Abstract
The National Cancer Institute (NCI) Human Tumor Cell Line Anti-Cancer Drug Screen has evaluated the cytotoxicity profiles of a large number of synthetic compounds, natural products, and plant extracts on 60 different cell lines. The data for each compound/extract can be assessed for similarity of cytotoxicity pattern, relative to a given test compound, using an algorithm called COMPARE. In applying a chemical biology approach to better understand the mechanism of eukaryotic protein synthesis, we used these resources to search for novel inhibitors of translation. The cytotoxicity profiles of 31 known protein synthesis inhibitors were used to identify compounds from the NCI database with similar activity profiles. Using this approach, two natural products, phyllanthoside and nagilactone C, were identified and characterized as novel protein synthesis inhibitors. Both compounds are specific for the eukaryotic translation apparatus, function in vivo and in vitro, and interfere with translation elongation. Our results demonstrate the feasibility of utilizing cytotoxicity profiles to identify new inhibitors of translation.
Keywords: protein synthesis inhibitor, COMPARE, phyllanthoside, nagilactone C
INTRODUCTION
Protein synthesis is a complex set of biochemical reactions involving the interaction of a large number of factors that catalyze the assembly of ribosomes, mRNA templates, and aminoacylated tRNAs. In this highly coordinated process, the ribosomes are assigned the task of correctly and efficiently producing all of the proteins in a cell. Regulation of this information pathway can be achieved at a number of levels, including the modulation of translation factor levels or activity (Raught et al. 2000), ribosome biogenesis (Ruggero and Pandolfi 2003), and small molecule/RNA interactions (Nahvi et al. 2002). Small molecule ligands that inhibit the process of translation have provided exquisite insight into ribosome function and translation factor activity in both prokaryotes and eukaryotes (Pestka 1977; Vazquez 1979). Inhibitors targeting a specific step of protein synthesis have enabled dissection of the translation pathway by allowing the characterization of events leading to the assembly of active polysomes, trapping intermediates of the initiation and elongation cycle, and providing insight into the molecular functions of protein factors (Pestka 1977; Vazquez 1979).
A number of studies indicate that deregulation of protein synthesis is a major contributor in cancer initiation (Watkins and Norbury 2002) and metastatic progression (Graff and Zimmer 2003). These include findings (1) that overexpression of some initiation factors can lead to malignant transformation, whereas down-regulation of these same factors can suppress the transformed phenotype (for review, see Hershey and Miyamoto 2000); (2) that components of the translation apparatus are overexpressed or mutated in cancers (Anand et al. 2002; Draptchinskaia et al. 1999); (3) that the tumor suppressor gene product pRB directly impacts on the translation process by affecting the levels of ribosomes (Cavanaugh et al. 1995; Ruggero and Pandolfi 2003), (4) of mutations impacting on translational efficiency within genes encoding important regulatory proteins (Meric and Hunt 2003); (5) of elevated translation rates as a consequence of aberrantly activated signal transduction pathways (e.g., PI3K/Akt pathway; Meric and Hunt 2003); and (6) that key components of anti-apoptotic pathways are translationally regulated (Holcik et al. 2000). As such, protein synthesis represents an interesting, underexplored target for chemotherapeutic intervention. Indeed, a few inhibitors of protein synthesis have been tested in clinical trials, with some of these demonstrating encouraging therapeutic effects (Ottenheijm and van den Broek 1988; Rinehart 2000; Kantarjian et al. 2001). Presumably, a therapeutic index is achieved due to the higher requirement of transformed cells for protein synthesis, as well as translation regulation of some of the proteins involved in cancer progression. Unfortunately, dose-limiting secondary effects have hampered further development of many of these compounds.
Surprisingly, little effort has been made to systematically search for new inhibitors of eukaryotic protein synthesis, with the majority of compounds currently used having been obtained from chemical screens performed several decades ago (Pestka 1977; Vazquez 1979). Their potential chemtherapeutic value, as well as their roles in better understanding the eukaryotic translation apparatus, has prompted us to undertake strategies aimed at identifying novel inhibitors. One such approach is based on a computer algorithm used to identify compounds within a given collection that demonstrate similar activity profiles. This program is called COMPARE (Paull et al. 1989). One reference source for activity profiles of different compounds is the data generated from the National Cancer Institute (NCI) Human Tumor Cell Line Anti-Cancer Drug Screen, in which a large number of agents (>41,000 synthetic and natural product compounds) have been evaluated for their cytotoxicity profiles on 60 tumor cell lines. The antiproliferative activity (GI50; concentration of compound required to inhibit growth by 50%) for each compound is stored in a database that can be analyzed by the COMPARE algorithm, as this program ranks all entries for similarity of cytotoxicity patterns to a reference compound (Paull et al. 1989). The degree of similarity between the cytotoxicity patterns is expressed numerically as a Pearson correlation coefficient. The COMPARE algorithm has been successfully used to identify a number of novel agents, including inhibitors of tubulin polymerization (Paull et al. 1992), inosine monophosphate (IMP) dehydrogenase (Gharehbaghi et al. 1994), dihydroororate dehydrogenase (Cleaveland et al. 1995, 1996), topoisomerase I (Kohlhagen et al. 1998), and cyclin-dependent kinases (Zaharevitz et al. 1999). In this report, we apply the COMPARE algorithm to search the data-ase of compounds tested in the NCI Human Tumor Cell Line Anti-Cancer Drug Screen for novel inhibitors of protein synthesis. In our approach, the cytotoxicity profiles (GI50) of 31 known protein synthesis inhibitors were used to identify two novel, eukaryotic-specific translation inhibitors.
RESULTS
Identification of potential protein synthesis inhibitors using the COMPARE algorithm
We used 31 known protein synthesis inhibitors as reference compounds (see Materials and Methods) in the COMPARE analysis. One method to assess whether our approach could be effective in identifying inhibitors is to evaluate how many of 31 protein synthesis inhibitors used as seeds identified one or more of the other reference compounds. The results indicated that 13 of the references compounds were identified by our approach (i.e., showed a Pearson correlation coefficient >0.5; data not shown). These results show that COMPARE can identify inhibitors of a multistep, complex biochemical pathway, such as eukaryotic protein synthesis, based on similarity of in vivo cytotoxicity data. Encouraged by these initial evaluations, we evaluated the “hits” obtained with the 31 seeds.
Compounds were grouped according to two arbitrarily chosen criteria. One group consisted of 203 compounds having a Pearson correlation coefficient greater than 0.7 that had been identified by one or two seeds. Seventy-six of these compounds were available for testing from the NCI and were added to ascites translation extracts to a final concentration of 25 μM to assess their potential to directly inhibit translation. None of these showed significant (greater than threefold) inhibition of protein synthesis (J. Mills and J. Pelletier, data not shown). The second group consisted of compounds having a Pearson correlation coefficient greater than 0.5 but identified by at least three seeds (Table 1 ▶). The activity of many compounds in this group was known, or could be deduced, and included known protein synthesis inhibitors, DNA-binding agents, and inhibitors of cytoskeletal function (Table 1 ▶). The ability of our approach to identify 14 known protein synthesis inhibitors, not used as initial references, provided a second validation of our approach (Table 1 ▶).
