Abstract
After deadenylation, most cytoplasmic mRNAs are decapped and digested by 5′ to 3′ exonucleases in Saccharomyces cerevisiae. Capped and deadenylated mRNAs are degraded to a lesser extent by 3′ to 5′ exonucleases. We have used a method, based on the electroporation of in vitro synthetised mRNAs, to study the relative importance of these two exonucleolytic pathways under stress conditions. We show that derepression of GCN4 upon amino acid starvation specifically limits the 5′-to-3′-degradation pathway. Because adenosine 3′-5′ biphosphate (pAp), which is produced by Met16p, inhibits this degradation pathway to a comparable extent, we were prompted to analyse the role of Met16p in this phenomenon. We show that the inhibitory effects of amino acid limitation on 5′ to 3′ mRNA degradation are absent in a met16 mutant. We therefore conclude that the GCN4 dependence of MET16 expression is responsible for the decrease in 5′ to 3′ digestion under stress conditions and that cells use pAp as a signal to limit 5′ to 3′ RNA degradation under stress conditions. Because 3′ to 5′ mRNA degradation is unaffected, the relative importance of this pathway in the decay of certain RNAs may be increased under stress conditions.
Keywords: Gcn4p, Met16p, pAp, Ski7p, Xrn1p
INTRODUCTION
The 5′-cap and the 3′-poly(A) structures stimulate translation initiation synergistically and are determinants of eukaryotic messenger RNA stability (Gerstel et al. 1992; Gallie 2001). The shortening of the poly(A) tail precedes two possible mRNA-decay pathways in Saccharomyces cerevisiae (Caponigro and Parker 1996). In the first, the 5′-capped RNA structure is removed, resulting in a 5′-monophosphorylated RNA (Beelman et al. 1996; LaGrandeur and Parker 1998), which is digested by the exonuclease Xrn1p in a rapid 5′ to 3′ processive reaction, referred to as 5′ degradation (Muhlrad et al. 1994; McCarthy 1998). In the second pathway, deadenylated mRNAs are degraded in the 3′ to 5′ direction, referred to as 3′ degradation (Muhlrad et al. 1995; Jacobs Anderson and Parker 1998). A complex known as the exosome, which exists in both the nucleus and the cytoplasm, is required for this mode of decay (Mitchell et al. 1997). The cytoplasmic form of the exosome has been characterized, and the majority of its essential components have homology to 3′ to 5′ exoribonucleases (Mitchell and Tollervey 2000).
Uncapped and/or deadenylated mRNAs undergo rapid elimination through either the 5′- or 3′-degradation pathway. Previous studies showed that mRNAs of the L-A and M double-stranded RNA viruses, which lack both the 5′ cap and the 3′ poly(A) tail, are well expressed (Bruenn and Keitz 1976; Hannig et al. 1984; Wickner 1996), despite being sensitive to exonucleolytic digestion. The SKI genes, first isolated on the basis of a higher level of expression of viral toxin encoding mRNAs in the ski mutant strains (Toh et al. 1978; Ridley et al. 1984; Wickner 1996), were shown to encode proteins necessary for 5′ or 3′ mRNA decay. Ski1p was shown to be the 5′-exonuclease Xrn1p required for 5′ degradation (Stevens 1980), and the Ski complex containing the Ski2p, Ski3p, and Ski8p proteins was shown to be specifically required for cytoplasmic 3′ degradation (Jacobs Anderson and Parker 1998; Brown et al. 2000). Ski7p was also shown to be necessary for 3′ degradation of deadenylated mRNAs (van Hoof et al. 2000), and Ski7p-mediated interactions between the exosome and the Ski complex appear essential for the cytoplasmic 3′-mRNA decay (Araki et al. 2001).
The effect of ski mutations on the translation of uncapped and non-poly(A) luciferase encoding mRNAs, resembling the 5′ and 3′ ends of viral mRNAs, was examined by electroporation of in vitro transcribed RNAs into yeast cells, and demonstrated a specific increase in non-poly(A) mRNA translation in ski mutants (Masison et al. 1995; Benard et al. 1998, 1999). A study of the effect of addition of a poly(G) sequence to the 3′ end of mRNAs further emphasized the role of the SKI genes in the degradation of non-poly(A) mRNAs (Brown and Johnson 2001), because poly(G) constitutes a block to exonucleolytic activity (Vreken and Raue 1992; Decker and Parker 1993). When placed at the 3′ end of non-poly(A) mRNAs, poly(G) tails were shown to enhance expression in a way usually associated with ski3 cells in which 3′ degradation is inactivated (Brown and Johnson 2001). Thus, 3′ digestion plays an important role in limiting new translational events on non-poly(A) mRNAs. The work presented here was inspired by this approach, suitable for analysis of changes in 3′ degradation in vivo and, as we show here for the first time, changes in 5′ degradation.
With this tool, we decided to test the effect of stress-mediated GCN4 derepression on both the 5′- and 3′-mRNA-degradation pathways. GCN4 encodes a transcription factor that is activated when cells are subjected to amino acid starvation (Hinnebusch 1997). We show that 5′ degradation decreases under stress conditions and that this decrease can also be triggered by constitutive Gcn4p overproduction. A similar decrease in 5′ degradation occurs in the presence of lithium, which affects the sulfate assimilation pathway. This motivated a study of the MET16 gene, whose function in methionine biosynthesis is related to sulfate assimilation, and whose expression requires Gcn4p under certain circumstances (Thomas and Surdin-Kerjan 1997). We show that adenosine 3′-5′ biphosphate (pAp), a product of the Met16p reaction and known to block the activity of 5′ exonucleases (Dichtl et al. 1997), is a potential regulator of 5′ degradation under stress conditions via its Gcn4p-dependent production.
RESULTS
The experimental approach: the rationale
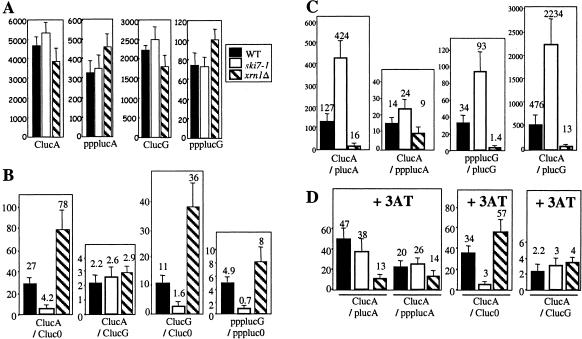
We decided to study variations in the 5′- and 3′-degradation pathways through the translation of luciferase-encoding mRNAs bearing features that affect the activity of exonucleases in cis. For example, non-poly(A) or poly(G) sequences at the 3′ end of mRNAs, which are sensitive or resistant to the activity of 3′ exonucleases, respectively, have already helped the characterization of the impact of 3′ degradation on luciferase expression (Brown and Johnson 2001). RNAs differing at their 5′ and 3′ ends were electroporated into yeast spheroplasts, and luciferase activity was measured in different mutant backgrounds. To avoid reproducibility problems when RNAs were electroporated into independent spheroplast preparations, we expressed the data as the ratio of a control RNA to an experimental RNA measured in the same spheroplast preparation. The control RNAs used for this study had either a cap or a triphosphate at the 5′ end and a poly(A) or a poly(G) tail at the 3′ end (Fig. 1A ▶). Because these features are known to limit the activity of exonucleases on mRNAs in cis (Stevens and Maupin 1987; Gerstel et al. 1992), luciferase expression from these RNAs was expected to remain relatively constant regardless of the genetic background. Indeed, luciferase levels were the same when these mRNAs were electroporated into wild-type cells, cells lacking the 5′ exonuclease, Xrn1p (xrn1Δ mutant), or cells disrupted for the 3′-degradation pathway (ski7 mutant; Fig. 1A ▶). The fact that highest luciferase expression was obtained with a capped and poly(A) mRNA shows that this method accurately reflects the cap and the poly(A) requirements for mRNA translation (Gallie 2001). We reasoned that a comparison of the ratio of the appropriate control RNA to its test RNA, in the wild-type strain and in strains inactivated for one or other of the exonuclease pathways, should provide a measure of the activity of the pathway in question.
