Abstract
Determination of haplotype frequencies (the joint distribution of genetic markers) in large population samples is a powerful tool for association studies. This is due to their greater extent of polymorphism since any two bi-allelic single nucleotide polymorphisms (SNPs) generate a potential four-allele genetic marker. Therefore, a haplotype may capture a given functional polymorphism with higher statistical power than its SNP components. The statistical estimation of haplotype frequencies, usually employed in linkage disequilibrium studies, requires individual genotyping for each SNP in the haplotype, thus making it an expensive process. In this study, we describe a new method for direct measurement of haplotype frequencies in DNA pools by allele-specific, long-range haplotype amplification. The proposed method allows the efficient determination of haplotypes composed of two SNPs in close vicinity (up to 20 kb).
INTRODUCTION
Haplotype analysis is becoming a common tool in association studies. The joint distribution of adjacent markers in a population, which is actually the haplotype frequency, represents the correlation between those markers. Several recent studies showed that haplotypes, if used as genetic markers, have higher statistical power than individual markers (1,2). A major difficulty in using haplotypes as genetic markers lies in determining the haplotype phase for individuals who are heterozygous for more than one marker. There are several approaches to overcome this difficulty. However, in order for an approach to be practical, it needs to meet the low cost and high throughput requirements. Only such approaches can potentially be used in studies using large samples and involving a large number of genetic markers.
Haplotype phase can be established by genotyping family members in order to infer parental chromosomes. This, however, requires the recruitment and genotyping of relatives, which may not be available or may be expensive to attain. Methods for chromosomal isolation (3), even though advantageous for long-range haplotypes, are currently applicable only for very small sample sizes due to the high costs involved.
Therefore, the most common approach presently used in association studies is the statistical estimation of the frequencies of various haplotype phases. In this approach, an algorithm estimates the most likely haplotype frequencies, given the genotypes distribution in a sample (4,5). Unfortunately, the process of attaining the primary data necessary for the statistical estimation requires numerous individual genotypings of all markers included in the hap-lotype, and this is expensive and time-consuming.
In this study, we describe a new method for haplotyping using DNA pools. Our approach is based on allele-specific PCR amplification from pooled DNA samples and quan-titative genotyping of the PCR products.
MATERIALS AND METHODS
DNA samples and SNP genotyping
DNA was extracted from blood samples using the Nucleon BACC kit (Amersham). Two SNP markers were selected from the APOE gene sequence, denoted SNP888 and SNP988 according to Martin et al. (6). Primers were designed using the Primer3 program (Whitehead Institute for Biomedical Research, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Long PCR amplification was performed using the Expand 20 kbplus PCR System (Roche Molecular Biochemicals), according to the manufacturer’s instructions. Quantitative SNP genotyping was performed by Pyro sequencing™, according to the manufacturer’s instructions. This methodology provides accurate estimates as sources of error, such as preferential amplification and allele drop-out, are rare (7). Each PCR amplification for the quantification reaction was repeated three times. The mean of the three measurements was taken as the quantitative result.
Pool assembly
Individual samples were genotyped at both SNPs. Individuals which were homozygous at least for one SNP were selected to assemble pools with known haplotype frequencies (Table 1). Optical density measurements for each individual sample were carried out in six replicates using a µQuant spectrophotometer (Bio-Tek Instruments). The DNA samples were then diluted to reach a set concentration of 10 ng/µl. These DNA samples were mixed in appropriate ratios to generate several pools, each with different known haplotype frequencies.
Table 1. Haplotype composition of the DNA template samples.
| DNA template | Sample size | Haplotype X888-X988 | Frequency (%) |
|---|---|---|---|
| Pools | |||
| P1 | 6 | T-T | 25 |
| T-C | 25 | ||
| C-T | 25 | ||
| C-C | 25 | ||
| P2 | 10 | T-T | 45 |
| T-C | 10 | ||
| C-T | 35 | ||
| C-C | 10 | ||
| P3 | 8a | T-T | 10 |
| T-C | 10 | ||
| C-T | 45 | ||
| C-C | 35 | ||
| P4 | 12 | T-T | 42 |
| T-C | 4 | ||
| C-T | 33 | ||
| C-C | 21 | ||
| Individuals | |||
| I1 | 1 | T-T | 50 |
| C-T | 50 | ||
| I2 | 1 | T-C | 50 |
| C-C | 50 | ||
| I3 | 1 | T-T | 50 |
| T-C | 50 | ||
| I4 | 1 | C-T | 50 |
| C-C | 50 | ||
| I5 | 1 | T-T | 50 |
| C-C | 50 |
aPool 3 was assembled from an equal amount of DNA from each of seven different individuals and a triple amount of DNA from an eighth individual.
