Abstract
To correct misactivation and misacylation errors, Escherichia coli valyl-tRNA synthetase (ValRS) catalyzes a tRNAVal-dependent editing reaction at a site distinct from its aminoacylation site. Here we examined the effects of replacing the conserved 3′-adenosine of tRNAVal with nucleoside analogs, to identify structural elements of the 3′-terminal nucleoside necessary for tRNA function at the aminoacylation and editing sites of ValRS. The results show that the exocyclic amino group (N6) is not essential: purine riboside-substituted tRNAVal is active in aminoacylation and in stimulating editing. Presence of an O6 substituent (guanosine, inosine, xanthosine) interferes with aminoacylation as well as posttransfer and total editing (pre- plus posttransfer editing). Because ValRS does not recognize substituents at the 6-position, these results suggest that an unprotonated N1, capable of acting as an H-bond acceptor, is an essential determinant for both the aminoacylation and editing reactions. Substituents at the 2-position of the purine ring, either a 2-amino group (2-aminopurine, 2,6-diaminopurine, guanosine, and 7-deazaguanosine) or a 2-keto group (xanthosine, isoguanosine), strongly inhibit both aminoacylation and editing. Although aminoacylation by ValRS is at the 2′-OH, substitution of the 3′-terminal adenosine of tRNAVal with 3′-deoxyadenosine reduces the efficiency of valine acceptance and of posttransfer editing, demonstrating that the 3′-terminal hydroxyl group contributes to tRNA recognition at both the aminoacylation and editing sites. Our results show a strong correlation between the amino acid accepting activity of tRNA and its ability to stimulate editing, suggesting misacylated tRNA is a transient intermediate in the editing reaction, and editing by ValRS requires a posttransfer step.
Keywords: tRNA, aminoacyl-tRNA synthetase, translation, aminoacylation, nucleoside analogs, translational editing
INTRODUCTION
In a two-step reaction that constitutes the first step of protein biosynthesis, each aminoacyl-tRNA synthetase esterifies a specific amino acid to the ribose of the universally conserved 3′-terminal adenosine of its set of cognate tRNA isoacceptors. The fidelity of this reaction is crucial for the accuracy of protein synthesis. Several aminoacyl-tRNA synthetases, however, have difficulty discriminating between cognate and structurally similar amino acids, resulting in misactivation of noncognate amino acids and misacylation of tRNA. To minimize errors and maintain translational fidelity, these enzymes catalyze a tRNA-dependent (Fersht and Kaethner 1976; Jakubowski and Fersht 1981) proofreading (editing) reaction that hydrolyzes either the incorrectly activated aminoacyl-adenylate (pretransfer editing) or the mischarged tRNA (posttransfer editing) (for review, see Jakubowski and Goldman 1992). Studies with the class Ia synthetases IleRS (Schmidt and Schimmel 1994; Lin et al. 1996), valyl-tRNA synthetase (ValRS; Lin and Schimmel 1996; Lin et al. 1996), and LeuRS (Chen et al. 2000; Cusack et al. 2000; Mursinna et al. 2001) showed that the aminoacylation and editing active sites of these enzymes are spatially separate; an ~200-amino-acid insert in the aminoacylation site, designated CP1, is responsible for the editing activity (Schmidt and Schimmel 1995; Lin et al. 1996; Nureki et al. 1998; Fukai et al. 2000, 2003; Lincecum et al. 2003). Cognate tRNAs specifically trigger transfer of misactivated amino acids from the synthetic active site to the editing active site, accounting for the tRNA dependence of the editing reaction (Silvian et al. 1999; Fukai et al. 2000; Bishop et al. 2002, 2003).
Chemical and enzymatic modification studies demonstrated the importance of the 3′-end of tRNA in the aminoacylation reaction (for review, see Sprinzl and Cramer 1979) and in translational editing (Tardif et al. 2001; Tardif and Horowitz 2002). Altering the 3′-terminal adenosine (Best and Novelli 1971; Tal et al. 1972; Rether et al. 1974; Von der Haar and Gaertner 1975) or its ribose moiety (Uziel and Jacobson 1974) results in complete loss or considerable decrease in aminoacylation efficiency, largely due to a decrease in kcat. 3′-end-modified tRNAs are also defective in editing (Tardif et al. 2001) and are readily misacylated (Tamura et al. 1994; Tardif et al. 2001; Nordin and Schimmel 2002).
Although these results imply that the universally conserved 3′-terminal adenosine is specifically recognized at both the aminoacylation and the editing sites of synthetases, recent experiments have shown that Escherichia coli tRNAVal (Liu and Horowitz 1994; Tamura et al. 1994; Tardif et al. 2001) and several other tRNAs (Liu et al. 1998) with base substitutions at the 3′-end are readily aminoacylated. 3′-end-modified tRNAVal, however, does not stimulate the editing activity of ValRS, nor can the modified tRNAVal, misacylated with threonine, be edited (Tardif et al. 2001), suggesting that the aminoacylation active site of the enzyme recognizes different features of the 3′-terminal nucleotide than the editing active site.
In this study we examined the role of functional groups on the 3′-terminal nucleoside of E. coli tRNAVal in specific recognition of the 3′-end of tRNAVal by ValRS. By replacing the universally conserved A76 with nucleoside analogs having different chemical substituents on the base or sugar, we probed the contribution of individual substituents to the specificity of recognition at the aminoacylation and editing active sites of the enzyme, and we compared the structural elements essential for function at each site.
RESULTS
Functional groups of the 3′-terminal base of tRNAVal recognized at the aminoacylation and editing sites of valyl-tRNA synthetase
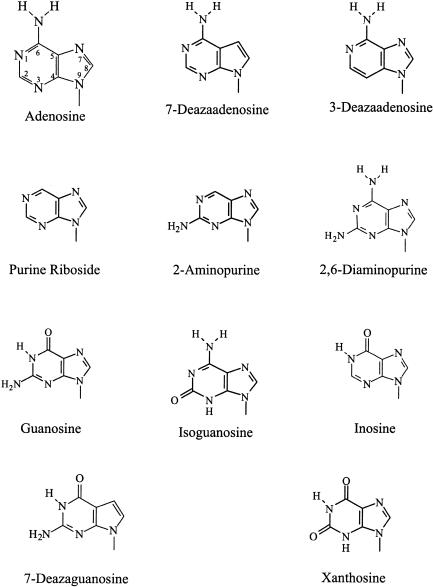
The universally conserved 3′-adenosine of E. coli tRNAVal was replaced with nucleotide analogs by using T4 RNA ligase to add bisphosphate derivatives of the analogs to tRNAVal transcripts lacking the terminal adenosine. After dephosphorylation with calf intestinal phosphatase, the product of a control ligation with pAp was fully active in both aminoacylation and editing (Tables 1 ▶, 3 ▶). Base analogs substituted for the 3′-terminal adenine differ from adenine either in the heterocyclic ring or in exocyclic substituents (see structures in Fig. 1 ▶) and thus have different reactivities and H-bonding characteristics. By determining the effect of substitutions on the ability of the modified tRNAs to accept valine and to stimulate translational editing, we were able to examine and compare the specific recognition of individual structural elements of the 3′-terminal nucleoside at the aminoacylation and the editing sites of ValRS.
TABLE 1.
Valine acceptance by tRNAVal with base analog substitutions at the 3′-end
| 3′-Analog substitution | Steady-state level of aminoacylation (pmole/A260) | kcat/Km (μM−1 s−1) | Relative aminoacylation efficiency |
| Adenosine | 1285 | 3.15 | (1.0) |
| 3-Deazaadenosine | 1471 | 2.01 | 0.64 |
| 7-Deazaadenosine | 1551 | 1.65 | 0.52 |
| Purine riboside | 1428 | 2.27 | 0.72 |
| Inosine | 1122 | 0.35 | 0.11 |
| 2-Aminopurine | 945 | 0.27 | 0.09 |
| 2,6-Diaminopurine | 1284 | 0.16 | 0.05 |
| Xanthosine | 1337 | 0.04 | 0.01 |
| Isoguanosine | 1022 | 0.55 | 0.18 |
| 7-Deazaguanosine | 849 | 0.07 | 0.02 |
| Guanosine | 1120 | 0.02 | 0.01 |
TABLE 3.