FIGURE 1.
Expression ratios of various luciferase-encoding mRNAs in strains defective in 3′ and 5′ degradations. (A) Expression of luciferase from mRNAs bearing features that limit the impact of 5′ and 3′ exonucleases on mRNA expression in vivo, chosen as controls for luciferase expression. Spheroplasted cells were electroporated with 2 μg of defined luciferase encoding mRNAs and assayed for luciferase activity. RNAs were introduced in wild-type cells (strain RV603 transformed by pRS3167), cells deficient for the 3′-degradation pathway (ski7-1 mutant, strain RV603 transformed by pRS316), or cells lacking the Xrn1p 5′-exonuclease activity (xrn1Δ mutant, strain B1510). Luciferase activity is expressed in relative light units (RLU)/μg of total protein. The blank had a luciferase activity of less than 0.02 RLU/μg of total protein. The abbreviations for luciferase mRNAs are as follows: luc, luciferase RNA; C, 5′-capped; ppp, 5′-triphosphorylated end; A, 3′-poly(A); G, 3′-poly(G). Solid bars represent the mean luciferase activity measured after three electroporations of a single spheroplast preparation of each strain. The error bar represents the standard deviation. (B,C,D) Bars represent the mean luciferase activity of the relative expression of the control RNA (chosen as a standard for expression; see A) to the test RNA and error bars represent the standard deviation measured from at least three independent experiments. The ratio measured is indicated below the corresponding bar. (B) 3′ degradation limits luciferase production in RNAs sensitive to the activity of 3′ exonucleases (i.e., Cluc0 and pppluc0 mRNAs; 0 refers to neither poly(A) nor poly(G) tail at the 3′ end). 3′ degradation is limited either by a 3′-poly(G) in cis or by ski7 mutation in trans. (C) 5′ degradation limits luciferase production in RNAs sensitive to the activity of 5′ exonucleases (i.e., plucA and plucG mRNAs; p refers to 5′-monophosphorylated RNA). 5′ degradation is limited either by a 5′ppp-end in cis or by xrn1Δ mutation in trans. (D) 3AT-mediated amino acid stress limits 5′ degradation. ClucA is used as the standard for luciferase expression in the presence of 3AT. Spheroplast preparations used in B and C were treated in parallel with 10 mM of 3AT at least 2 h before cell electroporation and during the additional hour until cell collection.
Effect of ski7 mutation on 3′ degradation of electroporated RNAs
To confirm that this experimental approach could be used to measure effects on 3′ degradation, we first measured luciferase activity produced from different RNAs under conditions known to affect the 3′ pathway. By comparing luciferase expression from a capped and polyadenylated encoding RNA (referred to as ClucA) to that from the same RNA with neither a poly(A) nor poly(G) tail (Cluc0), in wild-type cells, we measured a combination of the translational defect due to the lack of a 3′ poly(A) tail in Cluc0 mRNAs and the sensitivity of its 3′ end to 3′ degradation. The ClucA/Cluc0 ratio was 27 in wild-type cells (Fig. 1B ▶). By comparing this ratio to that measured in ski7-1 cells (ClucA/Cluc0 = 4.2), we were able to quantify the specific increase in luciferase expression from non-poly(A) mRNAs in this mutant (Fig. 1B ▶; Benard et al. 1999). The ClucA/ClucG ratio (ClucG refers to the capped and poly(G) luc mRNA) is of interest because 3′ degradation is limited by a poly(G) track in cis (Parker et al. 1991; Brown and Johnson 2001). The ClucA/ClucG ratio was almost identical in wild type (2.2) and in ski7-1 cells (2.6; Fig. 1B ▶). Thus, we conclude that the difference in ClucA/Cluc0 ratio observed in wild type versus ski7-1 cells is due to the increased sensitivity of Cluc0 to 3′ degradation, in agreement with the fact that Ski7p protein operates in the cytoplasmic 3′-to-5′ mRNA-decay pathway (van Hoof et al. 2000). Very similar results were obtained by others in ski3Δ cells, in which 3′ degradation is also inactivated (Brown and Johnson 2001). The difference in 3′ degradation between wild-type and ski7-1 cells was also observed with the ClucG/Cluc0 and ppplucG/pppluc0 ratios (ppplucG refers to the 5′-triphosphorylated luc mRNA and poly(G) luc mRNA and pppluc0 to the 5′-triphosphorylated with neither a poly(A) nor poly(G) tail), in which the contribution of translational activation from the poly(A) tail, or both the cap and the poly(A) tail, respectively, has been removed (Fig. 1B ▶). All of these results taken together argue that differences in ratio between these strains are due to effects on mRNA turnover rather than on translation.
When the same experiments were performed in the xrn1Δ mutant, in which 5′ digestion is disrupted (Fig. 1B ▶), the effect of 3′ degradation on the luciferase expression from Cluc0 or pppluc0 mRNA is particularly visible (see high ClucG/Cluc0 or ppplucG/pppluc0 ratios in xrn1Δ cells vs. wild type). This is true despite the fact that most cellular mRNAs have to be degraded by the 3′-exonuclease pathway and one might expect effects due to titration. The apparent increase in 3′ degradation when the 5′ pathway is blocked suggests that either a compensatory mechanism exists in the cell or the mRNA itself becomes more susceptible to 3′ degradation under these conditions.
Demonstration of 5′ degradation with electroporated RNAs
Initially this system, which was shown to permit detection of effects on 3′ decay (Brown and Johnson 2001), was reported to be insensitive to the influence of 5′ decay on uncapped mRNAs (Masison et al. 1995). We attributed this to the presence of a 5′-triphosphate (5′-ppp) group on the in vitro transcribed mRNA, which is known to limit Xrn1p action in cis (Stevens and Maupin 1987; Hsu and Stevens 1993). We therefore modified the in vitro RNA synthesis protocol to produce primarily 5′-monophosphorylated (5′-p) RNAs, mimicking decapped mRNAs better in vivo (see Materials and Methods). We then measured the ClucA/plucA ratio (plucA denotes 5′-p and poly(A) mRNAs) expecting that it would reflect a combination of the translational defect due to the lack of a cap structure in plucA and the sensitivity of its 5′ end to 5′ degradation (Fig. 1C ▶). The ClucA/plucA ratio fell from 127 to 16 when wild-type cells were compared to xrn1Δ cells. This is likely to be the consequence of a defective 5′ degradation that allows better luciferase expression from 5′-p RNAs in the mutant. Consistent with this idea, the decrease in expression ratio is significantly less (14 vs. 9 for ClucA/ppplucA ratio) when the 5′-p end is replaced by a 5′-ppp moiety (ppplucA refers to the 5′-triphosphorylated and poly(A) luc mRNA), limiting Xrn1p activity in cis (Fig. 1C ▶).