TABLE 1.
Small molecule ligands identified by the COMPARE algorithm
| Compound classa | Compound (NSC no.) | Name | No. of seedsb | Tested for protein synthesis inhibitionc | Comments |
| Translation inhibitor | 325014 | Bactobolin | 9 | Y(I.C.90 < 10 μM) | Related to actinobolin |
| 368672 | SUN 2071 | 7 | N | Quassinoid | |
| 269754 | 7 | Y(I.C.90 > 10 μM) | Trichothecene derivative | ||
| 269753 | 5 | Y(I.C.90 > 10 μM) | Trichothecene derivative | ||
| 267709 | Undulatone | 5 | N | Quassinoid | |
| 269760 | 4 | Y(I.C.90 > 10 μM) | Trichothecene derivative | ||
| 368671 | SUN 0237 | 4 | Y(I.C.90 < 10 μM) | Quassinoid | |
| 278571 | 3 | N | T-2 Toxin analog | ||
| 331120 | 3 | N | Trichothecene derivative | ||
| 292463 | 3 | N | Verrucarin derivative | ||
| 327993 | 3 | N | Verrucarin derivative | ||
| 65104 | Septacidin | 3 | Y(I.C.90 > 50 μM) | Aminoacyl nucleoside | |
| 126765 | Holacanthone | 3 | N | Quassinoid | |
| 163501 | Acivicin | 4 | Y(I.C.90 > 50 μM) | Amino acid analog | |
| Nucleic acid binding | 337766 | Bisantrene | 9 | Y(I.C.90 > 10 μM) | |
| 58514 | Chromomycin A3 | 9 | Y(I.C.90 > 50 μM) | ||
| 267469 | Deoxydoxorubicin | 9 | N | ||
| 82151 | Daunorubicin | 9 | Y(I.C.90 > 50 μM) | ||
| 123127 | Doxorubicin | 8 | Y(I.C.90 > 10 μM) | ||
| 164011 | 8 | N | |||
| 143020 | Mithramycin | 6 | Y(I.C.90 > 50 μM) | ||
| 526417 | Echinomycin | 6 | Y(I.C.90 > 50 μM) | ||
| 339281 | 4 | N | |||
| 3053 | Dactinomycin | 3 | Y(I.C.90 > 50 μM) | ||
| 76411 | Olivomycin | 4 | Y(I.C.90 > 50 μM) | ||
| 349174 | Oxanthrazole | 4 | Y(I.C.90 < 10 μM) | ||
| 353076 | Ellipticine der. | 3 | N | ||
| 359449 | Ellipticine der. | 3 | N | ||
| 355644 | Anthrapyrazole der. | 3 | Y(I.C.90 > 10 μM) | ||
| 354646 | Morpholino-adr. | 3 | N | ||
| 141540 | Etoposide | 3 | Y(I.C.90 > 50 μM) | ||
| 122819 | Teniposide | 3 | N | ||
| Cytoskeletal poisons | 67574 | Vincristine sulfate | 8 | Y(I.C.90 > 50 μM) | |
| 49842 | Vinblastine sulfate | 7 | Y(I.C.90 > 50 μM) | ||
| 125973 | Paclitaxel | 7 | N | ||
| 153858 | Maytansine | 8 | Y(I.C.90 > 50 μM) | ||
| 332598 | Rhizoxin | 5 | Y(I.C.90 > 50 μM) | ||
| 330500 | Macbecin II | 3 | N | ||
| Misc./unknown | 328426 | Phyilanthoside | 8 | Y(I.C.90 < 10 μM) | |
| 694330 | 6 | N | |||
| 237020 | 6 | Y(I.C.90 > 50 μM) | |||
| 211500 | Nagilactone C | 5 | Y(I.C.90 < 10 μM) | ||
| 77471 | Mitocromin | 5 | Y(I.C.90 > 50 μM) | ||
| 685703 | 5 | N | |||
| 673790 | 4 | N | |||
| 665806 | 4 | N | |||
| 139105 | 4 | N | |||
| 634791 | 4 | Y(I.C.90 > 50 μM) | |||
| 681236 | 4 | N | |||
| 640085 | 3 | Y(I.C.90 > 50 μM) | |||
| 630176 | 3 | N | |||
| 673792 | 3 | N | |||
| 682345 | Aurantimycin B | 3 | N | ||
| 673352 | 3 | N | |||
| 364170 | 15-Deactylsergeolide | 3 | N | ||
| 157365 | 3 | Y(I.C.90 > 50 μM) | |||
| 73754 | Fluorpan | 3 | Y(I.C.90 > 50 μM) | ||
| 267213 | 3 | Y(I.C.90 > 50 μM) | |||
| 333856 | Tetrocarcin A | 3 | Y(I.C.90 > 10 μM) | ||
| 366241 | 3 | N | |||
| 409962 | Carmustin | 3 | Y(I.C.90 > 50 μM) | ||
| 142982 | 3 | Y(I.C.90 > 50 μM) |
aCompounds are grouped according to known or predicted biological activities.
bThe number of initial seeds used in the COMPARE algorithm that identified a particular compound is given.
cWhether a compound was tested depended on its availabilty from the NCI-DTP or a commercial supplier. Y indicates that the compound was tested for translation inhibition activity in a Krebs ascites extract as described in the Materials and Methods. N indicates that the compound was not available for testing. Compounds showing an I.C.90 < 10 μM in Krebs extracts are highlighted in bold.
Titrations of known translation inhibitors (didemnin B, cephaeline, bouvardin, baccharinol, bruceantin, verucarrin, anisomycin, homoharringtonine, emetine, sparsomycin, puromycin) in Krebs extracts indicated that the I.C.90 for these were below 10 μM (J. Pelletier, unpubl. data). We therefore used this as our cutoff to identify novel potential protein synthesis inhibitors. Five compounds met this criteria: (1) two compounds structurally related to known protein synthesis inhibitors (bactobolin [NSC#325014] and SUN 0237 [NSC#368671]), (2) a DNA-binding agent (oxanthrazole [NSC#349174]), (3) phyllanthoside (NSC#328426), and (4) nagilactone C (NSC#211500). We have assessed the ability of a large number of intercalators to inhibit protein synthesis, and our results indicate that this class of compounds can inhibit both translation initiation and elongation (A. Malina and J. Pelletier, in prep.). The inhibitory effects of nucleic acid binding agents on protein synthesis, as well as possible explanations as to why these and cytoskeletal poisons were identified by the COMPARE algorithm, are discussed below (see Discussion). No experiments documenting the effects of phyllanthoside and nagilactone C (Fig. 1A ▶) on protein synthesis have been previously reported; hence the biological activity of these compounds was investigated.