Estimating haplotype frequencies: experimental procedure
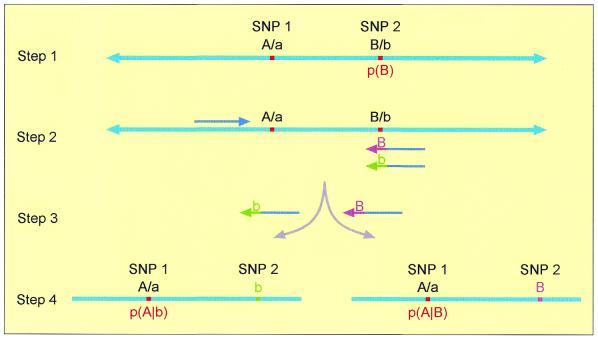
The key component of our technique resides in measuring the allele frequencies at SNP 1, given the allele at SNP 2. This is carried out through selective amplification of a DNA segment that includes both SNPs, according to a principle reported for individual haplotype genotyping, in the context of a bi-allelic Alu deletion, by Michalatos-Beloin et al. (8). In each of two alternate reactions, we use an allele-specific forward primer for one of the two alleles of SNP 2 (the 3′ nucleotide of each primer is either of the polymorphic nucleotides) and a com-mon reverse primer, located beyond SNP 1 (Fig. 1). The template to be amplified is a DNA pool. The distribution of SNP 1 in each of the reaction products and the distribution of SNP 2 in the original (unamplified) pool is then quantified.
Figure 1.
Estimating haplotype frequencies of two markers, SNP 1 and SNP 2, using DNA pooling. (Step 1) Measurement of the allele frequencies of SNP 2 [probability p(B)]. (Step 2) Design of two allele-specific forward primers ending at the polymorphic base of SNP 2 and a common reverse primer beyond SNP 1. (Step 3) Two separate PCR amplifications are carried out, resulting in alternative amplicons, each carrying a different allele of SNP 2. (Step 4) Quantitative genotyping of the PCR products from step 3 for measurement of allele frequencies at SNP 1 [probabilities p(A|B) and p(A|b)].
It is recommended to use a double heterozygous individual as a control template. Any individual can be referred to as a natural pool of two haplotypes with known frequencies. A double heterozygote is a useful control for various stages of the process: the specificity of the allele-specific primers, the validity of the long-range PCR and the quantitative genotyping reaction at both SNPs.
Estimating haplotype frequencies: statistical procedures
Consider a pair of markers included in a haplotype. The term ‘haplotype frequency’ is synonymous with the term ‘joint distribution of the markers’, i.e. the joint distribution of a pair of bi-allelic markers is described by a 2 × 2 table and the haplotype frequencies are the entries in that table. There are three degrees of freedom in the determination of the entries to the table (since the entries sum to 1). Thus, it is sufficient to measure three independent parameters in order to reconstruct the complete table, e.g. the marginal distribution of SNP 2, the conditional distribution of SNP 1 given one allele at SNP 2 and the conditional distribution of SNP 1 given the other allele at SNP 2. Since the joint distribution can be expressed by the conditional distribution via the equation:
joint distribution = (conditional distribution) × (marginal distribution)
it is possible to calculate the joint distribution of both SNPs from these three measurements using equations 1–4:
p(A,B) = p(A|B) · p(B)1
p(a,B) = [1 – p(A|B)] · p(B)2
p(A,b) = p(A|b) · [1 – p(B)]3
p(a,b) = [1 – p(A|b)] · [1 – p(B)]4
where A,a and B,b are the alleles for SNP 1 and SNP 2, respectively.
Measurement of the marginal distributions is attained by direct quantitative genotyping. Measurement of conditional distributions is enabled by quantitative genotyping of selectively amplified samples, as described above.