We next examined the luciferase expression from 5′-p mRNAs independent of the translational effect due to the presence of both the cap and the poly(A) tail in cis, by measuring the ppplucG/plucG ratio (plucG for 5′-p and poly(G) mRNAs). This ratio, which was 34 in wild-type cells, dropped to 1.4 in xrn1Δ cells, where expression from 5′-p and from 5′-ppp mRNAs was almost identical due to disruption of the 5′-degradation pathway (Fig. 1C ▶). Thus, the ppplucG/plucG ratio reflects the activity of Xrn1p. This approach was also performed in ski7 cells (Fig. 1C ▶), in which the blocking of 3′ degradation led to a supplementary increase in the ppplucG/plucG ratio (i.e., an increase in 5′ degradation) compared to wild-type cells (93 vs. 34). This increase was also observed in the ClucA/plucA and ClucA/plucG ratios (Fig. 1C ▶). The response to the inactivation of 3′ degradation could either be an increase in cellular 5′ degradation or an alteration to the susceptibility of the luciferase mRNA to 5′ degradation.
Imbalance in 5′- and 3′-degradation pathways under stress conditions
The experiments described above show that this method reproduces the effects of the 5′ cap and poly(A) tails on mRNA stability described in other systems. We next asked whether the rate of mRNA degradation might change as a function of growth conditions, such as during amino acid starvation. To mimic such a stress, we prepared spheroplasts in the presence of 3-amino-1,2,4-triazole (3AT). This drug causes a rapid histidine starvation by inhibiting the product of the HIS3 gene. This causes enhanced translation of GCN4 mRNAs, mediated by low quantities of eIF2-GTP, visible within the first hour of drug addition (Hinnebusch 1997).
We used the ClucA, ClucG, or ppplucG contructs as control RNAs because the corresponding luciferase production did not vary in wild type, xrn1Δ and ski7-1 cells under stress conditions (data not shown). The ClucA/plucA ratio was used to measure 5′ degradation in the presence of 3AT (Fig. 1D ▶). Wild-type cells grown in the presence of 3AT showed a decrease in ClucA/plucA ratio (from 127 to 47), particularly perceptible in a ski7-1 mutant (from 424 to 38; compare ClucA/plucA ratio in Fig. 1D ▶ to Fig. 1C ▶). Because the replacement of the 5′-p end of mRNA by a 5′-ppp end or the use of a xrn1Δ strain, limiting 5′ degradation in cis and in trans, respectively, were sufficient to abolish the 3AT-mediated effect, we conclude that the differences observed are due to 5′-mRNA decay rather than translation (compare Figs. 1D ▶ and 1C ▶ and note the small changes in ClucA/ppplucA ratio in all three genetic backgrounds, and in ClucA/plucA ratio in xrn1Δ cells). Thus 5′ degradation is reduced during amino acid stress.
We next investigated the impact of 3AT on 3′ degradation through analysis of ClucA/Cluc0 and ClucA/ClucG ratios. All of the ratios in Figure 1D ▶, measured in wild-type or xrn1Δ cells in which 3′ degradation is active, remain relatively constant in comparison to Figure 1B ▶. Thus, unlike 5′ degradation, 3′ degradation is not affected under amino acid stress conditions. As a control in these experiments, the blocking of the 3′-decay pathway in trans using a ski7-1 mutant resulted in little variation in the ClucA/Cluc0 ratio, in the presence or absence of 3AT (Fig. 1D ▶ and Fig. 1B ▶, respectively). Likewise, the addition of a 3′-poly(G) track, blocking 3′ degradation in cis, and leaving luciferase expression controlled principally by translational effects, had a very limited impact on relative expression in wild-type and mutant cells in the presence of 3AT (ClucA/ClucG in Fig. 1D ▶). Because 5′ degradation decreases whereas 3′ degradation remains unchanged in the presence of 3AT, the relative importance of what is often described as a secondary pathway in RNA degradation is increased under these conditions.
GCN4 derepression specifically reduces 5′ degradation
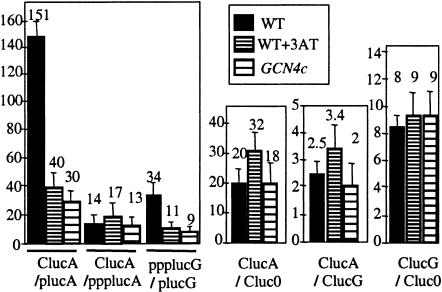
We considered the possibility that side effects from the use of 3AT might be responsible for the effect on 5′ degradation described above. Hence, we examined the effect of a direct constitutive overproduction of Gcn4p (GCN4c) from a plasmid on expression ratios. In these conditions, the ClucA/plucA ratio fell, showing that the marked decrease in 5′ degradation also occurs upon GCN4c expression (Fig. 2 ▶). No change was observed in 3′ degradation, determined by the ClucA/Cluc0 ratio. The fact that the 5′-ppp mRNA, which is resistant to 5′ degradation, showed unaltered luciferase expression under conditions of GCN4 derepression confirms that this decrease is due to a decrease in 5′ degradation rather than translational effects related to the cap structure (Fig. 2 ▶). We propose that the difference in the ratio of ppplucG to plucG mRNAs, which are independent of both cap and poly(A) translational activation, provides an accurate measure of 5′ degradation effects.
FIGURE 2.
Overexpression of Gcn4p reduces 5′ degradation. See legend to Figure 1 ▶ for abbreviations for mRNAs and ratios. ClucA, ClucG, and ppplucG are used as standards for luciferase expression. Gcn4p is overproduced by the addition of 10 mM of 3AT or by constitutive overproduction of Gcn4p, noted GCN4c. Wild-type cells correspond to the ski7-1 strain RV603 transformed by pRS3157 (SKI7 LEU2 CEN) and p16 (URA3 CEN). GCN4c cells (GCN4c) correspond to RV603 transformed by pRS3157 and plasmid p238 (GCN4c URA3 CEN).