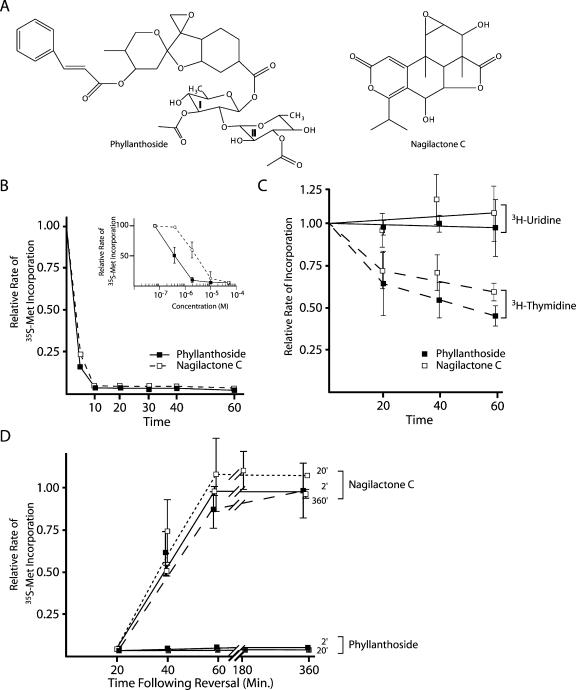
FIGURE 1.
Effect of phyllanthoside and nagilactone C on synthesis of protein, RNA, and DNA in vivo. (A) Structures of phyllanthoside and nagilactone C. (B) Effect of phyllanthoside and nagilactone C on protein synthesis in HeLa cells. The rate of protein synthesis (TCA precipitable counts per minute for a 5-min labeling period) obtained in the presence of compound was normalized to the rate obtained in the presence of DMSO and is plotted for 10 μM phyllanthoside and 50 μM nagilactone C. Results are shown for two experiments and the standard error is too small to be visualized. The relative rate of 35S-met incorporation of HeLa cells exposed to 600 μM cycloheximide (positive control) for 20 min was 0.04 (data not shown). The inset graph plots the relative rate of 35S-met incorporation as a function of phyllanthoside or nagilactone C concentration in HeLa cells. The rate of protein synthesis in the control reactions (containing DMSO vehicle) averaged 250,000 cpm per 10-min labeling. (C) Effect of phyllanthoside and nagilactone C on DNA (dashed lines) and RNA (solid lines) synthesis in HeLa cells. The rate of nucleic acid synthesis (TCA precipitable counts per minute for a 10-min labeling period) obtained in the presence of compound was normalized to the rate obtained in the presence of DMSO and is plotted for 10 μM phyllanthoside and 50 μM nagilactone C. Results are shown for two experiments and the standard error is shown. The rate of RNA and DNA synthesis in the control reactions (containing DMSO vehicle) averaged 13,000 cpm/10-min labeling and 5,000 cpm/10-min labeling, respectively. The relative rate of 3H-uridine incorporation of HeLa cells exposed to 50 μM actinomycin (positive control for inhibition of RNA synthesis) for 20 min was 0.02 (data not shown). (D) Assessment of reversal of inhibition of protein synthesis by phyllanthoside and nagilactone C after various periods of exposure to the compounds. HeLa cells were preincubated with 10 μM phyllanthoside or 50 μM nagilactone C for the indicated period of time, after which cells were washed three times and fresh media (lacking compound) was added to the cells. Ten minutes before harvesting at the indicated time points, 35S-methionine was added to the media. The relative rate of protein synthesis was determined by comparing to the rate obtained in control cells exposed to DMSO. Washout of cells exposed to 10 μM anisomycin for 20 min indicated that protein synthesis was back to 77% of control levels 40 min after reversal (data not shown).
Phyllanthoside and nagilactone C inhibit protein synthesis in vivo
To assess the activity of phyllanthoside and nagilactone C on cellular processes, we measured protein, RNA, and DNA synthesis in HeLa cells following exposure to these agents. Pulse-labeling experiments with 35S-methionine in the presence of increasing concentrations of compound yielded an IC50 of ~0.4 μM and 3 μM for phyllanthoside and nagilactone C, respectively (Fig. 1B ▶, see inset). Both compounds dramatically inhibited protein synthesis in HeLa cells within 5 to 10 min of their addition to the culture media, but did not significantly influence RNA metabolism (Fig. 1C ▶). DNA synthesis was inhibited by 50% in Hela cells after exposure to compounds for 1 h (Fig. 1C ▶).
The inhibition of protein synthesis in HeLa cells by 50 μM nagilactone C could be readily reversed in cells that had been preincubated for 2, 30, or 120 min with compound (Fig. 1D ▶). However, the inhibition observed with phyllanthoside (when added to HeLa cells for 2 or 20 min) could not be reversed, even within 6 h after removal of the compound from the media (Fig. 1D ▶). These results indicate that phyllanthoside and nagilactone C inhibit protein synthesis by different mechanisms.
Phyllanthoside and nagilactone C are eukaryotic-specific translation inhibitors
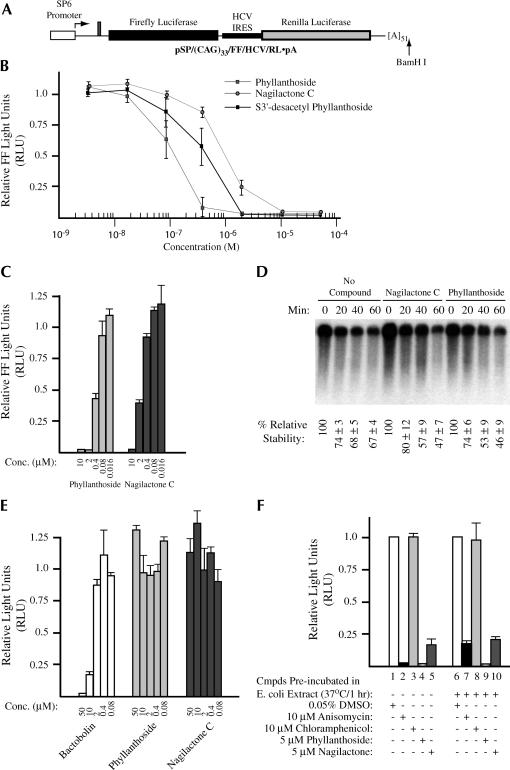
Titration of phyllanthoside and nagilactone C into translation extracts prepared from Krebs ascites cells indicated that they are potent inhibitors of eukaryotic protein synthesis in vitro (Fig. 2B ▶). Phyllanthoside displayed an IC50 of 0.15 μM, S3′-desacetyl phyllanthoside, which lacks an acetyl moiety on sugar II, showed an IC50 of 0.4 μM, and nagilactone C yielded an IC50 of 1 μM in Kreb’s extracts. Similar IC50 results were obtained if renilla luciferase was measured or when phyllanthoside and nagilactone C were titrated into rabbit reticulocyte lysate (data not shown). Both phyllanthoside and nagilactone C also inhibited translation in wheat germ extracts, yielding an IC50 of ~0.4 μM and ~1.5 μM, respectively, indicating that plant ribosomes are also sensitive to inhibition by both compounds (Fig. 2C ▶). One explanation that could account for the observed inhibition of mRNA translation in the presence of phyllanthoside and nagilactone C is if these harbored or induced nucleolytic activity. To directly address this, we performed a kinetic analysis of (CAG)33/FF/HCV/Ren mRNA stability when translated in wheat germ extracts in the absence or presence of 50 μM phyllanthoside or nagilactone C (Fig. 2D ▶). Reisolation of radiolabeled (CAG)33/FF/HCV/Ren transcripts from programmed translation extracts, followed by fractionation on formaldehyde agarose gels, revealed that the stability of (CAG)33/FF/HCV/Ren mRNA is similar in the absence or presence of compound during the time course of the experiment (Fig. 2D ▶). Thus, the observed effects of these compounds on translation cannot be attributed to an associated ribonucleolytic activity.
