated from yeast, wheat, and mouse, three organisms representing different eukaryotic kingdoms, shows that the complementary sequences were inaccessible for chemical modification by single-strand-specific reagents. The lack of reactivity seems not to depend on a general shielding of the investigated rRNA regions as neighboring bases were accessible to single-strand-specific modification (Figs. 2 ▶, 3 ▶). Furthermore, both putative complementary regions in yeast and wheat were accessible for cleavage by RNase V1. In mouse, RNase V1 cleavage was only seen in the ES6 sequence element, whereas no cleavages were detected in the complementary ES3 sequence (Figs. 2 ▶, 3 ▶). Thus, the structural analysis shows that the putative single strand bases in ES3 and ES6 that, according to the sequence analysis, have the potential to form a 9-bp helix are inaccessible to modification by single-strand-specific reagents but are accessible to cleavage by RNase V1. This indicates that these bases must be stacked or involved in base-pairing. It is therefore possible that the two complementary sequences found in ES3 and ES6 are involved in a tertiary interaction in intact ribosomes. The observation that the main species-dependent size variation in ES3 is seen in the terminal hairpin (Wuyts et al. 2002) indicates that the sequence capable of interacting with ES6 is located at a fixed position relative to the common rRNA core in all ES3-containing organisms. Similarly, the position, vis-à-vis the common rRNA core, of the complementary sequence in the apical loop of helix H23_14 is relatively fixed in all eukaryotic species with the possible exception of some Euglenozoans, (Wuyts et al. 2000; Busse and Preisfeld 2002). Thus, the two putative interacting sequences could have a similar spatial arrangement in all eukaryotic ribosomes despite considerable species-related length variation in the ES3 and ES6 sequences. Interestingly, the proposed tertiary interaction is compatible with the latest structure models for ES3 and ES6 generated by phylogenetic sequence comparisons (Cannone et al. 2002; Wuyts et al. 2002).
As mentioned above, ES3 has been mapped to the so-called left foot on the 40S ribosomal subunit, where it is juxtaposed to sequences most probably derived from the 3′-half of ES6 (Spahn et al. 2001). Figure 3D ▶ shows a structural model of the two interacting ES3 and ES6 sequences superimposed on the left foot region of the yeast 40S ribosome (adapted from Spahn et al. 2001) with ES3 shown in blue, helix H23_14 in ES6 shown in green, and the putative interacting ES3 and ES6 sequences in red. A high-resolution structure of the small eukaryotic ribosome will be needed to determine the exact position of the putative interacting sequences. The functional role of the left foot and the putative tertiary interaction also remains to be elucidated.
MATERIALS AND METHODS
Materials
Dimethyl sulfate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluensulfate (CMCT) were from Sigma-Aldrich. Ribonuclease V1 and cDNA oligonucleotides were from Amersham Pharmacia Biotech. Superscript reverse transcriptase was from Life Technologies, Inc. RNasin was from Promega.
Preparation of ribosomes
Mouse liver monosomes were prepared as previously described (Sloma and Nygård 2001). The isolated salt-washed monosomes were dissolved in Ribosome buffer (30 mM HEPES-KOH at pH 7.6, 70 mM KCl, 0.25 M sucrose, 2 mM MgCl2, 5 mM mercaptoethanol) at a concentration of 7.8 mg/mL and stored at −80°C in aliquots.
Wheat monosomes were prepared from wheat germ cell-free extracts (Clemens 1984; Seal et al. 1986). The extracts were treated with DOC and Triton X-100, final concentration 1% (w/v) each, and layered on a discontinuous sucrose gradient, containing 10 mL of 10% (w/v) sucrose in Buffer A (70 mM KCl, 5 mM MgCl2, 20 mM Tris-HCl at pH 7.6, 1 mM DTT) superimposed on 5 mL of 28% (w/v) sucrose in Buffer A. The gradients were centrifuged in an SW 27 rotor for 16 h at 90,000gav. The pelleted monosomes were dissolved in Ribosome buffer and stored at −80°C in aliquots.