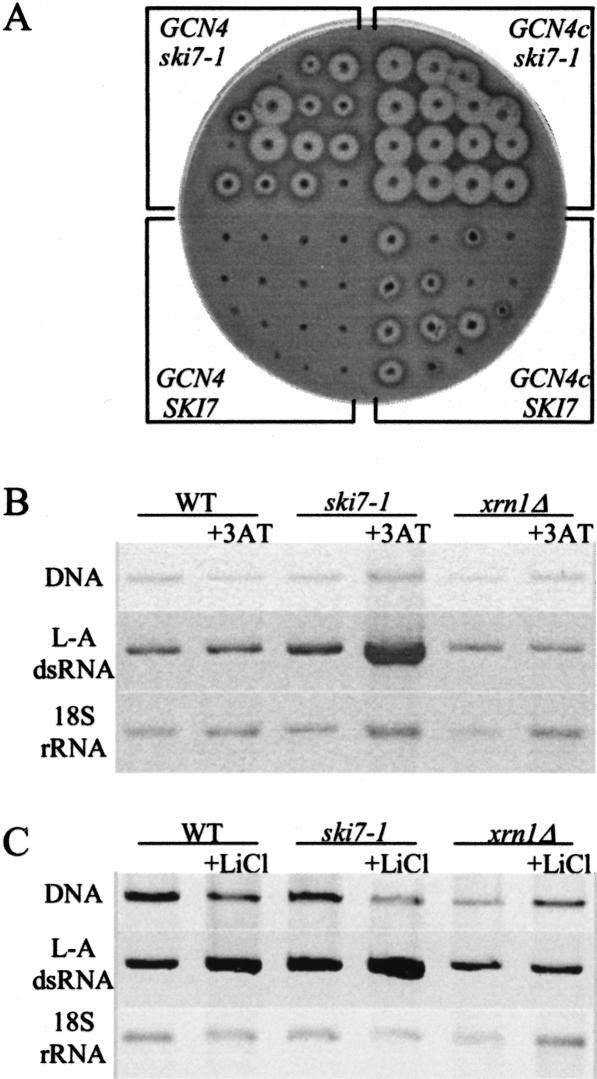
Propagation of viruses sensitive to 5′ exonuclease activity is increased under stress conditions
Yeast viruses produce uncapped and non-poly(A) mRNAs in the cytoplasm (for review, see Wickner 1996) and constitute a system that is physiologically relevant to the phenomenon described above. We wondered whether amino acid stress would have an effect on viral propagation. M2 viruses, whose Killer toxin expression and RNA replication depend on the integrity of the viral mRNA, were examined. M2 viruses are usually lost in wild-type cells grown at 32°C (SKI7 GCN4 in Fig. 3A ▶). Constitutive overproduction of Gcn4p, GCN4c, increased the maintenance of M2 viruses, even in ski7 cells in which 3′ degradation is inactivated (compare GCN4 ski7-1 to GCN4c ski7-1 cells). This is consistent with the hypothesis that 5′ degradation is limited upon GCN4 derepression, allowing exonuclease-sensitive M2 viruses to propagate better. The levels of L-A double-stranded RNAs (L-A dsRNA), which depend on the expression of uncapped and poly(A) viral mRNAs, were also analyzed in the presence of 3AT (Fig. 3B ▶). The addition of 3AT to ski7 cells resulted in a particularly significant increase in L-A dsRNA levels, relative to the amount of 18S rRNA or chromosomal DNA. On the contrary, the relative levels of L-A dsRNA diminished slightly in xrn1Δ cells in the presence of 3AT. We thus propose that the 3AT-mediated inhibition of the 5′ degradation is primarily responsible for large amounts of dsRNA observed in ski7 cells. This is in agreement with our observation that ClucA/plucA ratios, the marker of 5′ degradation, fell sharply in ski7 cells in the presence of 3AT (Fig. 1 ▶, cf. C and D). We also propose that the 3′ degradation, which is unaffected by stress conditions if we consider ClucA/Cluc0 ratios shown in Figure 1, B and D ▶, efficiently limits levels of L-A dsRNA in wild-type and in xrn1 cells in either the absence or presence of 3AT (Fig. 3B ▶).
FIGURE 3.
Maintenance of M2 killer viruses under constitutive overproduction of Gcn4p (A) and levels of L-A double-stranded RNA (dsRNA) in the presence of 3AT (B) or LiCl (C). (A) The killer M2 phenotype was tested on plates. Isolated colonies, obtained from different transformants of the Killer RV603 strain, were seeded with a lawn of the toxin-sensitive tester strain 5X47 (Ridley et al. 1984). RV603 was transformed by pRS316 or p238 (GCN4c) and pRS315 or pRS3157 (SKI7). (B,C) Agarose gels, shown in negative, stained with ethidium bromide. Strains were cured of M2 viruses for analysis of L-A dsRNA. L-A dsRNA levels are shown relatively to 18S rRNA and chromosomal DNA amounts, used as two standards of the nucleic acid preparations (see legend to Figure 1 ▶ for strains and Materials and Methods).
Inhibition of 5′ degradation by lithium
Other conditions that limit 5′-exonuclease activity have been described previously in the literature, such as the presence of lithium in the growth medium (Dichtl et al. 1997). We asked whether we could demonstrate this inhibition using our experimental approach and whether 3′ degradation might also be affected, because no information was available on this point. As shown in Figure 4 ▶, the addition of LiCl to cell cultures inhibits the enzymatic activity of Met22p/Hal2p and leads to an accumulation of adenosine 3′–5′ biphosphate (pAp), which in turn inhibits the 5′-exonuclease activity of Xrn1p (Dichtl et al. 1997). First, the relative levels of L-A dsRNA were analyzed in the presence of LiCl. As in the presence of 3AT, LiCl increased the relative amount of dsRNA in ski7-1 cells in which 3′ degradation is inactivated. Conversely, inactivation of the 5′-degradation pathway prevented the LiCl-mediated effect. Relative L-A dsRNA levels were even lower in xrn1Δ cells (Fig. 3C ▶). We correlated these observations to the lithium-mediated inhibition of 5′ degradation. We suggest that the 3′-degradation pathway remains fully functional in wild-type or xrn1 cells upon the addition of LiCl, as L-A dsRNA levels did not increase under these conditions (Fig. 3C ▶).
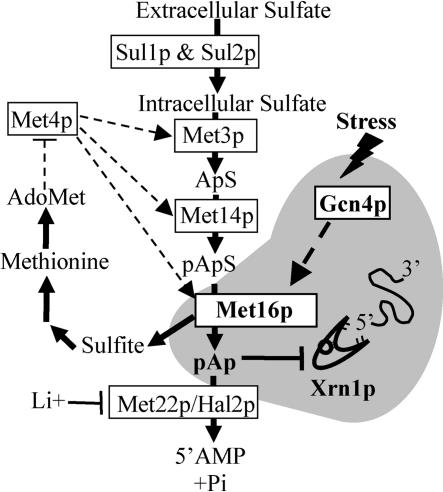
FIGURE 4.
The sulfate assimilation pathway shares steps with the methionine biosynthesis pathway (adapted from Thomas and Surdin-Kerjan 1997; Dichtl et al. 1997). Steps of sulfate assimilation are indicated by plain dark arrows. Sulfate enters the cell via the Sul1p and Sul2p transporters. Activities of Met3p, Met14p, and Met16p contribute to sulfate assimilation by producing sulfite. Sulfite is used in the production of methionine. Control of sulfate assimilation by methionine is ensured by subsequent production of AdoMet, which in turn represses the activity of Met4p (in dashed lines). Met4p participates in the transcriptional activation of MET3, MET14, and MET16 genes (dashed arrows). Consequently, production of both adenosine 5′-phosphosulfate (ApS) and of 3′-phosphoadenosine 5′-phosphosulfate (pApS) is greater in cells starved for methionine. Reduction of pApS by Met16p releases adenosine 3′-5′ biphosphate (pAp), which is metabolized into AMP and inorganic phosphate by Met22p/Hal2p. pAp inhibits the activity of the 5′-exonuclease Xrn1 p. Alternatively, pAp can accumulate because of the inhibition of Met22p/Hal2p in the presence of LiCl (Li+; Dichtl et al. 1997). Independently of Met4p, Gcn4p overproduction activates MET16 expression (dashed arrows).