Transfer RNA-dependent stimulation of editing by E. coli ValRS
| Analog substitution | Rate of ATP hydrolysis (pmole/min)a | Relative rate of ATP hydrolysis | Steady-state level of misacylation with threonine (pmole/A260) |
| Adenosine | 845 | (1.0) | 0 |
| 3-Deazaadenosine | 823 | 0.97 | 6 |
| 7-Deazaadenosine | 654 | 0.77 | 12 |
| Purine Riboside | 675 | 0.80 | 14 |
| Inosine | 17 | 0.02 | 153 |
| 2-Aminopurine | 20 | 0.02 | 429 |
| 2,6-Diaminopurine | 208 | 0.25 | 9 |
| Xanthosine | 10.2 | 0.01 | 34 |
| Isoguanosine | 169 | 0.20 | 381 |
| 7-Deazaguanosine | −48 | −0.06 | 12 |
| Uridine | −25 | −0.03 | 1628 |
| Cytidine | −88 | −0.10 | 1312 |
| Guanosine | −29 | −0.03 | 36 |
| No tRNA | −51 | −0.06 | — |
aNegative values are the result of small variations in the background rate of ATP hydrolysis.
FIGURE 1.
Chemical structures of nucleobase analogs inserted at the 3′-end of tRNAVal replacing the normal A76.
Recognition of functional groups at the aminoacylation site of ValRS
The importance of the heterocyclic ring nitrogens of the 3′-terminal adenosine of tRNAVal for aminoacylation by ValRS was tested by inserting 7-deazaadenosine (7-deazaA) and 3-deazaadenosine (3-deazaA), which have, respectively, the N7 and N3 ring nitrogens replaced by carbon, at the 3′-end of tRNAVal. E. coli tRNAVal terminating in 7-deazaA is a good substrate for ValRS: it is fully aminoacylated with valine with an efficiency (kcat/Km) half that of wild-type tRNAVal(A76) (Table 1 ▶). Similarly, tRNAVal with 3′-terminal 3-deazaA is completely charged with valine, with aminoacylation efficiency 64 percent that of tRNAVal(A76) (Table 1 ▶). Evidently the N7 and N3 ring nitrogens of the 3′-terminal adenosine are not essential for effective tRNA recognition at the aminoacylation site of ValRS.
The exocyclic 6-amino group of adenosine (N6) is also not a positive determinant for aminoacylation of tRNAVal by ValRS. E. coli tRNAVal with purine riboside, which lacks the exocyclic 6-amino group of adenosine (Fig. 1 ▶) in place of the normal 3′-terminal adenosine is fully charged by ValRS (Table 1 ▶). It is 72% as efficient as the wild-type tRNA in accepting valine (Table 1 ▶), with little change in kcat and only a twofold increase in Km (Table 2 ▶).
TABLE 2.
Kinetic parameters for valine acceptance by tRNAVal with base analog substitutions at the 3′-end
| Analog substitution | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Relative (kcat/Km) |
| Adenosine | 4.3 | 13.90 | 3.23 | (1) |
| Purine riboside | 8.8 | 14.4 | 1.63 | 0.50 |
| 2-Aminopurine | 5.0 | 0.68 | 0.14 | 0.043 |
| Isoguanosine | 2.0 | 0.99 | 0.50 | 0.15 |
| Inosine | 4.8 | 1.06 | 0.22 | 0.068 |
Replacing the 3′-A76 with inosine decreases aminoacylation efficiency more than ninefold compared to wild-type tRNAVal (Table 1 ▶), due largely to a 13-fold decrease in kcat (Table 2 ▶); 3′-inosine-substituted tRNAVal can, however, be fully charged with valine (Table 1 ▶). Inosine differs from adenosine at both the 1- and 6-positions of the purine ring; it has a 6-carbonyl (O6) substituent in place of the 6-NH2 group of adenosine, and its N1-nitrogen is protonated (Fig. 1 ▶). These chemical differences alter the H-bonding properties of the base, and either or both may account for the observed inhibition of aminoacylation (see Discussion).
A 2-amino substituent (N2) on the 3′-terminal base greatly reduces the efficiency of aminoacylation of tRNAVal. Substituting A76 of E. coli tRNAVal with 2,6-diaminopurine, which differs from adenosine in having an additional exocyclic amino group at the 2-position of the purine ring (Fig. 1 ▶), lowers the efficiency of valine acceptance 95% (Table 1 ▶); however, this tRNAVal variant can be fully aminoacylated with valine (Table 1 ▶). The adverse effect of an exocyclic 2-amino group on aminoacylation is also evident when 2-aminopurine, an analog of adenine in which the amino group on the purine ring is shifted from the 6– to the 2-position (Fig. 1 ▶), replaces A76 of tRNAVal. The efficiency of valine charging is reduced eightfold compared to that of the purine nucleoside-substituted tRNA (and 11-fold relative to wild-type tRNAVal; Table 1 ▶), largely the result of a 20-fold decrease in kcat (Table 2 ▶).
To test the combined effect of both an exocyclic 6-keto group (with a protonated N1-nitrogen) and a 2-NH2 substituent, A76 was replaced with G76. Although tRNAVal terminating in 3′-guanosine is completely charged with valine, it is a very poor substrate for ValRS; the efficiency of valine acceptance is two orders of magnitude lower than that of wild-type tRNAVal(A76) (Table 1 ▶; also see Liu and Horowitz 1994; Tamura et al. 1994; Tardif et al. 2001), primarily the result of a 300-fold decrease in kcat (Liu and Horowitz 1994). Presence of the 2-NH2 group reduces the efficiency of aminoacylation of tRNA(G76) 11-fold relative to that of tRNAVal terminating in 3′-inosine (Table 1 ▶), and the O6 substituent lowers valine charging efficiency by ninefold compared to that of 2-aminopurine-substituted tRNAVal (Table 1 ▶).
A keto group at the 2-position (O2) of the purine ring also adversely affects recognition of the 3′-terminal base of tRNAVal at the aminoacylation site of ValRS. This is best exemplified by experiments with E. coli tRNAVal terminating with 3′-isoguanosine, which resembles adenosine in having an exocyclic 6-amino group (N6) and an unprotonated N1 ring nitrogen (Fig. 1 ▶), but has a carbonyl at the 2-position (O2; see Fig. 1 ▶). Although the aminoacylation level of 3′-isoguanosine-substituted tRNAVal reaches 1022 pmole per A260 unit (Table 1 ▶), it is aminoacylated at only 18% the rate of wild-type tRNAVal (A76) (Table 1 ▶), due mainly to a 14-fold decrease in kcat (Table 2 ▶).
The inhibitory effect of an O2 substituent on aminoacylation is substantiated by studies with E. coli tRNAVal terminating in 3′-xanthosine. Xanthosine, which like inosine has a carbonyl group at position 6 (O6) and a proton on the N1-nitrogen, has an additional keto substituent at the 2-position (O2) of the purine ring (Fig. 1 ▶). Although E. coli tRNAVal terminating in 3′-xanthosine can be charged to high levels (1337 pmole/A260), it is a very poor substrate for aminoacylation by ValRS, having an aminoacylation efficiency an order of magnitude lower than that of tRNAVal with a 3′-terminal inosine (and almost two orders of magnitude lower than that of the wild-type tRNA; Table 1 ▶).