For preparation of yeast ribosomes, pelleted yeast cells were dissolved in a buffer (20 mM HEPES-KOH at pH 7.6, 2 mM Mg(CH3COO)2, 100 mM KCl, 1 mM DTT, and 1 mM PMSF) containing 4000 units RNasin/mL. An equal volume of glass beads was added, and the cells were homogenized by vigorous shaking on a vortex mixer. The glass beads were removed by centrifugation for 5 min at 8000g, and the supernatant was treated with DOC and Triton X-100 at a final concentration of 1% (w/v) each. The samples were centrifuged for 20 min at 15,000g, and the supernatants were collected and treated with KCl at a final concentration of 0.5 M. The supernatants were layered onto a 0.75 M sucrose cushion in Buffer A and the salt-washed ribosomes were pelleted by centrifugation for 4 h at 150,000gav. The pelleted ribosomes were dissolved in Ribosome buffer and stored at −80°C in aliquots.
Modification of rRNA in ribosomes
Chemical modification and enzymatic cleavage of rRNA in wheat and mouse 80S ribosomes were as previously described (Larsson and Nygård 2001). For chemical modification, 30 pmoles of ribosomes in final volumes of 50 μL were incubated at 37°C in the presence of 20 or 100 mM DMS or for 15 min in the presence of 50 or 100 mM CMCT. Incubation was for 5 min (DMS) or 15 min (CMCT). The ribosomes were precipitated with 2.5 volumes of ethanol and pelleted by centrifugation. The pellets were dissolved in 50 mM Tris-HCl (pH 7.6) and 0.5% (w/v) SDS, and the RNA was extracted with phenol (Brawerman et al. 1972).
For modification with RNase V1, 30 pmoles of ribosomes in final volumes of 50 μL were incubated for 5 min at 37°C in the presence of 0.2, 0.4, and 1.0 units of RNase V1. After incubation, the reactions were diluted with an equal volume of 0.1 M Tris-HCl (pH 7.6) and 1% (w/v) SDS, and the RNA was extracted with phenol (Brawerman et al. 1972).
Control samples were identically treated but without the modifying reagent.
Identification of modification sites
The positions of the modified bases/cleavage sites were identified using primer extension as previously described (Holmberg et al. 1992). The primers used were GTAATTTGCGCGCC, complementary to nucleotides 429–442, 433–446, and 479–492 in yeast, wheat, and mouse 18S rRNA; TCGGCATAGTTTATGG, complementary to nucleotides 1020–1034 in yeast 18S rRNA; and CG GCATCGTTTATGG, complementary to nucleotides 1026–1040 and 1080–1094 in wheat and mouse 18S rRNA. The primer extension products were analyzed on 4.75% (w/v) acrylamide sequencing gels in an Applied Biosystems 377 DNA Sequencer as described (Larsson and Nygård 2001).
Secondary and tertiary structure predictions
Secondary structure models for ES3 and ES6 were constructed based on the modification data using the Vienna RNA package (Hofacker et al. 1994).
A three-dimensional model of the ES3–ES6 interaction was constructed using the RNA modeling software ERNA–3D (Mueller et al. 1995).
Acknowledgments
We are indebted to Birgit Lundberg for technical assistance and to Thomas Olsson for computer support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
CMCT, 1-cyclohexyl-3-(morpholinoethyl) carbodiimide metho-p-toluene sulfonate
DMS, dimethyl sulfate
DTT, dithiothreitol
ES, expansion segment
DOC, sodium deoxycholate
PMSF, phenylmethylsolfonylfluoride
SDS, sodium dodecyl sulfate
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5135204.
REFERENCES
- Alkemar, G. and Nygard, O. 2003. A possible tertiary rRNA interaction between expansion segments ES3 and ES6 in eukaryotic 40S ribosomal subunits. RNA 9: 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, F., Ehresmann, C., Romby, P., Mougel, M., Colin, J., Lempereur, L., Bachellerie, J.P., Ebel, J.P., and Ehresmann, B. 1987. Higher-order structure of domain III in Escherichia coli 16S ribosomal RNA, 30S subunit and 70S ribosome. Biochimie 69: 1081–1096. [DOI] [PubMed] [Google Scholar]
- Brawerman, G., Mendecki, J., and Lee, S.Y. 1972. A procedure for the isolation of mammalian messenger ribonucleic acid. Biochemistry 11: 637–641. [DOI] [PubMed] [Google Scholar]
- Busse, I. and Preisfeld, A. 2002. Unusually expanded SSU ribosomal DNA of primary osmotrophic euglenids: Molecular evolution and phylogenetic inference. J. Mol. Evol. 55: 757–767. [DOI] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. 2002. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2; http://www.rna.icmb.utexas.edu/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, C.P., Hancock, J.M., Hwang, U.W., and Kim, W. 1999. Analysis of the primary sequence and secondary structure of the unusually long SSU rRNA of the soil bug, Armadillidium vulgare. J. Mol. Evol. 49: 798–805. [DOI] [PubMed] [Google Scholar]
- Clemens, M.J. 1984. Translation of eukaryotic messenger RNA in cell-free extracts. In Transcription and translation—A practical approach (eds. B.D. Hames and S.J. Higgins), pp. 231–270. IRL Press Limited, Oxford, UK.