RESULTS AND DISCUSSION
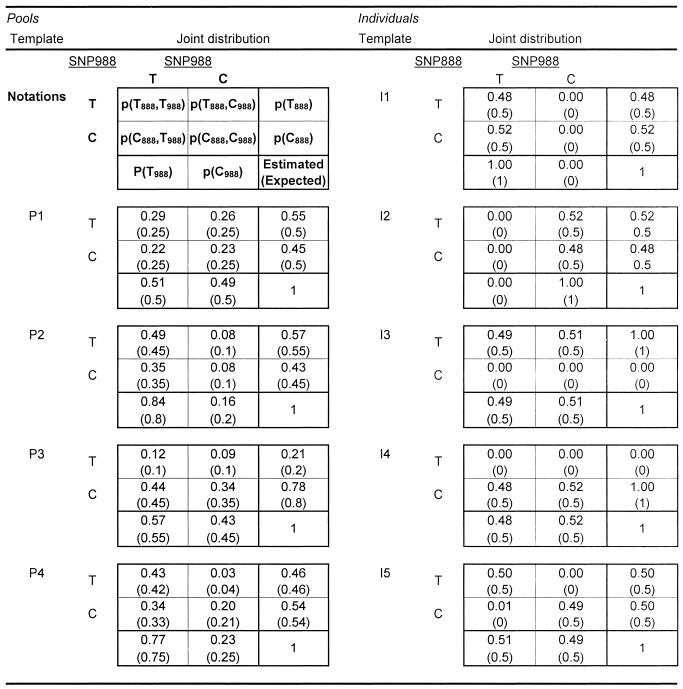
The three parameters measured were p(T988) (the marginal distribution of SNP988), p(T888|C988) (the conditional distribution of SNP888 given allele C at SNP988) and p(T888|T988) (the conditional distribution of SNP888 given allele T at SNP988), shown in Table 2. Our aim was to reach a highly specific amplification of the segment containing both SNPs, in a manner that will discriminate between segments with allele C at SNP988 and segments with allele T. We performed both amplifications on a number of DNA template samples, as shown in Table 1, and calculated the haplotype frequencies using equations 1–4. We then compared the estimated and expected frequencies for each template.
Table 2. Quantitative genotyping results.
| Template | p(T988) | p(T888|T988 ) | p(T888|C988 ) |
|---|---|---|---|
| P1 | 0.510 | 0.569 | 0.538 |
| P2 | 0.841 | 0.579 | 0.523 |
| P3 | 0.566 | 0.218 | 0.206 |
| P4 | 0.775 | 0.561 | 0.137 |
| I1 | 1.000 | 0.485 | |
| I2 | 0.000 | 0.521 | |
| I3 | 0.488 | 0.998 | 1.000 |
| I4 | 0.484 | 0.003 | 0.000 |
| I5 | 0.506 | 0.990 | 0.000 |
The method’s ability to identify and measure haplotypes is well illustrated if we compare the results for I5 (a double heterozygote) and for P1. Both templates have the same allele frequency in each marker (the marginal distributions in Table 3). Following the discriminating amplifications, however, it is evident that I5 presents only two haplotypes (T888-T988 and C888-C988), while P1 is composed equally of all four possible haplotypes.
Table 3. Estimated and expected joint distributions of SNP988 and SNP888 in the template samples.
When comparing estimated and expected haplotype frequencies for all templates, similar results were obtained (Table 3). The difference between the estimated and expected value ranged from 0 to 0.04, with an average of 0.007 for the individuals and 0.018 for the pools. This error rate is not significantly higher than estimating standard allele frequencies of SNPs in DNA pools (9,10). It should be noted that the pools were constructed using a small number of individuals (six to twelve) and, consequently, our results suffer from the unavoidable inaccuracies associated with the assembly of small pools. These inaccuracies will be avoided in practical experiments where the pools will usually consist of a relatively large number of individuals (11). In addition, a small number of replications can increase accuracy further. Thus the accuracy achieved for the described technology should allow its reasonable use for various haplotyping purposes. This includes primarily: (i) a method to rapidly assess common haplotypes across the genome in different populations; (ii) association analysis with haplotypes to identify the genetic basis of complex traits. In this context, the method suggested combines the advantages of case– control genetic association, haplotype analysis and DNA pooling for linkage disequilibrium mapping.