Functional group requirements for editing by ValRS
Mutational analysis has shown that replacing the 3′-terminal adenosine of tRNAVal affects aminoacylation and editing differently (Liu and Horowitz 1994; Tamura et al. 1994; Tardif et al. 2001). To compare the functional group requirements at the 3′-end of tRNAVal for aminoacylation by ValRS with those for editing, the effects of analog-substituted tRNAVal on proofreading were examined by following the tRNA-dependent hydrolysis of ATP (in the presence of the noncognate amino acid threonine) that is diagnostic of the overall editing reaction (the sum of pre- and posttransfer editing).
By inserting 3-deazaA and 7-deazaA at the 3′-end of E. coli tRNAVal assessed the contribution of the N7 and N3 ring nitrogens to tRNA recognition at the editing site of ValRS. Both the 3′-3-deazaA and the 3′-7-deazaA derivatives of tRNAVal are very active in promoting the editing reaction of ValRS. They are, respectively, 97% and 77% as efficient as wild-type tRNAVal and are, therefore, not readily misacylated with threonine (Table 3 ▶). These results indicate that the N7 and N3 ring nitrogens of the 3′-terminal purine are not necessary for productive recognition of tRNAVal at the editing site of ValRS.
3′-Purine riboside-substituted tRNAVal is also active in stimulating editing and is not misacylated with threonine (Table 3 ▶). Because purine riboside lacks an exocyclic amino group, the N6 substituent on the 3′-terminal base of E. coli tRNAVal, like the N7 and N3 functional groups, is not essential for tRNA recognition at the editing site.
E. coli tRNAVal with a 3′-inosine substitution fails to stimulate the editing activity of ValRS (Table 3 ▶), and as a result is misacylated with threonine to a level of 153 pmole/A260 (Table 3 ▶). Transfer RNAs terminating in guanosine, 7-deazaguanosine, and xanthosine, which, like inosine, have an exocyclic carbonyl group at the 6-position (O6) and a protonated N1-nitrogen, are also inactive in stimulating the editing reaction of ValRS (Table 3 ▶). However, previous studies showed that ValRS can deacylate mischarged Thr-tRNAVal(G76) (posttransfer editing), although at a much slower rate than mischarged wild-type tRNAVal(A76) (Tardif et al. 2001).
An exocyclic amino group at position-2 of the 3′-terminal purine (guanine, 2-aminopurine, and 2,6-diaminopurine) reduces the ability of tRNAVal to promote editing. E. coli tRNAVal terminating in 3′- 2-aminopurine riboside is inactive in stimulating ATP hydrolysis, and is stably misacylated with threonine to a level of more than 400 pmole/A260 (Table 3 ▶). Introducing a second exocyclic amino group into the 3′-terminal purine of tRNAVal by replacing the normal 3′-adenine (6-amino purine) with 3′-2,6-diaminopurine inhibits editing activity by 75% (Table 3 ▶); relatively little misacylation of this tRNA with threonine is observed (Table 3 ▶). Additional evidence of the inhibitory effect of a 2-NH2 substituent on the 3′-terminal nucleotide is the observation that the editing activity of 3′-guanosine-substituted tRNAVal is lower than that of the 3′-inosine-substituted tRNA (Table 3 ▶).
Experiments with E. coli tRNAVal terminating with 3′-isoguanosine or 3′-xanthosine show that an O2 substituent inhibits the editing-stimulatory activity of the tRNA. Addition of a keto group at position-2 of the 3′-terminal adenine, yielding isoguanine, reduces editing activity of tRNAVal by 80%, and consequently, 3′-isoguanosinesubstituted tRNAVal is misacylated with threonine to a level of almost 400 pmole/A260 (Table 3 ▶). The tRNAVal with a 3′-xanthosine substitution is less active than tRNA terminating in 3′-inosine (Table 3 ▶), presumably due to addition of the keto substituent at the 2-position.
Recognition of 3′-terminal hydroxyl groups by valyl-tRNA synthetase
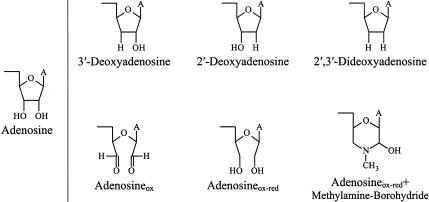
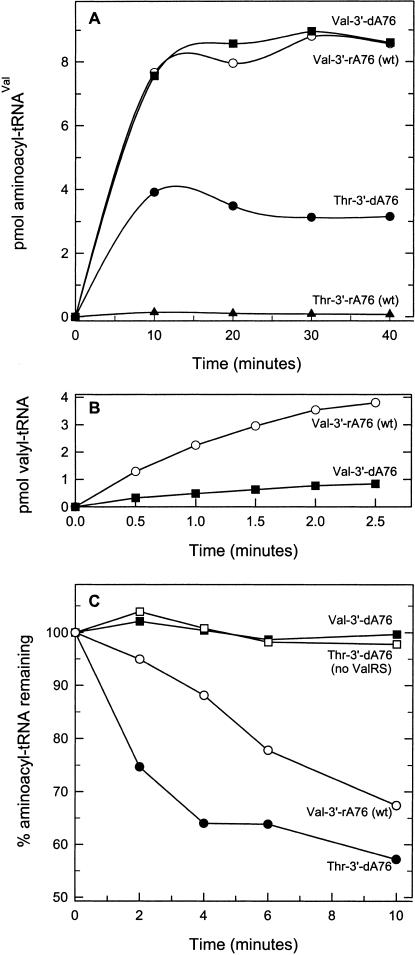
To explore the role of 3′-terminal hydroxyl groups in aminoacylation and editing by ValRS, the 3′-adenosine of tRNAVal was replaced with 2′-deoxyadenosine, 3′-deoxyadenosine, and 2′, 3′-dideoxyadenosine (structures shown in Fig. 2 ▶). ValRS initially esterifies valine onto the 2′-hydroxyl group of the 3′-terminal ribose (Hecht and Chinault 1976). As expected, at high concentrations of ValRS (1–2 μM), 3′-deoxyadenosine-substituted tRNAVal is charged with valine to the same level as wild-type tRNAVal (Fig. 3A ▶). At lower enzyme levels (1 nM), where the time course of valine acceptance can be more readily determined, substitution of the 3′-OH of tRNAVal with a hydrogen significantly lowers aminoacylation activity. 3′-deoxyadenosine-substituted tRNAVal is charged at a rate less than 20% that of wild-type tRNAVal (Fig. 3B ▶), suggesting that the 3′-hydroxyl group of the 3′-terminal nucleotide does contribute to tRNA recognition at the aminoacylation site of ValRS.
FIGURE 2.
Chemical structures of the modified ribose derivatives substituted at the 3′-end of tRNAVal.
FIGURE 3.
(A) Aminoacylation and misacylation of wild-type and 3′-deoxyribose-substituted tRNAVal: wild-type valyl-3′rA76 (○), valyl-3′dA76 (▪), threonyl-3′dA76 (•), threonyl-3′rA76 (▴). ValRS concentration is 1.5 μM. (B) Time course of aminoacylation of wild-type and 3′-deoxyribose-substituted tRNAVal with valine: wild-type 3′rA76 (○), 3′dA76 (▪). ValRS concentration is 1 nM. (C) Posttransfer editing by valyl-tRNA synthetase: wild-type valyl-3′rA76 (○), valyl-3′dA76 (▪), threonyl-3′dA76 (•), No ValRS (with threonyl-3′dA76) (□). ValRS and aminoacyl-tRNA concentrations were 0.3 μM and 2 μM, respectively.
Derivatives of tRNAVal with 3′-terminal 2′-deoxyA76, 3′-deoxyA76, and 2′,3′-dideoxyA76 do not stimulate the tRNA-dependent editing reaction, as measured by ATP hydrolysis (Table 4 ▶), even at a high (5 μM) tRNA concentrations (data not shown). As a result, 3′-deoxyA76 tRNAVal is stably misacylated with threonine (Fig. 3A ▶; Table 4 ▶). The maximum level of charging with threonine is, however, only about 25% that of valine acceptance (Fig. 3A ▶; Table 4 ▶).