FIGURE 2.
(A) Schematic diagram of pSP/(CAG)33/FF/HCV/Ren. To generate mRNA for in vitro transcriptions, the plasmid was linearized with BamHI. The firefly luciferase coding region is denoted by a black box, whereas the renilla coding region is denoted by a gray box. The SP6 promoter is denoted by an open box. The HCV IRES (indicated by a thickened line) allows for internal initiation upstream of the renilla ATG. (B) Titration of phyllanthoside, nagilactone C, and 3′-desacetyl phyllanthoside in Krebs extracts. Translations were performed in the presence of the indicated amounts of compound and at a final mRNA and K+ concentration of 5 μg/mL and 100 mM, respectively. Because the final concentration of DMSO in the compound additions was 0.5%, control translation reactions contained 0.5% DMSO. The obtained firefly luciferase activities were normalized to the activity obtained in the control translations (which were set at one). Only the firefly luciferase data is shown, but similar results were obtained with renilla luciferase (data not shown). Each data point represents the average of seven translations and the standard error of the mean is shown. (C) Titration of phyllanthoside and nagilactone C in wheat germ translation extracts. Translation reactions were performed as indicated by the manufacturer’s recommendation (Promega Corp.) at an mRNA concentration of 5 μg/mL and a final K+ concentration of 150 mM. Because the HCV IRES is inactive in wheat germ extracts, only firefly luciferase activity was assessed and is plotted relative to the values obtained from control translations containing 0.5% DMSO. Each data point represents the average of two translations and the standard error is shown. (D) Relative stability of (CAG)33/FF/HCV/Ren mRNA in wheat germ extracts. Following reisolation from the translation extracts, radioactive mRNA was fractionated on a 1.4% agarose/formaldehyde gel and quantitated on a Fuji BAS2000 with a Fuji imaging screen. A representative autoradiograph from one experiment is shown. The percent relative stability of each time point (and standard error) is indicated below each lane and is an average from three experiments. Values were set relative to the value obtained at the beginning of the experiment (T = 0 min). The time points and the presence or absence of 50 μM phyllanthoside or nagilactone C in the translation reaction is indicated above the panel. (E) Translation reactions utilizing E. coli S30 extracts (Promega; for linear templates) were programmed with a lux AB mRNA transcript (Szittner and Meighen 1990), which had been produced by in vitro transcription reactions from pT7-5 (Dr. Ted Meighen, McGill University) utilizing T7 RNA polymerase, followed by treatment with DNAse I to remove any remaining plasmid DNA. Translations in E. coli S30 extracts were performed at a final mRNA concentration of 115 μg/mL and the luciferase activity of the product was measured as previously described (Szittner and Meighen 1990). Values are plotted relative to the light values from lux AB mRNA translated in the presence of vehicle (0.5% DMSO). The average of duplicate translations is shown as well as the standard error. (F) Phyllanthoside and nagilactone C are not modified to inactive derivatives in E. coli S30 extracts. Compounds (100 μM anisomycin, 100 μM chloramphenicol, 50 μM phyllanthoside, 50 μM nagilactone C) were incubated in E. coli S30 extracts for 1 h at 37°C. An aliquot (1 μL) was then taken and added to a Krebs ascites translation extract (9 μL) programmed with (CAG)33/FF/HCV/Ren mRNA (lanes 6–10). Lanes in which compounds were preincubated in E. coli S30 extracts, as well as the nature and final concentration of compounds in the ascites extracts, are indicated below the panel. (Lanes 1–5) Translations performed in ascites extracts in which the compounds were not preincubated in bacterial S30 extracts. The translational efficiency is expressed relative to the values obtained in the absence of compounds (lanes 1 and 6). The average of duplicate translations is shown as well as the standard error.
Escherichia coli translation extracts are not inhibited by phyllanthoside or nagilactone C (Fig. 2E ▶). Neither compound had a significant effect on translation in E. coli extracts programmed with lux AB mRNA when present up to 50 μM (Fig. 2E ▶). As a positive control, we utilized bactobolin—identified in our initial COMPARE screen and a known inhibitor of prokaryotic (and eukaryotic) translation (Adachi et al. 2002). To ensure that the bacterial extract did not contain an activity that modified the compounds, rendering them inactive for inhibition, they were preincubated in E. coli S30 extracts for an hour, then added to a programmed Krebs ascites extract (Fig. 2F ▶). As controls, we used anisomycin—a potent inhibitor of eukaryotic protein synthesis (Fig. 2F ▶, bars 2 and 7) and chloramphenicol—a prokaryotic specific inhibitor (Fig. 2F ▶, bars 3 and 8). Preincubation of anisomycin in E. coli S30 extracts slightly reduced its effectiveness to inhibit eukaryotic protein synthesis (Fig. 2F ▶, cf. bars 2 and 7). Preincubating phyllanthoside or nagilactone C in E. coli S30 extracts for 1 h, followed by their addition to programmed ascites translation extracts showed that both compounds retained their inhibitory properties (Fig. 2F ▶, Cf. bars 4,5 and 9,10). When the experiment was performed with chloramphenicol, no inhibition of translation in Krebs extracts was observed following preincubation in E. coli S30 extracts, indicating that no trans-inhibitor was being transferred from the E. coli S30 extract to the Krebs translation mix (Fig. 2B ▶, cf. bars 3 and 8).