We have described the method for haplotypes composed of two SNPs. However, it can also be used for haplotyping pairs of polymorphisms where only one of them is a SNP and the other is any kind of polymorphism that can be genotyped quantitatively, e.g. microsatellite, Ins/Del. The method can also be extended to haplotypes comprising more than two markers through a series of nested PCR amplifications. It should be noted that the long-range, allele-specific PCR required for this method is technically more difficult than a standard PCR and may need some optimization to reach the required high specificity. The final result depends on the discrimination achieved. Yield reduction caused by a high GC content or low quality of the DNA will not have a significant effect on the results as long as the specificity of the primers ensures a proportional amplification of segments containing each allele of SNP 1. Additionally, a very low concentration of DNA is actually needed for the quantitative genotyping of that SNP in the next step. Low proofreading ability in the amplification is not crucial either, since the only amplified nucleotides that could affect the results are the two SNPs. The most important factor is the allele-specific primer design. Since they have to end at a specific nucleotide and have to fit a certain melting temperature, not every pair of SNPs would have a suitable sequence for allele-specific primers. The achievement of highly discriminating primers at the SNP is essential for this method to work. It should be mentioned, though, that the allele-specific primers can be located on either of the two SNPs. Furthermore, since any three independent parameters can be employed, it is sufficient to have only one good allele-specific primer. The other two parameters can be, for example, allele probabilities of both SNPs in the original pool.
The presented method uses DNA pools for haplotype frequency estimation and therefore significantly reduces the number of genotyping reactions necessary for haplotyping. For example, determining haplotype frequencies of a two-SNP haplotype in a sample population of 100 individuals would normally require 200 SNP genotyping reactions. Note that even when individual genotyping is performed, a statistical algorithm is applied which also introduces some error. In contrast, with the method suggested here, only five reactions are needed: two allele-specific amplifications and three genotyping reactions. If sample size is increased to, say, 1000, under individual genotyping the number of reactions is increased accordingly, whereas under the method presented here the number of reactions remains fixed. Therefore, this method may provide an efficient solution to the growing need for haplotype data collection.
Acknowledgments
ACKNOWLEDGEMENTS
This study was supported by the FIRST foundation of the Israeli Academy of Science.
REFERENCES
- 1.Akey J., Jin,L. and Xiong,M. (2001) Haplotypes vs single marker linkage disequilibrium tests: what do we gain? Eur. J. Hum. Genet., 9, 291–300. [DOI] [PubMed] [Google Scholar]
- 2.Zollner S. and von Haeseler,A. (2000) A coalescent approach to study linkage disequilibrium between single-nucleotide polymorphisms. Am. J. Hum. Genet., 66, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasko J.E., Battini,J.L., Kruglyak,L., Cox,D.R. and Miller,A.D. (2000) Precise gene localization by phenotypic assay of radiation hybrid cells. Proc. Natl Acad. Sci. USA, 97, 7388–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fallin D. and Schork,N.J. (2000) Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data. Am. J. Hum. Genet., 67, 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Excoffier L. and Slatkin,M. (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol., 12, 921–927. [DOI] [PubMed] [Google Scholar]
- 6.Martin E.R., Lai,E.H., Gilbert,J.R., Rogala,A.R., Afshari,A.J., Riley,J., Finch,K.L., Stevens,J.F., Livak,K.J., Slotterbeck,B.D., Slifer,S.H., Warren,L.L., Conneally,P.M., Schmechel,D.E., Purvis,I., Pericak-Vance,M.A., Roses,A.D. and Vance,J.M. (2000) SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am. J. Hum. Genet., 67, 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neve B., Froguel,P., Corset,L., Vaillant,E., Vatin,V. and Boutin,P. (2002) Rapid SNP allele frequency determination in genomic DNA pools by pyrosequencing. Biotechniques, 32, 1138–1142. [DOI] [PubMed] [Google Scholar]
- 8.Michalatos-Beloin S., Tishkoff,S.A., Bentley,K.L., Kidd,K.K. and Ruano,G. (1996) Molecular haplotyping of genetic markers 10 kb apart by allele-specific long-range PCR. Nucleic Acids Res., 24, 4841–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germer S., Holland,M.J. and Higuchi,R. (2000) High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res., 10, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giordano M., Mellai,M., Hoogendoorn,B. and Momigliano-Richiardi,P. (2001) Determination of SNP allele frequencies in pooled DNAs by primer extension genotyping and denaturing high-performance liquid chromatography. J. Biochem. Biophys. Methods, 47, 101–110. [DOI] [PubMed] [Google Scholar]
- 11.Jawaid A., Bader,J.S., Purcell,S., Cherny,S.S. and Sham,P. (2002) Optimal selection strategies for QTL mapping using pooled DNA samples. Eur. J. Hum. Genet., 10, 125–132. [DOI] [PubMed] [Google Scholar]