TABLE 4.
Stimulation of editing by E. coli tRNAVal with modified 3′-terminal hydroxyl groups
| 3′-Modified tRNAVal | Rate of ATP hydrolysis (pmole/min)a | Relative rate of ATP hydrolysis | Steady-state level of misacylation with threonine (pmole/A260) |
| Ribose derivative | |||
| Adenosine | 745 | (1.0) | 10 |
| 3′-Deoxyadenosine | −14 | −0.02 | 408 |
| 2′-Deoxyadenosine | 1.8 | 0 | 151 |
| 2′,3′-Dideoxyadenosine | 48 | 0.06 | 163 |
| tRNAVal (−A) | −33 | −0.04 | NDb |
| No tRNA | −0.07 | 0 | — |
| Chemically modified ribose | |||
| Adenosineox | −8.0 | −0.03 | ND |
| Adenosineox-red | −1.3 | −0.004 | ND |
| Adenosineox-red + methylamine-borohydride | −9.6 | −0.03 | ND |
| No tRNA | −3.7 | −0.01 | ND |
aNegative values are the result of small variations in the background rate of ATP hydrolysis.
bND, not determined.
Although tRNAVal terminating in 3′-deoxyA76 [tRNAVal (3′dA76)] fails to stimulate editing (ATP hydrolysis) under our conditions, mischarged Thr-tRNAVal(3′dA76) can be deacylated by ValRS (posttransfer editing; Fig. 3C ▶). Absence of the terminal 3′-OH group, however, decreases the editing efficiency of the enzyme; the rate of deacylation of Thr-tRNAVal(3′dA76) is much slower than that of misacylated wild-type Thr-tRNAVal(3′rA76). Although we could not directly compare the deacylation rate of Thr-tRNAVal (3′dA76) with that of Thr-tRNAVal(3′rA76) because the latter is deacylated too rapidly by wild-type ValRS to be isolated, Nordin and Schimmel (2002), using an editing-deficient mutant of ValRS to prepare Thr-tRNAVal(3′rA76), showed that it is deacylated 10 times more rapidly than Thr-tRNAVal(3′dA76). The ability of ValRS to slowly hydrolyze Thr-tRNAVal(3′dA76) explains the observed reduced levels of stable misacylation of this tRNAVal variant with threonine (Fig. 3A ▶; Table 4 ▶), because plateau charging levels represent a steady state resulting from competing aminoacylation and deacylation reactions.
Further evidence that absence of the 3′-OH inhibits recognition of aminoacyl-tRNA at the editing site of ValRS comes from examination of the deacylation of correctly charged valyl-tRNAVal. Aminoacyl-tRNA synthetases are weak deacylases of their cognate aminoacyl-tRNAs (e.g., Schreier and Schimmel 1972; Igloi et al. 1977), and E. coli ValRS hydrolyzes correctly charged wild-type valyl-tRNAVal (Fig. 3C ▶). However, valyl-tRNAVal lacking the 3′-OH [Val-tRNAVal(3′dA76)] is not deacylated (Fig. 3C ▶).
Our results indicating that the hydroxyl groups at the 3′-end of tRNAVal play a significant role in the recognition of tRNAVal at the aminoacylation and editing sites of ValRS and in minimizing ValRS-catalyzed misacylation of tRNAVal are corroborated by chemical modification of the 3′-terminal ribose of E. coli tRNAVal. Periodate oxidation of the 3′-terminal ribose, to form the dialdehyde (see structures in Fig. 2 ▶) eliminates the valine-accepting activity (data not shown) and the editing-stimulatory activity of tRNAVal (Table 4 ▶). Moreover, borohydride reduction of the oxidized tRNAVal to yield primary hydroxyls at C2′ and C3′ of the 3′-terminal nucleotide (Fig. 2 ▶) does not restore the valine-accepting activity or the editing activity of the tRNA (Table 4 ▶). Tal et al. (1972) also observed that neither periodate-oxidized or oxidized-reduced tRNAVal is able to accept valine, whereas several other tRNAs recover accepting activity after the oxidized tRNA is reduced. Thus, whereas the integrity of the covalent bond between C2′ and C3′ of the 3′-terminal adenosine plays only a minor role in the aminoacylation of some tRNAs, it is clearly an important recognition element for the aminoacylation of E. coli tRNAVal by ValRS. Finally, treatment of oxidized tRNAVal with methylamine and borohydride chemically and structurally modifies the 3′-terminal ribose of tRNAVal (Fig. 2 ▶), abolishing its aminoacylation and editing-stimulatory activity (Table 4 ▶).
DISCUSSION
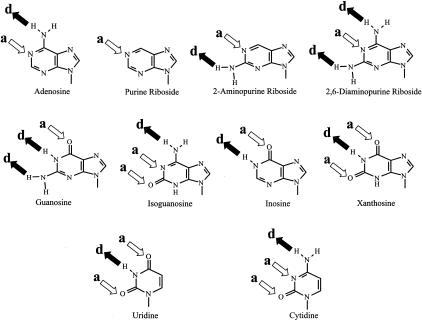
The 3′-end of tRNAVal plays an essential role in aminoacylation and editing by ValRS (see Introduction). It is correctly positioned at the synthetic and editing active sites of the enzyme by a variety of molecular interactions including hydrogen bonding between donor (d) and acceptor (a) groups on the terminal nucleotide and appropriately positioned amino acids, and by hydrophobic or stacking interactions with nearby protein side chains. Our previous results showed that replacing A76 of E. coli tRNAVal with pyrimidines (U76 or C76) yields mutant tRNAs that can be actively aminoacylated (Liu and Horowitz 1994), but these tRNAs do not stimulate the tRNA-dependent editing reaction of ValRS (Table 3 ▶; Tardif et al. 2001). As a result, 3′-pyrimidine-substituted tRNAVal is stably misacylated with threonine (Tamura et al. 1994; Liu et al. 1998) and several other noncognate amino acids (Tardif et al. 2001). Moreover, ValRS is unable to hydrolyze these misacylated tRNAs (posttransfer editing; Tardif et al. 2001). Conversely, although 3′-guanosine-substituted tRNAVal [tRNAVal (G76)] is aminoacylated only with difficulty (Liu and Horowitz 1994; see Table 1 ▶), misacylated tRNAVal(G76) is hydrolyzed by ValRS (Tardif et al. 2001). Evidently, only 3′-terminal purines are recognized at the editing site, whereas the aminoacylation site recognizes pyrimidines as well as adenine. Proteins frequently recognize adenine through H-bond interactions between amino acid side chains and the proton acceptor (a) N1 and the proton donor (d) N6 sites (Fig. 4 ▶). A similar a•d pattern of hydrogen bond donor and acceptor groups is present in cytidine and uridine, but not in guanosine (Fig. 4 ▶). This may explain why cytidine and uridine, but not guanosine, can functionally substitute for A76 of tRNAVal in the aminoacylation reaction of ValRS, and suggests that the exocyclic amino group (N6) and/or the ring N1 of 3′-terminal adenosine are specifically recognized at the synthetic (aminoacylation) active site.
FIGURE 4.
Hydrogen bond acceptor (a) and donor (d) sites of purine and pyrimidine bases. Filled arrows, donor groups; open arrows, acceptor sites.
Such hydrogen bonding has been observed at the editing active site of LeuRS (like ValRS a class Ia aminoacyl-tRNA synthetase), where the N1 and N6 positions of a nonhydrolyzable sulfamoyl analog of norvalyl-adenylate (pretransfer editing complex) and of an analog of the 3′-terminal adenosine of norvalyl-tRNA (posttransfer editing complex) are both H-bonded to the α-NH and α-CO groups, respectively, of Leu329, and the adenine ring stacks on a conserved hydrophobic amino acid side chain (Ile337; Lincecum et al. 2003). The recently solved crystal structure of Thermus thermophilus ValRS, complexed with tRNAVal (with its 3′-end at the editing site) and a Val-AMP analog (Fukai et al. 2000), shows that the N1 atom and 6-NH2 group of A76 form H-bonds with the α-NH and α-CO groups, respectively, of Glu261. The purine ring of A76 is sandwiched between the side chains of Phe264 and Leu269 by van der Waals contacts. Such an extensive network of H-bonds and hydrophobic interactions emphasizes the importance of a purine, particularly adenosine, at the 3′-end of tRNA and would explain why substitution of A76 has a major perturbing effect on the editing reaction.