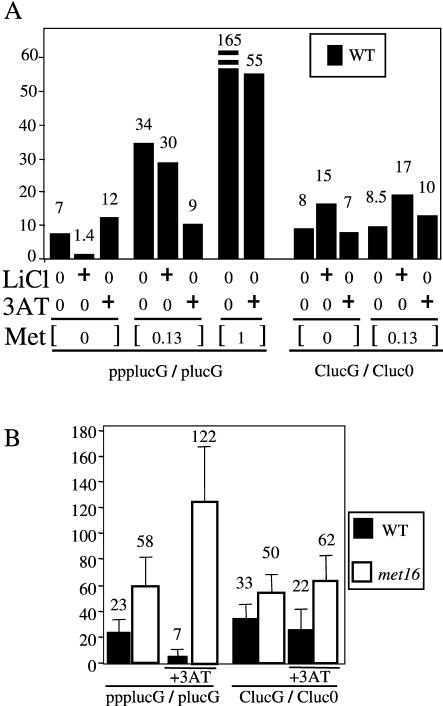
We thus anticipated correctly from these observations that the ppplucG/plucG ratio, which we have established as a marker of 5′ degradation, would decrease upon addition of LiCl to the culture (Fig. 5A ▶). These experiments were done in the absence of methionine, because the production of pAp is connected to the sulfate assimilation pathway. This pathway shares steps with the methionine biosynthesis pathway, and the lithium-induced accumulation of pAp is expected to be abolished by the presence of methionine (Fig. 4 ▶; Murguia et al. 1995; Thomas and Surdin-Kerjan 1997). For instance, the presence of 0.13 mM methionine in the growth medium reduces both adenosine 5′-phosphosulfate (ApS) and 3′-phosphoadenosine 5′-phosphosulfate (pApS) production, and a fortiori pAp synthesis, which occurs in downstream enzymatic steps (Jakubowski and Goldman 1993; Adams et al. 1997). Consequently, the effect of LiCl on the ppplucG/plucG ratio was abolished upon methionine addition, as shown in Figure 5A ▶. These experiments suggest that pAp accumulation is responsible for the decrease in 5′ degradation observed. 3′ degradation was not decreased by the addition of LiCl; on the contrary, the ClucG/Cluc0 ratios increased a little following LiCl addition (Fig. 5A ▶).
FIGURE 5.
Role of sulfate assimilation in 5′-to-3′ RNA degradation. See legend to Figure 1 ▶ for all the abbreviations for mRNAs and ratios. (A) Extracellular methionine affects 5′ degradation. Cells were grown in SD minimal media supplemented with tryptophan, isoleucine, arginine, valine, and lysine. In normal and spheroplast cell cultures, methionine was present in media when indicated. LiCl and 3AT were added 2 h before and 1 h after electroporation. Met indicates concentration of methionine (0, 0.13, 1 mM). Li+, lithium chloride: Absence (0) or addition (+ is 5 mM) of LiCl to block 5′-exonuclease activity through the accumulation of pAp (see Fig. 4 ▶; Dichtl et al. 1997). Absence (0) or addition (+ is 10 mM) of 3AT. Bars represent the mean calculated from two independent experiments. (B) Mutation in MET16 gene reverses the 3AT-mediated effect on the ppplucG/plucG ratio. The met16 mutant is strain CC362-2A and wild-type strain C130 (Thomas et al. 1990). Error bars represent the standard deviation measured from three independent experiments.
A concentration of 1 mM methionine is known to ensure a better inhibition of sulfate assimilation than 0.13 mM. As a consequence, only traces of ApS, pApS, and a fortiori pAp can be detected in cells at the higher methionine concentration (Fig. 4 ▶; Jakubowski and Goldman 1993; Murguia et al. 1995, 1996). We therefore expected that the highest ppplucG/plucG ratio would be detected in the presence of 1 mM of methionine, because of the increased activity of Xrn1p on 5′-monophosphorylated mRNAs. This was indeed the case as shown in Figure 5A ▶. We conclude that level of extracellular methionine also greatly influences 5′ degradation.
Reduction of 5′ degradation in response to 3AT depends on MET16
We noticed that the ppplucG/plucG ratio remained constant upon 3AT addition under conditions of a total absence of extracellular methionine (Fig. 5A ▶). The fact that the reduction in 5′ degradation under conditions of general amino acid stress was dependent on the presence of methionine suggested that the mechanisms may be linked and this motivated us to analyze the role of the MET16 gene in this process. The Met16p reductase participates in the biosynthesis of methionine and produces sulfite from pApS with pAp as a by-product (Fig. 4 ▶; Thomas et al. 1990). Furthermore, inactivation of GCN4 is known to affect expression of MET16 (Thomas et al. 1990). Conversely, the derepression of GCN4 by tryptophan limitation was also shown to increase the steady-state level of MET16 mRNA. We thus postulated that direct Gcn4p overexpression, or 3AT-mediated GCN4 derepression, activated MET16 expression. As a consequence, a temporary accumulation of pAp would limit 5′ degradation. The ppplucG/plucG ratio in Figure 5B ▶ shows that 5′ degradation is no longer decreased by amino acid stress in met16 cells. The CluG/Cluc0 ratio increases slightly under the same conditions. Thus, unlike 5′ degradation, 3′ decay is not inhibited by pAp accumulation. These results strongly support a model in which the GCN4 derepression activates MET16 expression, leading to a temporary accumulation of pAp, which in turn inhibits 5′ degradation (Fig. 4 ▶).
DISCUSSION
The introduction of in vitro synthesised luciferase encoding mRNAs into S. cerevisiae by electroporation has previously been used to evaluate 3′-exonucleolytic activity on mRNAs (Brown and Johnson 2001). What these authors found using ski3 cells is reproduced here with ski7 cells. We demonstrate through a similar experimental procedure that the use of 5′-p RNAs and 5′-ppp RNAs supplies additional information on 5′-exonucleolytic activity. We go on to exploit this system to show that an inhibition of 5′ degradation during the general stress response is likely due to GCN4-dependent expression of MET16. Met16p produces pAp, a well-known inhibitor of 5′ degradation as a by-product of its enzymatic activity.
The fact that in vitro synthesized and electroporated mRNAs must be translated amplifies effects on degradation and can lead to variations of up to 30-fold in the production of luciferase from 5′-monophosphorylated mRNAs. This sensitivity constitutes an advantage over conventional methods for the detection of potentially small effects on 5′ degradation. Electroporation of uncapped and/or poly(A)-deficient mRNAs also eliminates complications related to Pol II transcription and to the export of such mRNAs from the nucleus to the cytoplasm. This method is at a disadvantage if the purpose is to analyze cellular mRNA–protein complexes (mRNPs) because these artificial RNAs do not undergo nuclear maturation and may not acquire the appropriate mRNP proteins. However, because our aim is to test the capacity of the exonucleolytic machinery to attack RNA under different growth conditions, we believed that this question can be more than adequately addressed using electroporation of artificial mRNAs.