- Crease, T.J. and Colbourne, J.K. 1998. The unusually long small-subunit ribosomal RNA of the crustacean, Daphnia pulex: Sequence and predicted secondary structure. J. Mol. Evol. 46: 307–313. [DOI] [PubMed] [Google Scholar]
- Dube, P., Bacher, G., Stark, H., Mueller, F., Zemlin, F., Van Heel, M., and Brimacombe, R. 1998. Correlation of the expansion segments in mammalian rRNA with the fine structure of the 80 S ribosome—A cryoelectron microscopic reconstruction of the rabbit reticulocyte ribosome at 21 Å resolution. J. Mol. Biol. 279: 403–421. [DOI] [PubMed] [Google Scholar]
- Favorova, O.O., Fasiolo, F., Keith, G., Vassilenko, S.K., and Ebel, J.P. 1981. Partial digestion of tRNA–aminoacyl-tRNA synthetase complexes with cobra venom ribonuclease. Biochemistry 20: 1006–1011. [DOI] [PubMed] [Google Scholar]
- Gerbi, S.A. 1996. Expansion segments: Regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In Ribosomal RNA: Structure, evolution, processing, and function in protein biosynthesis (eds. R.A. Zimmermann and A.E. Dahlberg), pp. 71–87. CRL Press, Boca Raton, FL.
- Gonzalez, I.L., Gorski, J.L., Campen, T.J., Dorney, D.J., Erickson, J.M., Sylvester, J.E., and Schmickel, R.D. 1985. Variation among human 28S ribosomal RNA genes. Proc. Natl. Acad. Sci. 82: 7666–7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, I.L., Sylvester, J.E., and Schmickel, R.D. 1988. Human 28S ribosomal RNA sequence heterogeneity. Nucleic Acids Res. 16: 10213–10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski, J.L., Gonzalez, I.L., and Schmickel, R.D. 1987. The secondary structure of human 28S rRNA: The structure and evolution of a mosaic rRNA gene. J. Mol. Evol. 24: 236–251. [DOI] [PubMed] [Google Scholar]
- Hancock, J.M. and Dover, G.A. 1988. Molecular coevolution among cryptically simple expansion segments of eukaryotic 26S/28S rRNAs. Mol. Biol. Evol. 5: 377–391. [DOI] [PubMed] [Google Scholar]
- Hancock, J.M. and Vogler, A.P. 1998. Modelling the secondary structures of slippage-prone hypervariable RNA regions: The example of the tiger beetle 18S rRNA variable region V4. Nucleic Acids Res. 26: 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, J.M., Tautz, D., and Dover, G.A. 1988. Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster. Mol. Biol. Evol. 5: 393–414. [DOI] [PubMed] [Google Scholar]
- Hofacker, I.L., Fontana, W., Stadler, P.F., Bonhoeffer, M., Tacker, P., and Schuster, P. 1994. Fast folding and comparison of RNA secondary structures. Monatshefte fur chemie 125: 167–188. [Google Scholar]
- Holmberg, L., Melander, Y., and Nygård, O. 1992. Ribosome-bound eukaryotic elongation factor 2 protects 5 S rRNA from modification. J. Biol. Chem. 267: 21906–21910. [PubMed] [Google Scholar]
- ———. 1994. Probing the structure of mouse Ehrlich ascites cell 5.8S, 18S and 28S ribosomal RNA in situ. Nucleic Acids Res. 22: 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houge, G. and Doskeland, S.O. 1996. Divergence towards a dead end—Cleavage of the divergent domains of ribosomal RNA in apoptosis. Experientia 52: 963–967. [DOI] [PubMed] [Google Scholar]
- Houge, G., Robaye, B., Eikhom, T.S., Golstein, J., Mellgren, G., Gjertsen, B.T., Lanotte, M., and Doskeland, S.O. 1995. Fine mapping of 28S rRNA sites specifically cleaved in cells undergoing apoptosis. Mol. Cell. Biol. 15: 2051–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, S. and Nygård, O. 2001. Proposed secondary structure of eukaryote specific expansion segment 15 in 28S rRNA from mice, rats, and rabbits. Biochemistry 40: 3222–3231.11258939 [Google Scholar]