Although these results suggest that specific recognition of the exocyclic amino group (N6) of the 3′-terminal adenosine is important for function, our base substitution experiments demonstrate that the N6 substituent is not a positive determinant for tRNA recognition at the synthetic (aminoacylation) site of E. coli ValRS; the same is true for the N3 and N7 ring nitrogens of A76. Variants of tRNAVal terminating in 3′-purine riboside, which lacks the exocyclic N6 amino group, or in 3′-terminal 3-deazaadenosine or 7deazaadenosine, which have, respectively, their N3 and N7 ring nitrogens replaced by carbons, retain almost full aminoacylation activity (Table 1 ▶).
Whereas the exocyclic amino group of adenosine is not important for recognition of the 3′-end of tRNAVal at the synthetic active site of ValRS, an O6 substituent interferes with aminoacylation. E. coli tRNAVal terminating in 3′-guanosine, 3′-inosine, or 3′-xanthosine is a poor substrate for aminoacylation by ValRS (Table 1 ▶). Either the 6-keto group acts as an antideterminant or, more likely because ValRS does not recognize substituents at the 6-position, changing the protonation state of N1 from an H-bond acceptor to a hydrogen bond donor (Fig. 4 ▶) inhibits function. The requirement for an unprotonated N1 may be a major positive determinant for recognition of the 3′-terminal nucleotide of tRNAVal.
Substituents at the 2-position are antideterminants for recognition of the 3′-terminal purine of tRNAVal, largely due to a decrease in kcat (Table 2 ▶). The aminoacylation activity of 2-aminopurine-substituted tRNAVal is lower than that of the purine riboside-substituted tRNA, as is that of the guanosine-substituted tRNA compared to 3′-inosine-substituted tRNAVal and of 2,6-diaminopurine-substituted tRNAVal relative to wild-type tRNAVal (A76) (Table 1 ▶), presumably due to the inhibitory effect of the 2-amino substituent.
A 2-keto (O2) substituent also inhibits the aminoacylation activity of ValRS; however, the 2-keto group is less inhibitory than the 2-amino group (Table 1 ▶). Although tRNAVal with a 3′-terminal isoguanine (2-keto,6-amino purine) is 20 times more efficient than the tRNA with a 3′-guanine (2-amino,6-keto purine) in aminoacylation (Table 1 ▶; interchanging positions of the exocyclic functional groups improves recognition of the 3′-end of the tRNA by the enzyme), the 3′-isoguanosine-substituted tRNAVal is only 20% as efficient as wild-type tRNAVal (A76) in accepting valine (Table 1 ▶), presumably due to the inhibitory effect of the O2 substituent. Furthermore, the aminoacylation efficiency of tRNAVal terminating in 3′-inosine is higher than that of the tRNA terminating in 3′-xanthosine (Table 1 ▶), again the result of the O2 substituent present in the latter.
The 3′-terminal hydroxyl groups of tRNAVal also contribute to tRNA recognition by the synthetic site of ValRS. E. coli tRNAVal terminating with 3′-deoxyadenosine can be fully aminoacylated, as expected, because ValRS is known to esterify the 2′-hydroxyl group (Hecht and Chinault 1976). This tRNAVal variant, however, is charged at only 17% the rate of wild-type tRNAVal (Fig. 3B ▶), demonstrating that the 3′-hydroxyl group improves the aminoacylation efficiency of the enzyme. Furthermore, an intact covalent bond between C2′ and C3′ of the 3′-terminal adenosine is important for aminoacylation of tRNAVal by ValRS, because E. coli tRNAVal terminating in adenosineox-red is not aminoacylated by the enzyme (Table 4 ▶).
Because our results indicate that transient misacylation of tRNA is a necessary prelude to editing (Tardif and Horowitz 2002; also see later discussion), it is difficult to identify functional groups specifically recognized at the editing site. However, because replacing A76 with purine riboside, 7-deazaA, and 3-deazaA has little or no effect on aminoacylation or total editing (Tables 1 ▶, 3 ▶), we can conclude that the exocyclic 6-amino group and the N7 and N3 ring nitrogens of A76 are not essential for triggering the editing reaction of ValRS. Furthermore, we have shown (Tardif et al. 2001) that ValRS deacylates mischarged Thr-tRNAVal (G76) (posttransfer editing) at a much slower rate than mischarged wild-type (A76) tRNAVal. Either the editing site preferentially recognizes an unprotonated N1, or the NH2 substituent at position-2 acts as a negative determinant, or both.
In addition to enhancing tRNA recognition at the aminoacylation site of ValRS, the 3′-hydroxyl group of A76 is a recognition element for translational editing. 3′-deoxyadenosine-substituted tRNAVal, misacylated with threonine on the 2′-OH, is deacylated by ValRS (posttranslational editing; Fig. 3B ▶; Nordin and Schimmel 2002) but at a rate 10 times slower than that of misacylated wild-type Thr-tRNAVal(3′rA76) (Nordin and Schimmel 2002). Analysis of the cocrystal structure of T. thermophilus ValRS with its cognate tRNA (Fukai et al. 2000) and that of the closely related T. thermophilus LeuRS/tRNALeu complex (Lincecum et al. 2003) provides insight into the structural role of the 3′-OH in promoting editing (the structure of the E. coli ValRS/tRNAVal complex has not been solved, and the 3′-end of the tRNA is not resolved in the structure of the T. thermophilus IleRS/tRNA complex). In both structures, a conserved threonine-rich sequence in the editing (CP1) domain, including Thr214 and Val215 in T. thermophilus ValRS (corresponding to Thr221 and Thr222 in E. coli ValRS) and Thr247 and Thr248 in T. thermophilus LeuRS (Lincecum et al. 2003), interacts with the hydroxyl groups of A76. The T. thermophilus ValRS cocrystal structure shows hydrogen bonds between the side chain OH of Thr214 and the 2′-OH of A76 and between the 3′-OH of A76 and the peptide NH of Val215 (Fukai et al. 2003). Molecular modeling studies, using Swiss-PdbViewer (Guex and Peitsch 1997) to convert Val215 of T. thermophilus ValRS to a Thr residue so that the sequence corresponds to that in E. coli ValRS, demonstrate that Thr215 can make two H-bonds to the 3′-OH of the ribose of A76 (data not shown). This is similar to the structure of the T. thermophilus LeuRS complex with a posttransfer editing analog, where the 3′-OH of the A makes three H-bonds with Thr247 and Thr248 (corresponding to Thr212 and Thr222 in E. coli ValRS). Such hydrogen bond interactions with the 3′-OH may serve to properly orient the mischarged tRNA in the editing site and thus explain the contribution of the 3′-OH to the efficiency of editing.
Our results regarding the effects of replacing the universally conserved 3′-adenosine of E. coli tRNAVal with nucleoside analogs that have a variety of substituents on the purine and ribose rings support and extend previous findings (Horowitz et al. 1999; Tardif and Horowitz 2002) that there is a close correlation between the aminoacylation efficiency of a tRNA and its effectiveness in stimulating the editing reaction of ValRS (Tables 1 ▶, 3 ▶). These results, together with our earlier findings (Tardif et al. 2001), and those of Nordin and Schimmel (2003), showing that misacylated tRNAVal variants that cannot be deacylated fail to promote overall editing by ValRS, strongly suggest that misacylated tRNAVal is a transient intermediate in the editing reaction, that is, that editing by ValRS requires a posttransfer step.