The relatively efficient translation of electroporated non-poly(A) or poly(G) mRNAs (Masison et al. 1995; Benard et al. 1998, 1999; Brown and Johnson 2001) may seem contradictory to the fact that deadenylated cellular mRNAs are known to be rapidly decapped and degraded by the 5′ mRNA decay pathway (McCarthy 1998). It should be remembered, however, that uncapped and unadenylated viral mRNAs are actually well expressed in yeast (Wickner 1996) and poly(A)-deficient mRNAs can be efficiently translated in certain mutant backgrounds (Proweller and Butler 1994, 1997). It has previously been suggested that the mRNP formed by electroporated in vitro transcribed RNAs differ from that of cellular mRNAs, in a way that renders them more resistant to decapping and hence 5′ degradation (Brown and Johnson 2001). One potential difference could be the Lsm1p–Lsm7p complex, which is required for decapping and only stably interacts with mRNAs following deadenylation (Tharun and Parker 2001). It has been suggested that the process of deadenylation itself triggers the interaction of Lsm1p–7p complex with the cellular mRNA. If this is true, then electroporation of non-poly(A) mRNAs may lead to an mRNP organization incapable of efficiently triggering a decapping step. This might explain why the Cluc0 mRNA is more highly translated than would be expected with an in vivo matured mRNA. Despite the uncertainty about whether the mRNP structures of electroporated RNAs are similar to that of a Pol II-transcribed mRNA, this system can nonetheless be exploited to provide valuable information about changes in exonucleolytic activity, as highlighted by the effect of ski mutations on luciferase expression from electroporated mRNAs by us and others (Brown and Johnson 2001). In this context, we noticed that disruption of one degradation pathway led to an apparent increase in the activity of the other. Another possibility is that disruption of one pathway somehow alters the susceptibility of the mRNA to the remaining decay pathway, although alterations in mRNP structure in response to inactivation of one of the degradation pathways has not yet been reported.
5′-exonucleolytic activity is decreased upon addition of 3AT to yeast spheroplasts or upon constitutive Gcn4p overproduction. The 3′-degradation pathway, often reported as secondary, seems to predominate in cells undergoing amino acid stress. Previous studies demonstrated that addition of lithium to cultures also inhibits 5′-exonuclease activity (Dichtl et al. 1997). We initially analyzed the inhibitory effect of lithium to validate our experimental approach. As expected, lithium inhibited the 5′ degradation on 5′-p mRNAs. Lithium causes pAp accumulation, via the inhibition of the Met22p/Hal2p nucleotidase activity that dephosphorylates pAp, which in turn blocks 5′-exonuclease activity (Fig. 4 ▶; Dichtl et al. 1997). Interestingly, pAp is an intermediate of the sulfate assimilation pathway, which shares steps with the methionine biosynthetic pathway (Thomas and Surdin-Kerjan 1997). Extracellular methionine limits sulfate assimilation (and pAp accumulation), via a mechanism that monitors the intracellular level of S-adenosylmethionine (AdoMet; Fig. 4 ▶). Consequently, the inhibition of 5′ degradation by lithium is totally abrogated by the presence of extracellular methionine.
MET16, like other methionine biosynthetic MET genes, is regulated by MET4 in response to methionine availability (Thomas and Surdin-Kerjan 1997). In media containing increasing methionine concentrations (from 0 mM to 1 mM), Met16p activity decreases, as does pAp production. pAp has been previously shown to directly inhibit the in vitro activities of the two 5′ exonucleases, Xrn1p and Rat1p (Dichtl et al. 1997). Consistent with this idea, we saw that the ppplucG/plucG ratio, the indicator of the level of 5′ decay, increased. The in vivo concentration of pAp was not determined in this study. Rather, we chose a genetic approach to study the possible involvement of pAp in this phenomenon, and all of the results above sustain a model in which Met16p-produced pAp is the key molecule responsible for decreasing 5′ degradation. In parallel, we showed that the activity of 3′ exonucleases, present in the exosome, and other cofactors like the Ski2p/Ski3p/Ski8p complex and Ski7p, are not inhibited under these stress conditions and a fortiori by pAp accumulation.
We wondered how a limitation in methionine might affect 3AT-mediated effects. The fact that absence of extracellular methionine did not result in an additional decrease of 5′ degradation in the presence of 3AT led us to suspect the existence of a link between methionine levels, pAp accumulation, and amino acid stress. The MET16 gene product was a good candidate for this connection, first, because its enzymatic activity generates sulfite and pAp from pApS (Thomas and Surdin-Kerjan 1997), and second, because GCN4-dependent expression of MET16 has been shown to occur during the general control response to tryptophan starvation (O’Connell et al. 1995). We thus analyzed the effect of a mutation in MET16 and showed that it reversed the 3AT-induced inhibition of 5′ degradation.
The absence of extracellular methionine was shown to dominate over the 3AT-induced inhibition of 5′ degradation. This can be explained by the fact that MET16 transcription can be activated by two independent mechanisms (O’Connell et al. 1995). The first is a specific control pathway that is triggered by MET4 when the level of extracellular methionine is low (O’Connell and Baker 1992). The second mechanism triggers MET16 expression in response to starvation for many different amino acids and involves GCN4 (O’Connell et al. 1995). Despite the involvement of general control in the modulation of MET16 expression, GCN4 is not essential for the methionine-specific (MET4-dependent) regulation of MET16 gene, because the response of MET16 gene to methionine withdrawal still occurs in the gcn4 mutant. The activation of MET16 by the general control mechanism in response to tryptophan starvation was shown not to be as effective as methionine limitation. Thus, 3AT does not have much effect during methionine limitation because the MET genes are already fully activated by MET4. Alternatively, the activation of MET16 by general control differs from the specific control pathway because it does not require MET4. Therefore, MET16 expression can easily be triggered by 3AT through GCN4 in cells growing in the presence of 0.13 mM methionine, in which MET genes are likely to be moderately expressed, but pApS levels are sufficient to allow generation of pAp (Jakubowski and Goldman 1993).
The control of the sulfate assimilation pathway by methionine led us to suppose that in cells growing in 1 mM methionine, only traces of pApS would be produced and, consequently, no pAp would accumulate. Because 5′ degradation is most active (i.e., the highest ppplucG/plucG ratio) in the presence of 1 mM methionine and reflects the absence of pAp, how does the inhibition of 5′ degradation occur during 3AT-induced stress? The enzymatic activity of the MET genes is not, in fact, as low as expected in conditions of high extracellular methionine (about 10% of activity remains; Thomas and Surdin-Kerjan 1997). We propose that 3AT-mediated stress, or GCN4 derepression, triggers the early phase of sulfate assimilation, and, as a consequence, increases intracellular pApS and pAp concentrations. Interestingly, MET4 contains multiple GCN4 sites and is upregulated by the general control response (Mountain et al. 1993). Therefore, even though the MET3 and MET14 gene promoters do not contain GCN4-binding sites, an indirect activation by Gcn4p is possible via Met4p. Furthermore, these genes and the SUL1 and SUL2 genes, encoding high-affinity sulfate transporters, have been shown to be targets of GCN4 by cDNA microarray analysis (Natarajan et al. 2001). We therefore propose that sulfate assimilation is boosted by the general control pathway and that the presence of 1 mM extracellular methionine does not limit pAp production under these stress conditions.
Sodium, like lithium, inhibits the MET22/HAL2 gene product, and causes pAp accumulation (Murguia et al. 1996). Part of the salt toxicity is due to the inhibition of ribosomal RNA processing, mostly through the direct salt inhibition of the essential MRP endonuclease activity, but also through inhibition of the activities of the Xrn1p and Rat1p 5′ exonucleases via pAp accumulation (Dichtl et al. 1997). Interestingly, it has been demonstrated that constitutive overproduction of Gcn4p causes a partial sodium hypersensitivity (Goossens et al. 2001). This may thus be partially explained by the fact that GCN4 derepression inhibits RNA degradation, through increased pAp accumulation, and inhibition of 5′ exonucleases.