Phyllanthoside and nagilactone C inhibit translation elongation
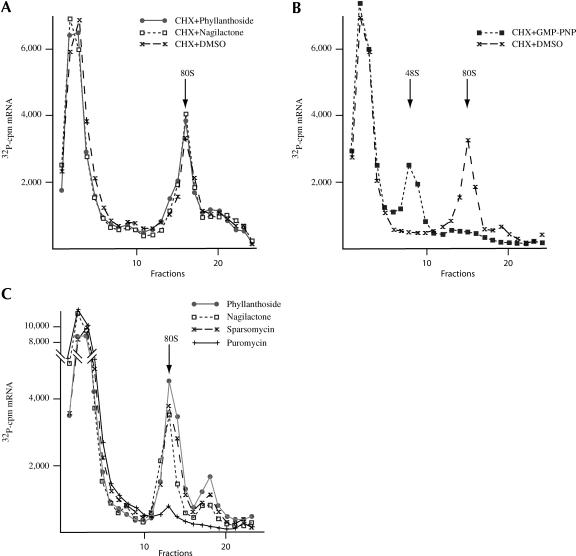
To assess whether either compound had an effect on the initiation process, ribosome-binding experiments were performed in the presence of compound and cycloheximide (which inhibits elongation; Fig. 3A ▶). Neither compound affected the formation of 80S ribosome/RNA initiation complexes on CAT mRNA in the presence of cycloheximide, indicating that they do not affect initiation of protein synthesis. As a positive control for these experiments, the inhibitor GMP-PNP was used. GMP-PNP, which inhibits the joining of the 60S ribosome subunit as well as release of eIF2, prevented the formation of 80S initiation complexes and trapped a 40S ribosome on the mRNA template (Fig. 3B ▶). To assess whether these compounds were capable of trapping an 80S/mRNA initiation complex, we performed ribosome-binding experiments in the presence of only sparsomycin, phyllanthoside, or nagilactone C (Fig. 3C ▶). In this experiment, the sedimentation time was decreased slightly from that used in Figure 3A ▶. We were able to detect the formation of 80S/mRNA complexes when phyllanthoside or nagilactone C was present in the in vitro initiation complex reactions (Fig. 3C ▶). As well, the heavier sedminenting peak likely represents disomes. These complexes were similar in mobility to those obtained with sparsomycin, consistent with these compounds being capable of inhibiting the elongation process (Fig. 3C ▶). Purmomycin on its own was not capable of trapping 80S initiation complexes, as expected (Fig. 3C ▶).
FIGURE 3.
Phyllanthoside and nagilactone C inhibit elongation of protein synthesis. (A) Phyllanthoside and nagilactone C do not prevent ribosome binding to mRNA templates. 32P-labeled CAT mRNA was incubated in rabbit reticulocyte lysates in the presence of 600 μM cycloheximide (CHX), 600 μM CHX and 10 μM phyllanthoside, or 600 μM CHX and 10 μM nagilactone C. Following centrifugation (SW40; 39,000 rpm/3.5 h), fractions of each sucrose gradient were collected using a Brandel Tube Piercer connected to an ISCO fraction collection, and were individually counted. Total counts recovered from each gradient and the percent mRNA bound to 80S complexes were: CAT mRNA/CHX, 41,008 cpm, 24% binding; CAT mRNA/phyllanthoside+CHX, 39,377 cpm, 28% binding; CAT mRNA/nagilactone C+CHX, 39,025 cpm, 27% binding. (B) Ribosome bindings performed with 32P-labeled CAT mRNA in rabbit reticulocyte lysates in the presence of 1 mM 5′-guanylylimidodiphosphate (GMP-PNP; to monitor 48S assembly). Initiation complexes were visualized as indicated above. Total counts recovered from each gradient and the percent mRNA bound were: CAT mRNA/CHX, 34,749 cpm, 15% binding; CAT mRNA/CHX+GMP-PNP, 31,705 cpm, 15% binding. (C) Phyllanthoside and nagilactone C can trap 80S complexes on mRNA templates. 32P-labeled CAT mRNA was incubated in rabbit reticulocyte lysates in the presence of 600 μM sparsomycin, 10 μM phyllanthoside, or 10 μM nagilactone C. Centrifugation was performed in an SW40 at 39,000 rpm for 3 h. Total counts recovered from each gradient and the percent mRNA bound to 80S complexes were: CAT mRNA/sparsomycin, 48,034 cpm, 15% binding; CAT mRNA/phyllanthoside, 48,033 cpm, 19% binding; CAT mRNA/nagilactone C, 47,662 cpm, 11% binding; CAT mRNA/puromycin, 46,093 cpm, 1% binding.
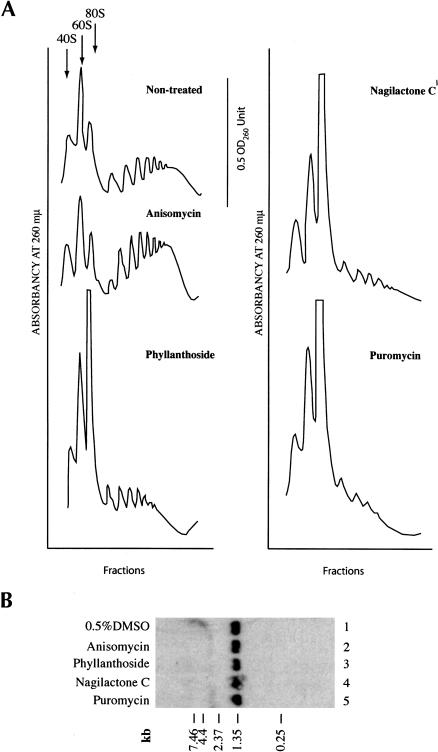
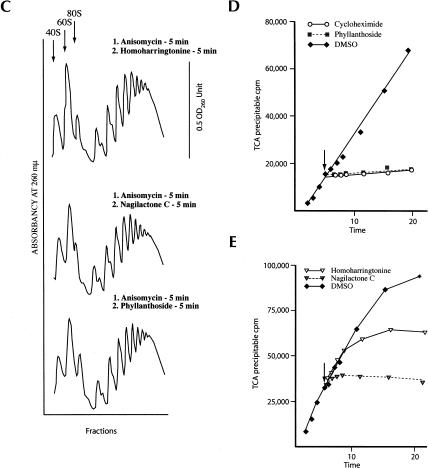
To analyze the effect of these translation inhibitors in vivo, they were added to a growing culture of HeLa cells for 20 min and the polyribosomes analyzed (Fig. 4A ▶). Anisomycin is known to stall ribosomes on mRNA templates and its addition causes a slight increase in the polyribosome profile, as previously documented (Fig. 4A ▶; Grollman 1967). Addition of puromycin, phyllanthoside, or nagilactone C to HeLa cells for 20 min significantly collapsed the polysome profile, with a concomitant increase in 80S and ribosomal subunits. The collapse of endogenous polysomes by phyllanthoside and nagilactone C could be the due to several possibilities: (1) degradation of RNA is induced in vivo, which, in turn, would lead to a reduction in the polysome content; (2) translating ribosomes are not sensitive to compound inhibition and run off the mRNA template; or (3) phyllanthoside and nagilactone C cause termination of protein synthesis and ribosome dissociation. To assess if the observed collapse of polysomes is the result of generalized mRNA degradation, stimulated or activated in the presence of phyllanthoside or nagilactone C, we analyzed mRNA integrity following treatment of HeLa cells with 50 μM compound for 20 min. Northern blot analysis of GAPDH mRNA revealed no differences in mRNA quality between compound-treated and DMSO-treated cells (Fig. 4B ▶, cf. lanes 1 and 2–5). To evaluate whether active protein synthesis was required for phyllanthoside or nagilactone C to cause polysome dissociation, we first pretreated HeLa cells with anisomycin for 5 min, followed by exposure to either homoharringtonine (an inhibitor of reinitiating ribosomes [Fresno et al. 1977]), phyllanthoside or nagilactone C for 5 min (Fig. 4C ▶). Polysomes isolated from these cells indicated that preblocking translation prevents the polysome dissociation mediated by phyllanthoside or nagilactone C (Fig. 4C ▶).