The question of which editing pathway, pre- or posttransfer, predominates has been extensively investigated for IleRS and ValRS. Based on comparison of the cocrystal structures of the homologous IleRS and ValRS complexed with a variety of substrates, and on modeling predictions, distinct but proximal subsites for binding the amino acids of pretransfer and posttransfer editing substrates were proposed, each involving a separate set of main chain amino acids (Nureki et al. 1998; Fukai et al. 2000). Separate sites would explain the ability of the tRNA synthetases to hydrolyze two very different substrates, a mixed anhydride (pretransfer editing) and an aminoacyl ester (posttransfer editing).
A model for editing by IleRS was recently proposed (Bishop et al. 2002) that explains many previous observations, including the finding that mutation of D342 in E. coli IleRS greatly diminishes total editing (pre- plus posttransfer) even though the Fukai two-subsite editing model indicates that this amino acid is part of the posttransfer editing site of IleRS but not of the pretransfer editing site. Bishop et al. (2002) suggested that the IleRS/tRNAIle complex catalyzes one round of posttransfer editing to prime the enzyme for subsequent rounds of pretransfer editing. The model implies that the 3′-end of tRNA and the adenylate moiety of the aminoacyl-adenylate can bind simultaneously to the editing site. One difficulty with this model, as Bishop et al. point out, is that pretransfer editing requires that IleRS successively misactivate two or more molecules of the noncognate amino acid, valine. This seems unlikely in vivo, because the enzyme binds the cognate isoleucine ~100 times more tightly than valine.
Recent results with LeuRS (Lincecum et al. 2003) shed new light on the editing mechanism. ValRS, IleRS, and LeuRS represent a closely related subfamily of class Ia aminoacyl-tRNA synthetases. All are large (100-kD) monomers and contain a sizable CP1 insert responsible for the editing reaction. Although there are some structural differences among the three enzymes (Cusack et al. 2000), their overall structures are quite similar (Nureki et al. 1998; Cusack et al. 2000; Fukai et al. 2000; Lincecum et al. 2003). The high structural and sequence homologies among the editing sites of these enzymes suggest that they may share a similar editing mechanism (Lincecum et al. 2003).
High-resolution crystallographic structures of LeuRS bound to pre- and posttransfer editing substrate analogs indicate that both bind to the same or largely overlapping sites in the editing domain (Lincecum et al. 2003). Analysis of these structures shows that the conformation of the ribose moiety of the pretransfer substrate differs from that of the posttransfer substrate, allowing the adenine ring and the amino acid of each to be recognized by a single editing site (Lincecum et al. 2003), which is able to hydrolyze both the pretransfer substrate (an aminoacyl-adenylate) and the posttransfer substrate (an aminoacyl-ester). This result is inconsistent with the two-subsite editing model for IleRS and ValRS proposed by Fukai et al. (2000), in which the amino acid of the pre- and posttransfer substrates is bound at different subsites of the editing domain. A possible reason for the difference is that in modeling threonyl-adenylate (pretransfer substrate) at the observed location of the 3′-terminal adenosine of tRNAVal (posttransfer substrate) at the editing site of ValRS, Fukai et al. assumed that the conformation of the ribose in the pre- and posttransfer substrates is identical, which is not the case for LeuRS. A consequence of the results with LeuRS, showing that the enzyme has a single editing pocket, is that the 3′-end of tRNA and the adenylate portion of the aminoacyl adenylate cannot simultaneously bind to the editing site as implied in the model for editing proposed by Bishop et al. (2002). Further studies will be required to determine whether the same or different editing mechanisms are operative in all three synthetases.
MATERIALS AND METHODS
Materials
Restriction endonucleases were purchased from either New England Biolabs or Promega. T4 RNA ligase and calf intestinal phosphatase were from New England Biolabs; inorganic pyrophosphatase was a product of Boehringer Mannheim Biochemicals. Homogeneous valyl-tRNA synthetase was prepared from E. coli GRB238/pHOV1 by the procedure of Chu and Horowitz (1991). T7 RNA polymerase was isolated from E. coli BL21/pAR1219 as reported by Zawadzki and Gross (1991). ATP(CTP):tRNA nucleotidyltransferase was purified from E. coli BL21(DE3) (kindly provided by Drs. N. Maizels and A.M. Weiner, University of Washington, Seattle) according to Shi et al. (1998).
Nucleotide triphosphates and guanosine 5′-monophosphate were the products of Sigma, Amersham Life Sciences, or United States Biochemical. Modified nucleosides: isoguanosine, 7-deazaadenosine, 7-deazaguanosine, purine riboside, inosine, 2,6-diaminopurine, and 2-aminopurine, were obtained from ChemGenes; 3-deazaadenosine was purchased from Southern Research Institute and xanthosine from Sigma. [3H]valine (23–32 Ci/mmole), [3H]threonine (15.8 Ci/mmole), and [γ-32P]adenosine-5′-triphosphate (3 Ci/mmole) were from Amersham Life Sciences.
Preparation and characterization of nucleoside bisphosphates
Nucleoside bisphosphates were synthesized by the method of Barrio et al. (1978) and chromatographically purified on a DEAE Sephacel column (30 × 1.5 cm) equilibrated with 50 mM TEAB (pH 8.0). The column was developed with a linear gradient from 50 mM to 400 mM TEAB (pH 8.0). This method yields mixtures of the 2′, 5′- and 3′, 5′-bisphosphates which were not separated because the 2′, 5′-bisphosphates do not interfere with ligation of the 3′, 5′-bisphosphates with tRNA (Barrio et al. 1978). Purified nucleoside bisphosphates (50 A260) were characterized by 31P NMR (Table 5 ▶) in a solution containing 0.84 M tetramethylammonium hydroxide (pH 10), 2 mM EDTA, 20% D2O. 31P NMR spectra were collected on a Bruker AC-200 spectrometer.
TABLE 5.
31P chemical shifts of nucleoside 3′(2′), 5′-bisphosphates
| Chemical shifta | |||
| Nucleoside bisphosphate | 3′-P | 2′-P | 5′-P |
| pAdenosine3′p | 4.883 | — | 4.589 |
| pAdenosine3′(2′)p | 4.885 | 4.302 | 4.590 |
| pGuanosine3′(2′)p | 4.922 | 4.134 | 4.577 |
| p2,6-Diaminopurine3′(2′)p | 4.995 | 4.284 | 4.643 |
| p2-Aminopurine3′(2′)p | 4.961 | 4.218 | 4.661 |
| pIsoguanosine3′(2′)p | 4.943 | 4.016 | 4.644 |
| pPurine riboside3′(2′)p | 5.127 | 4.331 | 4.637 |
| pInosine3′(2′)p | 4.971 | 4.205 | 4.612 |
| p7-Deazaadenosine3′(2′)p | 5.016 | 4.022 | 4.607 |
| p7-Deazaguanosine3′(2′)p | 4.973 | 3.997 | 4.561 |
| p3-Deazaadenosine3′(2′)p | 5.039 | 4.278 | 4.669 |
| pXanthosine3′(2′)p | 4.893 | 3.930 | 4.457 |
| pCytidine3′(2′)p | 4.806 | 4.312 | 4.556 |
aSpectra were recorded in 0.84 M tetramethylammonium hydroxide (pH 10), 2 mM EDTA, 20% D2O.
Preparation of tRNA
Wild-type tRNAVal was transcribed in vitro by T7 RNA polymerase as reported previously (Chu and Horowitz 1989) from a DNA template derived from the recombinant phagemid pFVAL119, which contains a T7 promoter directly upstream from a tRNAVal gene and a FokI restriction site directly downstream of the tRNAVal gene (Liu and Horowitz 1993). Transcripts were purified by HPLC (Liu and Horowitz 1993). Valine tRNA lacking the 3′-terminal adenosine [tRNAVal(-A)] was transcribed from a deletion mutant of pFVAL119 prepared by PCR site-directed mutagenesis using the QuikChange kit (Stratagene) as described by the manufacturer. The mutagenic oligonucleotides 5′-CATCACCCACCGGATC CAGTCATC-3′ and its complement were synthesized by the Nucleic Acid Facility at Iowa State University. Mutants were selected by restriction endonuclease digestion patterns, and the sequence was confirmed by automated dideoxy sequence analysis performed by the Nucleic Acid Facility.