We propose that amino acid stress decreases 5′-exonucleolytic activity to reduce the destabilization of decapped mRNAs. Independent of stress, the chemical half-life of full-length mRNAs increases substantially when the 5′-exonuclease Xrn1p is inactivated. It has been observed that decapped and non-poly(A) mRNAs accumulate under these conditions (Hsu and Stevens 1993). The expression of uncapped mRNA, which is albeit 10-fold lower than capped species, and the presence of non-poly(A) mRNAs in the polysome fraction has already been described (Proweller and Butler 1994; Lo et al. 1998), but because the cap is required for efficient translation initiation, the uncapped mRNAs are not likely to be as competent for translation as other mRNA species. Therefore, stress-regulated 5′ degradation may not lead to a major modification of the general protein pattern. This may explain why the possibility that a decapped cellular mRNA is expressed more abundantly, because of its specific stabilization during the general control response, has not yet been shown. This regulation by stress could be of general significance for particular classes of cellular mRNAs, as we showed for yeast viral mRNAs, and may also be extended in the future to noncoding RNAs.
We conclude that GCN4 derepression limits the 5′-mRNA-degradation pathway, which is likely to be conserved in all eukaryotes. These observations may be attributed in the future to many stresses, because Gcn4p is considered as a master regulator of gene expression under diverse conditions (Hinnebusch and Natarajan 2002). Another conserved system is the 3′-to-5′-degradation pathway, which seems to be biochemically resistant to pAp accumulation. Therefore, an analysis of 3′-to-5′ exonuclease activity during lithium, sodium, or other toxic stresses could be of future interest to the eukaryotic world.
MATERIALS AND METHODS
Media
Yeast were grown in media based on Hartwell’s synthetic complete (HC), or synthetic minimum media (SD) as described elsewhere (Adams et al. 1997). Two percent glucose was used as the carbon source. For spheroplast preparations and cultures, 1 M of sorbitol was systematically added. For cell cultures HC-uracil-histidine was used for all of the experiments shown in Figures 1 ▶ and 5B ▶. HC-uracil-histidine-leucine synthetic media was used for the experiments described in Figure 2 ▶ (Adams et al. 1997). For cell cultures described in Figure 5A ▶, minimal medium SD was completed with tryptophan, isoleucine, arginine, valine, and lysine in concentration based on HC media to minimize GCN4 expression. Induction of the GCN4 regulatory mechanism upon histidine starvation was achieved by addition of the antimetabolite 3-amino-1,2,4-triazole, 3AT, to a final concentration of 10 mM in all liquid cultures. 3AT, LiCl, and methionine are used when indicated. Yeast transformations were performed by the LiAc procedure (Adams et al. 1997).
Yeast strains
The ski7-1 strain RV603 (MATa ura3 leu2 ski7-1 mkt1; L-A-HN M2) transformed by pRS3167 (CEN URA3 SKI7; Benard et al. 1999) or by pRS3157 (CEN LEU2 SKI7) was used in this study as a wild-type strain. RV603 was crossed with xrn1Δ strain 4602-3C (MATalpha trp1 ura3 his3 xrn1 :: URA3) to give strain B1510 (MATa ura3 leu2 mkt1 xrn1 :: URA3; L-A-HN M2) after three successive back crosses. This was the xrn1Δ mutant strain used for all this study. The met16 mutant strain is CC362-2A and isogenic wild-type strain is C130 (Thomas et al. 1990).
Plasmids
Plasmid pRS3157 (SKI7 LEU2 CEN) was created by subcloning the HindIII-SpeI region containing SKI7 into the HindIII-SpeI sites of pRS315 (LEU2 CEN). Plasmid pT7-lucG30 (3′-poly(G)) was constructed from pT7-luc (Gallie et al. 1991) by substitution of XbaI-SmaI DNA fragment with annealed oligonucleotides OlbG (5′(Gx29)CCC-3′) and OlbC (5′-GGG(C)30TAG-3′). All inserts and junctions were confirmed by sequencing. Plasmid p16 (URA3 CEN) and plasmid p238, which overproduces Gcn4p (GCN4c URA3 CEN), were generous gifts from A. Hinnebusch (NIH, Bethesda, MD; Rolfes and Hinnebusch 1993).
In vitro transcription of luciferase mRNA
The luciferase mRNA expression plasmids pT7-luc (no 3′-poly(A)) and pT7-lucG30 (3′-poly(G)) were linearized with SmaI. pT7-luc50A (50mer poly(A) tail) was linearized with DraI. Plasmids pT7-luc and pT7-luc50A are generous gifts from D. Gallie (University of California, Riverside; Gallie et al. 1991). 5′-Capped or 5′-triphosphorylated RNAs were synthesized with an Ambion MEGAscript kit as previously described (Masison et al. 1995). We modified the Ambion protocol for the synthesis of 5′-monophosphorylated mRNAs by adding GMP (Sigma) in a concentration identical to ATP, UTP, and CTP, and lowering GTP concentration by fourfold. Like Cap analog incorporation, efficient GMP incorporation is characterized by at least a threefold reduction in RNA yield.
Expression of luciferase from mRNAs
The preparation of spheroplasts was performed as described previously with minor modifications (Everett and Gallie 1992). The last step of the treatment consists of a 2-h incubation at 30°C in HC or SD completed media, with 3AT or LiCl, when indicated. Cells were resuspended and washed in 1 M sorbitol for electroporation. Two micrograms of luciferase mRNAs, within the linear range of the luciferase assay (data not shown), were used for electroporation. After 1 h of growth at 30°C in corresponding media, with 3AT or LiCl when indicated, cells were pelleted and frozen in dry ice. Cells were assayed for luciferase activity as previously described (Masison et al. 1995) except for the use of a lysis buffer (Promega luciferase assay kit buffer) and a freezing step in dry ice just before melting and mixing with the luciferin reaction mix (Promega) for a gain in luciferase activity.
M2 killer phenotype test
M2 dsRNA is a killer toxin-encoding satellite of the L-A virus. MKT1 gene product is only needed for M2 replication or maintenance above 30°C. Loss of M2 viruses is total in an mkt1 strain, such as RV603 strain transformed by pRS3157 (referred to as the wild-type strain). The ski7-1 mutation in the mkt1 strain RV603 allows maintenance of M2 viruses at 32°C (Benard et al. 1999 and references therein). Each transformant was streaked for single colonies at 32°C on solid HC medium lacking uracil, leucine, and histidine. Thirty colonies were pooled and streaked again. Isolated colonies were finally tested for their M2 killer phenotype (Ridley et al. 1984).
Analysis of double-stranded RNA
Because M2 compete with L-A viruses for replication, RV603 and B1015 strains cured of M2 viruses were analyzed to facilitate observations on L-A dsRNA levels. Strains were grown on solid HC media lacking uracil and histidine, in the presence of 10 mM 3AT when indicated, or on solid SD minimal media complemented with tryptophan, isoleucine, arginine, valine, and lysine, in the presence of 0.2 M LiCl when indicated. Nucleic acids were prepared as previously described (Fried and Fink 1978). Four micrograms were loaded in each lane, separated on a 1% agarose gel, and stained with ethidium bromide.
Acknowledgments
I thank Reed Wickner, D. Gallie, A. Hinnebusch, T. Dever, and D. Thomas for plasmids and strains. I am very grateful to P. Stragier for his hospitality to facilitate the development of the project, and to R. Kuras for providing the apparatus for electroporation. I would like to thank my Yeast Group colleagues, especially A. Morillon, P. Lesage, and M. Springer, for technical support and stimulating discussions. Special thanks go to C. Condon for helpful discussions and critical reading of the manuscript. This work was supported by a grant from the CNRS (UPR 9073). The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5183804.