FIGURE 4.
(A) Effect of phyllanthoside and nagilactone C in vivo on polyribosomes. In each case shown, 10 μM anisomycin, phyllanthoside, or nagilactone C or 1 mM puromycin was added to a culture of HeLa cells for 20 min and the polyribosomes collected and analyzed as described in Materials and Methods. The polysome profile tracing is shown for each of the compounds tested and the scale bar corresponds to 0.5 OD260 units. (B) Northern blot analysis of RNA from HeLa cells exposed to translation inhibitors. HeLa cells were exposed to 0.5% DMSO (lane 1), 50 μM anisomycin (lane 2), 50 μM phyllanthoside (lane 3), 50 μM nagilactone C (lane 4), or 1 mM puromycin (lane 5), for 20 min and total RNA isolated using Trizol (Invitrogen). Seven micrograms of total RNA were fractionated on a 1.0% formaldehyde/agarose gel, transferred to Hybond N+, and hybridized with radiolabeled GAPDH DNA. Prehybridizations and hybridizations were performed in Church buffer. Two washes were performed with 2× SSC/0.1% SDS for 30 min at 65°C and one wash in 0.2× SSC/0.1%SDS for 30 min at 65°C. (C) Effect on polysomes of pretreating cells with anisomycin (5 min), followed by treatment with 10 μM homoharringtonine, 10 μM phyllanthoside, or 10 μM nagilactone C for 5 min. The polysome profile tracing is shown for each set of conditions tested and the scale bar corresponds to 0.5 OD260 units. (D,E) Effect of adding phyllanthoside and nagilactone C to an actively translating extract. Translations in Krebs extracts were initiated in the presence of 35S-methionine, and aliquots for TCA precipitation were removed at various time points. Five minutes after initiation of translation, compound (600 μM cycloheximide, 100 μM homoharringtonine, 10 μM phyllanthoside, or 50 μM nagilactone C) or vehicle (0.5% DMSO were added to the reaction (represented by a downward arrow) and the translation was continued for another 15 min, with aliquots for TCA precipitation being removed at the indicated times. Each data point is the average of two TCA precipitations.
To assess whether actively translating ribosomes were sensitive to inhibition by phyllanthsoide or nagilactone C, each inhibitor was added to an actively translating Krebs system (Fig. 4D,E ▶). When 10 μM phyllanthoside or 50 μM nagilactone C were added 5 min after the start of the in vitro translation reaction, immediate inihibition of protein synthesis was observed (Fig. 4D,E ▶). The same phenomenon was also observed with cycloheximide (Fig. 4D ▶). On the other hand, homoharringtonine showed a delay (~5 min) in inhibiting protein synthesis, consistent with actively translating ribosomes being able to complete their cycle of elongation (Fig. 4E ▶). These results suggest that both phyllanthoside and nagilactone C immediately inhibit elongation and do not allow ribosomes to terminate their current round of elongation.
We characterized the events during elongation that might be affected by phyllanthoside and nagilactone C by analyzing various steps of the process. We assessed the ability of these compounds to interfere with eEF-1α-dependent aminoacyl-tRNA delivery, peptidyl transferase, and eEF-2-dependent translocation. The effect of phyllanthoside and nagilactone C upon eEF-1α-dependent aminoacyl-tRNA delivery was investigated by monitoring the amount of [14C]Phe-tRNA bound to salt-washed ribosomes, following incubation with eEF-1α and GMP-PNP (Table 2 ▶). Inhibition of eEF-1α-dependent [14C]Phe-tRNA binding to the A site of ribosomes was observed with both phyllanthoside and nagilactone C (Table 2 ▶). In addition, nagilactone C inhibited nonenzymatic binding of [14C]Phe-tRNA by 50%. Didemnin B, a known inhibitor of the eukaryotic elongation cycle that acts by preventing eukaryotic elongation factor-2 (eEF-2)-dependent translocation (SirDeshpande and Toogood 1995), did not inhibit [14C]Phe-tRNA delivery (Table 2 ▶), as expected (SirDeshpande and Toogood 1995; Ahuja et al. 2000).
TABLE 2.
Effects of phyllanthoside and nagilactone C on eEF-2 dependent assays
| [14C]Phe-tRNA bound (pmole) | ||||
| No compound | Phyllanthoside | Nagilactone C | Didemnin B | |
| [14C]Phe-tRNA bindinga | ||||
| eEF-1 dependenta | 4.1 | 1.35 | 0.5 | 4.1 |
| Nonenzymaticb | 24.4 | 17.2 | 12.8 | n.t. |
| Translocationc | ||||
| eEF-2 dependent (Preloaded with eEF-1, [14C]Phe-tRNA, GMP-PNP) | 2.8 | 0.38 | n.t.d | 0.96 |
| eEF-2 dependent ([14C]Phe-tRNA preloaded nonenzymatically) | 15.6 | 7.3 | n.t.d | n.t. |
| Nonenzymatic ([14C]Phe-tRNA preloaded nonenzymatically) | 7.8 | 7.7 | n.t.d | n.t. |
aEukaryotic eEF-1-dependent Phe-tRNA binding was monitored using 10-pmoles [14C]Phe-tRNA, 89 pmoles of ribosomes, and 0.2 μg poly(U). Results shown are the average of two independent determinations and 50 μM compound was used in each reaction.
bNonenzymatic Phe-tRNA binding was performed with 20 pmoles [14C]Phe-tRNA, 177 pmoles of ribosomes, and 40 μg poly(U). Results shown are the average of four independent determinations and 50 μM compound was used in each reaction. (n.t.) not tested.
cTranslocation assays were performed as described in the Materials and Methods, either with or without eEF-1 (enzymatic and nonenzymatic binding, respectively). In both cases, phyllanthoside was added just prior to the addition of eEF-2. Results shown are the average of two independent determinations and 50 μM compound was used in each reaction. (n.t.) not tested. The obtained values are corrected for P site reactive [14C]Phe-tRNA present at the start of the translocation assay.
dNagilactone C could not be tested in the translocation assay because it inhibits peptidyl transferase activity which is required to measure the amount of P site reactive [14C]Phe-tRNA.