Preparation of 3′-end-substituted tRNAVal
Valine tRNAs with modified bases at the 3′-end were prepared by ligating nucleoside bisphosphates to the 3′-terminus of tRNAVal lacking the 3′-terminal adenosine [tRNAVal (-A)] with T4 RNA ligase, as described by Paulsen and Wintermeyer (1984). 3′- (and 5′)-terminal phosphates were removed with calf intestinal phosphatase in a reaction mixture containing 50 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 2 u/μL calf intestinal phosphatase, and 0.8 μg/μL tRNA. Modified RNAs were separated from unligated starting RNA by electrophoresis on 8% denaturing polyacrylamide gels (20 × 45 × 0.2 cm) for 18 h at a constant current of 15 mA. RNA was extracted from the gel with 0.5 M NH4(OAc), 10 mM Mg(OAc)2, 1 mM EDTA (pH 8.0), and 0.1% SDS, using the crush and soak method (Maniatis et al. 1989). Gel-purified RNAs were ethanol-precipitated and dissolved in 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2.
Because of difficulties synthesizing nucleoside bisphosphates of adenine nucleosides having sugar modifications, valine tRNAs with modified sugars were prepared by ligating the nucleoside triphosphates ATP, 2′-dATP, 3′-dATP, and 2′,3′-ddATP to tRNAVal (-A) with E. coli nucleotidyltransferase, as described by Francis et al. (1983). The resulting modified tRNAs were purified by chromatography on a Toyopearl DEAE-650S HPLC column (Liu and Horowitz 1993), followed by electrophoretic purification as described previously.
The 3′-terminal ribose of tRNAVal was chemically modified by periodate oxidation, reduction with sodium borohydride, and reaction with methylamine, as described by Fahnestock and Nomura (1972).
Aminoacylation assays
Maximum (plateau) aminoacylation levels of tRNA transcripts were determined at 37°C as described (Kintanar et al. 1994) in a 60-μL reaction mix containing 100 mM HEPES-KOH (pH 7.5), 10 mM KCl, 15 mM MgCl2, 7 mM ATP, 1 mM DTT, 100 μM L-[3H]valine (23–32 Ci/mmole), 1–4 μg of tRNA, and 2–4 μg of ValRS (an excessive amount). Levels of misacylation with threonine were assayed as described for valine, with a ValRS concentration of 1.5 μM and 83 μM L-[3H]threonine (15.8 Ci/mmole). The time course of aminoacylation, under kcat/Km conditions, was followed in reactions using 1 nM ValRS and a low (1 μM) tRNA concentration. Steady-state aminoacylation kinetics, to determine individual kcat and Km values (Table 2 ▶), were measured under similar conditions with transfer RNA concentrations ranging from 0.5 to 6.0 μM. Reactions were initiated by addition of 1 nM of purified ValRS. Km and kcat values were calculated from a least-squares fit of the double reciprocal plot of the data using the Enzfitter computer program (Elsevier-Biosoft). Values reported are the average of two or more experiments.
Editing assay
Total editing activity (pre- plus posttransfer) was determined by an ATP hydrolysis assay essentially as described by Schmidt and Schimmel (1994). Reaction mixtures of 60 μL containing 150 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 37 mM threonine, 3 mM [γ-32P]ATP (20 to 30 cpm/pmole), 1 μM tRNA, and 2 μM ValRS were incubated at 25°C. At intervals over 30 min, 10 μL of the reaction was quenched with 25 volumes of 7% HClO4, 10 mM sodium pyrophosphate, and 3% activated charcoal (Sigma). [32P]pyrophosphate released as a result of ATP hydrolysis was separated from charcoal-bound ATP/AMP by centrifugation. Radioactivity in a 50-μL sample of the supernatant was determined by liquid scintillation counting. Results reported are the average of two or more determinations. Under conditions of the experiment, the rate of ATP hydrolysis is proportional to the concentration of tRNA (Tardif and Horowitz 2002).
Deacylation of aminoacyl-tRNA (posttransfer editing)
To measure the rate of deacylation of aminoacylated tRNA (posttransfer editing), the RNA was aminoacylated, as described above, with a [3H]-labeled amino acid and purified by chromatography on Sephadex G-25 (coarse grade, Pharmacia) at pH 5.0 (5 mM potassium acetate) to remove residual free radioactive amino acid. Purified aminoacyl-tRNAs were stored in 5 mM potassium acetate (pH 5.0).
Deacylation of aminoacyl-tRNA was followed as described by Lin and Schimmel (1996) at 37°C in a 60-μL reaction mixture containing 150 mM Tris-HCl (pH 7.5), 150 mM KCl, 10 mM MgCl2, and 2 μM [3H]-labeled aminoacyl-tRNA. The reaction was started by addition of 0.3 μM ValRS. Samples of 10 μL were removed at the indicated times, and remaining labeled aminoacylated tRNA was determined (Liu et al. 1998). Spontaneous hydrolysis of the aminoacyl bond was followed under the same conditions in the absence of ValRS.
Acknowledgments
We thank Dr. Jack C.-H. Liu for helpful discussions, and Diane Shogren, Lene Larsen, Dan Spielbauer, and Tuhina Dayal for their technical support. This research was supported in part by a grant from the National Science Foundation (MCB 95-13932).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5166704.