REFERENCES
- Adams, A., Gottschling, D.E., Kaiser, C.A., and Stearns, T. 1997. Methods in yeast genetics: A cold spring harbor laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Araki, Y., Takahashi, S., Kobayashi, T., Kajiho, H., Hoshino, S., and Katada, T. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20: 4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646. [DOI] [PubMed] [Google Scholar]
- Benard, L., Carroll, K., Valle, R.C., and Wickner, R.B. 1998. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 18: 2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard, L., Carroll, K., Valle, R.C., Masison, D.C., and Wickner, R.B. 1999. The ski7 antiviral protein is an EF1-α homolog that blocks expression of non-Poly(A) mRNA in Saccharomyces cerevisiae. J. Virol. 73: 2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.T. and Johnson, A.W. 2001. A cis-acting element known to block 3′ mRNA degradation enhances expression of polyA-minus mRNA in wild-type yeast cells and phenocopies a ski mutant. RNA 7: 1566–1577. [PMC free article] [PubMed] [Google Scholar]
- Brown, J.T., Bai, X., and Johnson, A.W. 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn, J. and Keitz, B. 1976. The 5′ ends of yeast killer factor RNAs are pppGp. Nucleic Acids Res. 3: 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G. and Parker, R. 1996. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 60: 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J. and Parker, R. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 7: 1632–1643. [DOI] [PubMed] [Google Scholar]
- Dichtl, B., Stevens, A., and Tollervey, D. 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16: 7184–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett, J.G. and Gallie, D.R. 1992. RNA delivery in Saccharomyces cerevisiae using electroporation. Yeast 8: 1007–1014. [DOI] [PubMed] [Google Scholar]
- Fried, H.M. and Fink, G.R. 1978. Electron microscopic heteroduplex analysis of “killer” double-stranded RNA species from yeast. Proc. Natl. Acad. Sci. 75: 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, D.R. 2001. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 75: 12141–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, D.R., Feder, J.N., Schimke, R.T., and Walbot, V. 1991. Posttranscriptional regulation in higher eukaryotes: The role of the reporter gene in controlling expression. Mol. Gen. Genet. 228: 258–264. [DOI] [PubMed] [Google Scholar]
- Gerstel, B., Tuite, M.F., and McCarthy, J.E. 1992. The effects of 5′-capping, 3′-polyadenylation and leader composition upon the translation and stability of mRNA in a cell-free extract derived from the yeast Saccharomyces cerevisiae. Mol. Microbiol. 6: 2339–2348. [DOI] [PubMed] [Google Scholar]
- Goossens, A., Dever, T.E., Pascual-Ahuir, A., and Serrano, R. 2001. The protein kinase Gcn2p mediates sodium toxicity in yeast. J. Biol. Chem. 276: 30753–30760. [DOI] [PubMed] [Google Scholar]
- Hannig, E.M., Thiele, D.J., and Leibowitz, M.J. 1984. Saccharomyces cerevisiae killer virus transcripts contain template-coded polyadenylate tracts. Mol. Cell. Biol. 4: 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, A.G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J. Biol. Chem. 272: 21661–21664. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A.G. and Natarajan, K. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1: 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C.L. and Stevens, A. 1993. Yeast cells lacking 5′ → 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson, J.S. and Parker, R. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski, H. and Goldman, E. 1993. Methionine-mediated lethality in yeast cells at elevated temperature. J. Bacteriol. 175: 5469–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur, T.E. and Parker, R. 1998. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, H.J., Huang, H.K., and Donahue, T.F. 1998. RNA polymerase I-promoted HIS4 expression yields uncapped, polyadenylated mRNA that is unstable and inefficiently translated in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison, D.C., Blanc, A., Ribas, J.C., Carroll, K., Sonenberg, N., and Wickner, R.B. 1995. Decoying the cap-mRNA degradation system by a double-stranded RNA virus and poly(A)-mRNA surveillance by a yeast antiviral system. Mol. Cell. Biol. 15: 2763–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, J.E. 1998. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 62: 1492–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2000. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7: 843–846. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466. [DOI] [PubMed] [Google Scholar]
- Mountain, H.A., Bystrom, A.S., and Korch, C. 1993. The general amino acid control regulates MET4, which encodes a methionine-pathway-specific transcriptional activator of Saccharomyces cerevisiae. Mol. Microbiol. 7: 215–228. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., and Parker, R. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes & Dev. 8: 855–866. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murguia, J.R., Belles, J.M., and Serrano, R. 1995. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267: 232–234. [DOI] [PubMed] [Google Scholar]
- ———. 1996. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J. Biol. Chem. 271: 29029–29033. [DOI] [PubMed] [Google Scholar]
- Natarajan, K., Meyer, M.R., Jackson, B.M., Slade, D., Roberts, C., Hinnebusch, A.G., and Marton, M.J. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21: 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell, K.F. and Baker, R.E. 1992. Possible cross-regulation of phosphate and sulfate metabolism in Saccharomyces cerevisiae. Genetics 132: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell, K.F., Surdin-Kerjan, Y., and Baker, R.E. 1995. Role of the Saccharomyces cerevisiae general regulatory factor CP1 in methionine biosynthetic gene transcription. Mol. Cell. Biol. 15: 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., Herrick, D., Peltz, S.W., and Jacobson, A. 1991. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 194: 415–423. [DOI] [PubMed] [Google Scholar]
- Proweller, A. and Butler, S. 1994. Efficient translation of poly(A) deficient mRNAs in Saccharomyces cerevisiae. Genes & Dev. 8: 2629–2640. [DOI] [PubMed] [Google Scholar]
- Proweller, A. and Butler, J.S. 1997. Ribosome concentration contributes to discrimination against poly(A)- mRNA during translation initiation in Saccharomyces cerevisiae. J. Biol. Chem. 272: 6004–6010. [DOI] [PubMed] [Google Scholar]
- Ridley, S.P., Sommer, S.S., and Wickner, R.B. 1984. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 4: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfes, R.J. and Hinnebusch, A.G. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: Implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13: 5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, A. 1980. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to 3′ mode of hydrolysis. J. Biol. Chem. 255: 3080–3085. [PubMed] [Google Scholar]
- Stevens, A. and Maupin, M.K. 1987. A 5′–3′ exoribonuclease of Saccharomyces cerevisiae: Size and novel substrate specificity. Arch. Biochem. Biophys. 252: 339–347. [DOI] [PubMed] [Google Scholar]
- Tharun, S. and Parker, R. 2001. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8: 1075–1083. [DOI] [PubMed] [Google Scholar]
- Thomas, D. and Surdin-Kerjan, Y. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61: 503–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D., Barbey, R., and Surdin-Kerjan, Y. 1990. Gene–enzyme relationship in the sulfate assimilation pathway of Saccharomyces cerevisiae. Study of the 3′-phosphoadenylylsulfate reductase structural gene. J. Biol. Chem. 265: 15518–15524. [PubMed] [Google Scholar]
- Toh, E.A., Guerry, P., and Wickner, R.B. 1978. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 136: 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Staples, R.R., Baker, R.E., and Parker, R. 2000. Function of the Ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol. 20: 8230–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken, P. and Raue, H.A. 1992. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3′-terminal part of the coding region. Mol. Cell. Biol. 12: 2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R.B. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 60: 250–265. [DOI] [PMC free article] [PubMed] [Google Scholar]