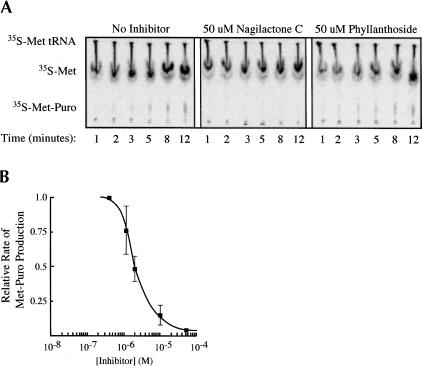
The effect of the compounds on ribosome-catalyzed peptide bond formation was studied using the puromycin reaction. In this approach, [35S]methionine-charged tRNA (Met-tRNAi) is bound to the P site of the 80S ribosomal complex. As an analog of the 3′ end of a charged tRNA, puromycin is capable of binding in the A site of the ribosome and reacting with the ester linkage of Met-tRNAi to produce a [35S]methionine–puromycin dipeptide. Hence, small molecule effects on peptidyl transferase are monitored by assessing the rates of [35S]methionine–puromycin dipeptide formation. Inhibition of Met-puromycin production was observed when nagilactone C was included in the puromycin assay (Fig. 5A,B ▶). Varying the concentration of nagilactone C in the puromycin assay yielded a dose–response curve (Fig. 5B ▶) in which the relative extent of inhibition of nagilactone C paralleled those observed in vivo and in vitro (Figs. 1 ▶, 2 ▶). Phyllanthoside did not inhibit peptidyl transferase when present up to 50 μM in this assay (Fig. 5A ▶).
FIGURE 5.
Nagilactone C inhibits peptidyl transferase. (A) Formation of the [35S]Met-puromycin dipeptide was followed using cation-exchange TLC. A kinetic analysis in which a single concentration (50 μM) of phyllanthoside and nagilactone C was used in the Met-puromycin assay is shown. The positions of migration of [35S]Met-tRNAi, free [35S]-methionine, and the [35S]Met-puromycin dipeptide product are indicated. (B) Dose response curve showing the effect of nagilactone C concentration on [35S]Met-puromycin production. Each point represents the average of three experiments, along with the standard deviation.
Translocation during the elongation cycle involves transfer of the A site-bound peptidyl-tRNA to the ribosomal P site. This process is catalyzed by eEF-2 and can be analyzed by first loading salt-washed ribosomes with [14C]Phe-tRNA in their A site enyzmatically by the addition of eEF-1α, GMP-PNP, and [14C]Phe-tRNA, or nonenzymatically by using high Mg++, then adding eEF-2 and GTP to initiate translocation, and then assessing the amounts of puromycin-reactive (P site-positioned) [14C]Phe-tRNA. Inhibition of eEF-2-dependent translocation was observed when Didemnin B was added to the reaction mix (Table 2 ▶), as previously documented (SirDeshpande and Toogood 1995). Inhibition of translocation was also observed when phyllanthoside was added after eEF-1α-mediated loading of [14C]Phe-tRNA into the A site (Table 2 ▶) or if it was present during eEF-1α-mediated binding of [14C]Phe-tRNA (data not shown). eEF-2-dependent translocation was also inhibited by phyllanthoside following nonenzymatic loading of [14C]Phe-tRNA to the ribosomal A site (Table 2 ▶; Ayuso and Heredia 1968; Carrasco et al. 1975). However, phyllanthoside did not inhibit nonenzymatic translocation (Table 2 ▶). We could not assess the effect of nagilactone C on EF-2-dependent reactions because puromycin reactivity is utilized to assess P site occupancy and nagilactone C inhibits this activity (Fig. 5 ▶).
DISCUSSION
By comparing the cellular cytotoxicity profiles (GI50 obtained on 60 human tumor cell lines) of a large number of compounds to those of known protein synthesis inhibitors, we identified two novel eukaryotic translation inhibitors: phyllanthoside and nagilactone C. A number of nucleic acid binding compounds and cytoskeletal poisons were identified in our analysis (Table 1 ▶). Inhibition of protein synthesis by nucleic acid intercalators has been previously reported (Weinstein and Finkelstein 1967; Birkmayer and Balda 1970; Momparler et al. 1976; Craig and Kostura 1983). Additionally, we have tested a number of these in ascites extracts and found that many can directly inhibit protein synthesis in vitro (A. Malina and J. Pelletier, data not shown). We tested the possibility that phyllanthoside and nagilactone C were intercalators by incubating with supercoiled DNA and monitoring potential mobility shifts by agarose gel electrophoresis (Burres and Clement 1989). Neither compound was able to cause a mobility shift of supercoiled DNA in this assay, suggesting that they are not intercalators (J. Chan and J. Pelletier, data not shown). Cytoskeletal poisons were also identified by the COMPARE algorithm, and indeed, these compounds would be expected to decrease in vivo protein synthesis, given the intimate link between the cytoskeleton and translation (for review, see Jansen 1999). Although some of the cytoskeletal poisons in Table 1 ▶ failed to show a significant effect on protein synthesis in vitro (Table 1 ▶), we have not tested their activity in vivo.
Phyllanthoside was first isolated from an ethanolic extract prepared from the Central American tree Phyllanthus acuminatus, based on bioassay guided cytotoxicity fractionation (Kupchan et al. 1977). Phyllanthoside is rapidly converted to an inactive metabolite, within ~2 min of intravenous injection in mice or dogs (Moore and Powls 1986). The metabolite, identified as the aglycone derivative of phyllanthoside resulting from cleavage of the ester bond linking the disaccharide moiety to phyllanthocin, is 6,000 times less potent that the parent compound (Chapman et al. 1989). Consistent with an important biological role for the carbohydrate residues, S3′-desacetyl phyllanthoside lacks one of the sugar residues and displays an approximate threefold higher IC50 compared to phyllanthoside in ascites extracts (Fig. 2B ▶). Nagilactone C was identified as a secondary metabolite from species of the genus Podocarpus and is cytotoxic against P-388 leukemia cells in vivo (Hayashi et al. 1975) and in vitro against HT-1080 fibrosarcomas and 26-L5 colon carcinoma cells (Shrestha et al. 2001). The compound also has insecticidal properties (Sing et al. 1978; Hayashi et al. 1992), is an inhibitor of plant growth, and displays antifungal properties (Kubo et al. 1993).
We have observed a partial inhibition (approximately twofold) on DNA synthesis when HeLa cells were exposed to nagilactone C and phyllanthoside for 1 h (Fig. 1C ▶). This could result from a primary effect on protein synthesis, because translation is required for concurrent synthesis of DNA in mammalian cells (Mueller et al. 1962; Lieberman et al. 1963). This effect may be due to changes in levels of ribonucleotide reductase (an enzyme that synthesizes dNTP—a limiting step in DNA synthesis), because regulation of this protein by a component of the translation apparatus has been previously shown to link protein synthesis and DNA replication (Abid et al. 1999). This same phenomenon has been reported for a number of protein synthesis inhibitors, including anisomycin (Grollman 1967), puromycin (Mueller et al. 1962), emetine (Grollman 1968), tylocrebrine (Huang and Grollman 1972), cycloheximide (Ennis 1966), pactamycin (Young 1966), and harringtonine (Huang 1975). The average rate of total RNA synthesis during the first hour of exposure was unaffected by 10 μM phyllanthoside or 50 μM nagilactone C (Fig. 1C ▶).