REFERENCES
- Barrio, J.R., Barrio, M.G., Leonard, N.J., England, T.E., and Uhlenbeck, O. 1978. Synthesis of modified nucleoside 3′,5′-bisphosphates and their incorporation into oligoribonucleotides with T4 RNA ligase. Biochemistry 17: 2077–2081. [DOI] [PubMed] [Google Scholar]
- Best, A.N. and Novelli, G.D. 1971. Studies with tRNA adenylyl(cytidylyl)transferase from Escherichia coli B. II. Regulation of AMP and CMP incorporation into tRNApCpC and tRNApC. Arch. Biochem. Biophys. 142: 539–547. [DOI] [PubMed] [Google Scholar]
- Bishop, A.C., Nomanbhoy, T.K., and Schimmel, P.R. 2002. Blocking site-to-site translocation of a misactivated amino acid by mutation of a class I tRNA synthetase. Proc. Natl. Acad. Sci. 99: 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A.C., Beebe, K., and Schimmel, P.R. 2003. Interstice mutations that block site-to-site translocation of a misactivated amino acid bound to a class I tRNA synthetase. Proc. Natl. Acad. Sci. 100: 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.F., Guo, N.N., Li, T., Wang, E.D., and Wang, Y.L. 2000. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry 39: 6726–6731. [DOI] [PubMed] [Google Scholar]
- Chu, W.-C. and Horowitz, J. 1989. 19F NMR of 5-fluorouracil-substituted transfer RNA transcribed in vitro: Resonance assignment of fluorouracil-guanine base pairs. Nucleic Acids Res. 17: 7241–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, W.-C. and Horowitz, J. 1991. Recognition of Escherichia coli valine transfer RNA by its cognate synthetase: A fluorine-19 NMR study. Biochemistry 30: 1655–1663. [DOI] [PubMed] [Google Scholar]
- Cusack, S., Yaremchuk, A., and Tukalo, M. 2000. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 15: 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahnestock, S.R. and Nomura, M. 1972. Activity of ribosomes containing 5S RNA with a chemically modified 3′-terminus. Proc. Natl. Acad. Sci. 69: 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht, A. and Kaethner, M. 1976. Enzyme hyperspecificity. Rejection of threonine by the valyl-tRNA synthetase by misacylation and hydrolytic editing. Biochemistry 15: 3342–3346. [DOI] [PubMed] [Google Scholar]
- Francis, T.A., Ehrenfeld, G.M., Gregory, M.R., and Hecht, S.M. 1983. Transfer RNA pyrophosphorolysis with CTP(ATP):tRNA nucleotidyltransferase. A direct route to tRNAs modified at the 3′-terminus. J. Biol. Chem. 258: 4279–4284. [PubMed] [Google Scholar]
- Fukai, S., Nureki, O., Sekine, S., Shimada, A., Tao, J., Vassylyev, D.G., and Yokoyama, S. 2000. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103: 793–803. [DOI] [PubMed] [Google Scholar]
- Fukai, S., Nureki, O., Sekine, S., Shimada, A., Vassylyev, D.G., and Yokoyama, S. 2003. Mechanism of molecular interactions for tRNAVal recognition by valyl-tRNA synthetase. RNA 9: 100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N. and Peitsch, M.C. 1997. Swiss-model and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hecht, S.M. and Chinault, C. 1976. Position of aminoacylation of individual Escherichia coli and yeast tRNAs. Proc. Natl. Acad. Sci. 73: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz, J., Chu, W.-C., Derrick, W.B., Liu, J.C.H., Liu, M., and Yue, D. 1999. Synthetase recognition determinants of Escherichia coli valine transfer RNA. Biochemistry 38: 7737–7746. [DOI] [PubMed] [Google Scholar]
- Igloi, G.L., von der Haar, F., and Cramer, F. 1977. Hydrolytic action of aminoacyl-tRNA synthetases from baker’s yeast. “Chemical proofreading” of Thr-tRNAVal by valyl-tRNA synthetase studied with modified tRNAVal and amino acid analogues. Biochemistry 16: 1696–1702. [DOI] [PubMed] [Google Scholar]
- Jakubowski, H. and Fersht, A. 1981. Alternative pathways for editing noncognate amino acids by aminoacyl-tRNA synthetases. Nucleic Acids Res. 9: 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski, H. and Goldman, E. 1992. Editing of errors in selection of amino acids for protein synthesis. Microbiol. Rev. 56: 412–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintanar, A., Yue, D., and Horowitz, J. 1994. Effect of nucleoside modifications on the structure and thermal stability of Escherichia coli valine tRNA. Biochimie 76: 1192–1204. [DOI] [PubMed] [Google Scholar]
- Lin, L. and Schimmel, P. 1996. Mutational analysis suggests the same design for editing activities of two tRNA synthetases. Biochemistry 35: 5596–5601. [DOI] [PubMed] [Google Scholar]
- Lin, L., Hale, S.P., and Schimmel, P. 1996. Aminoacylation error correction. Nature 384: 33–34. [DOI] [PubMed] [Google Scholar]
- Lincecum Jr., T.L., Tukalo, M., Yaremchuk, A., Mursinna, R.S., Williams, A.M., Sproat, B.S., van Den Eynde, W., Link, A., van Calenbergh, S., Grøtli, M., et al. 2003. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell 11: 951–963. [DOI] [PubMed] [Google Scholar]
- Liu, M. and Horowitz, J. 1993. In vitro transcription of transfer RNAs with 3′-end modifications. Biotechniques 15: 264–266. [PubMed] [Google Scholar]
- Liu, M. and Horowitz, J. 1994. Functional transfer RNAs with modifications in the 3′-CCA end: Differential effects on aminoacylation and polypeptide synthesis. Proc. Natl. Acad. Sci. 91: 10389–10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.C.-H., Liu, M., and Horowitz, J. 1998. Recognition of the universally conserved 3′-CCA end of tRNA by elongation factor EF-Tu. RNA 4: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Mursinna, R.S., Lincecum Jr., T.L., and Martinis, S.A. 2001. A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry 40: 5376–5381. [DOI] [PubMed] [Google Scholar]
- Nordin, B.E. and Schimmel, P. 2002. Plasticity of recognition of the 3′-end of mischarged tRNA by class I aminoacyl-tRNA synthetases. J. Biol. Chem. 277: 20510–20517. [DOI] [PubMed] [Google Scholar]
- Nordin, B.E. and Schimmel, P. 2003. Transiently misacylated tRNA is a primer for editing of misactivate adenylates by class I aminoacyl-tRNA synthetases. Biochemistry 42: 12989–12997. [DOI] [PubMed] [Google Scholar]
- Nureki, O., Vassylyev, D.G., Tateno, M., Shimada, A., Nakama, T., Fukai, S., Konno, M., Hendrickson, T.L., Schimmel, P., and Yokoyama, S. 1998. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 280: 578–582. [DOI] [PubMed] [Google Scholar]
- Paulsen, H. and Wintermeyer, W. 1984. Incorporation of 1,N6-ethanoadenosine into the 3′ terminus of tRNA using T4 RNA ligase. Eur. J. Biochem. 138: 117–123. [DOI] [PubMed] [Google Scholar]
- Rether, B., Gangloff, J., and Ebel, J.-P. 1974. Studies on tRNA nucleotidyltransferase from baker’s yeast. Replacement of the terminal CCA sequence in yeast tRNAPhe by several unusual sequences. Eur. J. Biochem. 50: 289–295. [DOI] [PubMed] [Google Scholar]
- Schmidt, E. and Schimmel, P. 1994. Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science 264: 265–268. [DOI] [PubMed] [Google Scholar]
- Schmidt, E. and Schimmel, P. 1995. Residues in a class I tRNA synthetase which determine selectivity of amino acid recognition in the context of tRNA. Biochemistry 34: 11204–11210. [DOI] [PubMed] [Google Scholar]
- Schreier, A.A., and Schimmel, P.R. 1972. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry 11: 1582–1589. [DOI] [PubMed] [Google Scholar]
- Shi, P.Y., Weiner, A.M., and Maizels, N. 1998. A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA 4: 276–284. [PMC free article] [PubMed] [Google Scholar]
- Silvian, L.F., Wang, J., and Steitz, T.A. 1999. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science 285: 1074–1077. [PubMed] [Google Scholar]
- Sprinzl, M. and Cramer, F. 1979. The C-C-A end of tRNA and its role in protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 22: 1–69. [DOI] [PubMed] [Google Scholar]
- Tal, J., Deutscher, M.P., and Littauer, U.Z. 1972. Biological activity of Escherichia coli tRNAPhe modified in its C-C-A terminus. Eur. J. Biochem. 28: 478–491. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Nobukazu, N., Tsunemi, H., Shimizu, M., and Himeno, H. 1994. Role of the CCA terminal sequence of tRNAVal in aminoacylation with valyl-tRNA synthetase. J. Biol. Chem. 269: 22173–22177. [PubMed] [Google Scholar]
- Tardif, K.D. and Horowitz, J. 2002. Transfer RNA determinants for translational editing by Escherichia coli valyl-tRNA synthetase. Nucleic Acids Res. 30: 2538–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif, K.D., Liu, M., Vitseva, O., Hou, Y.M., and Horowitz, J. 2001. Misacylation and editing by E. coli valyl-tRNA synthetase: Evidence for two tRNA binding sites. Biochemistry 40: 8118–8125. [DOI] [PubMed] [Google Scholar]
- Uziel, M. and Jacobson, K.B. 1974. Aminoacylation of Escherichia coli valine transfer RNA after oxidation and reduction. Biochim. Biophys. Acta 366: 182–187. [DOI] [PubMed] [Google Scholar]
- von der Haar F. and Gaertner, E. 1975. Phenylalanyl-tRNA synthetase from baker’s yeast: Role of 3′-terminal adenosine of tRNAPhe in enzyme-substrate interaction studied with 3′-modified tRNAPhe species. Proc. Natl. Acad. Sci. 72: 1378–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki, V. and Gross, H.J. 1991. Rapid and simple purification of T7 RNA polymerase. Nucleic Acid Res. 19: 1948–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]