- Lowman, H.B. and Draper, D.E. 1986. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 261: 5396–5403. [PubMed] [Google Scholar]
- Moazed, D. and Noller, H.F. 1986. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47: 985–994. [DOI] [PubMed] [Google Scholar]
- Morgan, D.G., Ménétret, J.-F., Radermacher, M., Neuhof, A., Akey, I.V., Rapoport, T.A., and Akey, C.W. 2000. A comparison of the yeast and rabbit 80S ribosome reveals the topology of the nascent chain exit tunnel, inter-subunit bridges and mammalian rRNA expansion segments. J. Mol. Biol. 301: 301–321. [DOI] [PubMed] [Google Scholar]
- Mueller, F., Doring, T., Erdemir, T., Greuer, B., Junke, N., Osswald, M., Rinke-Appel, J., Stade, K., Thamm, S., and Brimacombe, R. 1995. Getting closer to an understanding of the three-dimensional structure of ribosomal RNA. Biochem. Cell Biol. 73: 767–773. [DOI] [PubMed] [Google Scholar]
- Musters, W., Boon, K., van der Sande, C.A., van Heerikhuizen, H., and Planta, R.J. 1990. Functional analysis of transcribed spacers of yeast ribosomal DNA. EMBO J. 9: 3989–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters, W., Concalves, P.M., Boon, K., Raue, H.A., van Heerikhuizen, H., and Planta, R.J. 1991. The conserved GTPase center and variable region V9 from Saccharomyces cerevisiae 26S rRNA can be replaced by their equivalents from other prokaryotes or eukaryotes without detectable loss of ribosomal function. Proc. Natl. Acad. Sci. 88: 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs, J.M. and De Wachter, R. 1990. A proposal for the secondary structure of a variable area of eukaryotic small ribosomal subunit RNA involving the existence of a pseudoknot. Nucleic Acids Res. 18: 5695–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent, D.L. and Sargent, M.L. 1991. An overview of the secondary structure of the V4 region of eukaryotic small-subunit ribosomal RNA. Nucleic Acids Res. 19: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal, S.N., Schmidt, A., and Marcus, A. 1986. The wheat germ protein synthesis system. Methods Enzymol. 118: 128–140. [Google Scholar]
- Sloma, M.S. and Nygård, O. 2001. Chemical accessibility of 18S rRNA in native ribosomal complexes: interaction sites of mRNA, tRNA and translational factors. Biol. Chem. 382: 661–668. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., and Frank, J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell 107: 373–386. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz, B. and Gerbi, S.A. 1991. Structural analysis of the peptidyl transferase region in ribosomal RNA of the eukaryote Xenopus laevis. J. Mol. Biol. 217: 93–112. [DOI] [PubMed] [Google Scholar]
- Sweeney, R., Chen, L., and Yao, M.C. 1994. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol. Cell. Biol. 14: 4203–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nues, R.W., Venema, J., Planta, R.J., and Raue, H.A. 1997. Variable region V1 of Saccharomyces cerevisiae 18S rRNA participates in biogenesis and function of the small ribosomal subunit. Chromosoma 105: 523–531. [DOI] [PubMed] [Google Scholar]
- Wuyts, J., De Rijk, P., Van de Peer, Y., Pison, G., Rousseeuw, P., and De Wachter, R. 2000. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res. 28: 4698–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts, J., Van de Peer, Y., Winkelmans, T., and De Wachter, R. 2002. The European database on small subunit ribosomal RNA. Nucleic Acids Res. 30: 183–185. [DOI] [PMC free article] [PubMed] [Google Scholar]