Protein synthesis in HeLa cells is decreased by 95% within 10 min of having been exposed to either nagilactone C or phyllanthoside (Fig. 1B ▶). The inhibition observed by nagilactone C in HeLa cells could be reversed after exposure to 50 μM nagilactone C by thoroughly washing the cells. After exposure of cells to nagilactone C for 2, 20, or 360 min, the rate of protein synthesis recovered to control levels within 1 h of washing out the compound (Fig. 1E ▶). On the other hand, a 2-min exposure to phyllanthoside was sufficient to produce an inhibition that was not reversible for up to 6 h after the compound had been washed from the cells (Fig. 1E ▶). This suggests either a strong noncovalent interaction with, or covalent binding to, the target. Alternatively, a high intracellular concentration of phyllanthoside may be maintained if the compound is modified in vivo to prevent its subsequent exit from the cell.
Nagilactone C inhibits eEF-1α-dependent loading of aminoacyl-tRNA into the ribosomal A site, as well as peptidyl transferase (as measured with P site reactive 35S-Met-tRNAi; Fig. 5 ▶; Table 2 ▶), whereas phyllanthoside inhibits eEF-1α-dependent aminoacyl-tRNA binding and eEF-2 mediated translocation (Table 2 ▶) but does not affect peptidyl transferase activity (at concentrations below 10 μM). The effects on eEF-2 activity are not a consequence of inhibition of eEF-1α events, because phyllanthoside was added to the reaction after Phe-tRNA had been loaded onto ribosomes. Our experiments suggest that these compounds interfere with A-site function.
The activity of nagilactone C and phyllanthoside appears different from known inhibitors of translation. Our working model is that they have two distinct biological activities—inhibition of elongation and stop-codon-independent ribosome dissociation. The first activity was revealed in ribosome-binding assays (Fig. 3 ▶) and indicates that newly initiated ribosomes are not “sensitive” to dissociation induced by phyllanthoside or nagilactone C. One speculation would be that conformational changes necessary for ribosome release by these compounds does not occur on newly initiated ribosomes. This could happen if the conformation of the A site of newly initiated ribosomes was different than that of ribosomes that had undergone a cycle of peptide bond formation. Along these lines, the observed molecular mimicry between eEF-1α ternary complex and eEF2•GDP has lead to the suggestion that when eEF2•GDP leaves the ribosome, it leaves an imprint of a binding site suitable for subsequent binding of the eEF-1α ternary complex (Liljas 1996). This imprint should be absent on a newly initiated ribosome that has yet to undergo a cycle of translocation.
Active translation is necessary for ribosome dissociation by phyllanthoside and nagilactone C, as preblocking elongation with anisomycin protects against polysome disruption by these compounds (Fig. 4C ▶). These observations are in contrast to other inhibitors of peptide chain elongation, such as emetine (Grollman 1968), anisomycin (Grollman 1967), cycloheximide (Ennis 1966), and tylocrebrine (Huang and Grollman 1972), which block protein synthesis quickly when added to an in vitro translation reaction, but do not collapse the polyribosome profile. Other inhibitors of elongation such as homoharringtonine collapse the polysome profile by allowing ribosomes to run off mRNA templates and blocking elongation of newly initiated ribosomes (Huang 1975; Fresno et al. 1977). One mechanism by which ribosome dissociation could be stimulated would be by recruiting eukaryotic release factor 1 in a stop-codon-independent fashion. Blocking A site access with anisomycin could explain the absence of ribosome release by phyllanthoside and nagilactone C. Clearly, additional experiments are required to better define the molecular mechanism of action of these compounds. In sum, our results demonstrate the feasibility of utilizing cellular cytotoxicity profiles to identify new inhibitors of translation.
MATERIALS AND METHODS
COMPARE analysis
The COMPARE algorithm was used as described (Zaharevitz et al. 2002). The seed compounds used in the analysis were three ipecac alkaloids (emetine [NSC#33669], cephaeline [NSC#32944], tubulosine [NSC#131547]), didemnin B [NSC#325319], three cephalotaxus alkaloids (homoharringtonine [NSC#141633], harringtonine [NSC#124147], and isoharringtonine [NSC#141634]), a quassinoid (bruceatin [NSC#165563]), deoxybouvardin [NSC#259969], baccharinol [NSC#269756], sparsomycin [NSC#59729], five trichothecenes (verrucarin A [NSC#126728], anguidine [NSC#141537], T-2 Toxin [NSC#138780], tricodermin [NSC#267033], tenuazonic acid [NSC#638261]), anisomycin [NSC#76712], two aminoacyl nucleosides (anthelmycin [NSC#337588], puromycin [NSC#3055]), two glutarimides (acetoxycycloheximide [NSC#32743], streptimidone [NSC#66645]), three amaryllidaceae alkaloids (haemanthamine [NSC#403140], narciclasine [NSC#266535], pretazettine [NSC#109808]), two phenanthroindolizidine alkaloids (tylocrebrine [NSC#60387], cryptopleurine [NSC#19912]), actinobolin [NSC#31083], pactamycin [NSC#52947], a chartreuse analogue (elsamicin [NSC#369327]), edeine [NSC#663297], and a pederine (mycalamide [NSC#626168]). The cytotoxicity data for the synthetic compound databases of the August 2000 and October 2002 releases were analyzed and are available to the public at the DTP Web site (http://dtp.nci.nih.gov/). The majority of compounds tested were obtained from the Drug Synthesis and Chemistry Branch of the NCI. They were dissolved in dimethyl sulfoxide and equivalent amounts of solvent were included in all in vivo and in vitro control reactions.
In vitro translations
In vitro translation assays were performed with a bicistronic mRNA reporter, (CAG)33/FF/HCV/Ren, in which the first cistron encodes the firefly (FF) luciferase (luc) protein and the second cistron encodes the renilla (Ren) luc protein (Fig. 2A ▶). Expression of the second cistron is driven by HCV IRES sequences, and 33 (CAG) trinucleotide repeats are present within the 5′ UTR of the FF luc cistron. Construction and details of this reporter will be presented elsewhere, but the essential details are shown in Fig. 2A ▶. In vitro transcriptions were performed as previously described (Harvey et al. 2002) using BamHI linearized templates. Translations were performed in Krebs extracts as previously reported (Carriere et al. 2002). Translations in rabbit reticulocyte lysates, wheat germ extracts, and E. coli S30 extracts were performed as recommended by the manufacturer (Promega Corp). Firefly and renilla luciferase activity (RLU) were measured on a Berthold Lumat LB 9507 luminometer as previously reported (Dyer et al. 2000).
RNA stability assays
To determine mRNA stability in the presence or absence of inhibitor, wheat germ translation extracts were programmed with 32P-labeled mRNA, except that 35S-methionine was replaced with 80 μM unlabeled methionine. At various time points, an aliquot was removed (10 μL), incubated with 50 μg Proteinase K for 30 min at 37°C, and the sample was phenol/chloroform extracted. Following ethanol precipitation, RNA samples were fractionated on a 1.4% agarose/formaldehyde gel. Gels were stained with SYBR gold (Molecular Probes) to ensure equal recovery of rRNA from the translation extracts (data not shown) and then dried and exposed to X-OMAT film (Kodak) at −70°C with an intensifying screen. Quantitations were performed on a Fujix BAS2000 with a Fuji imaging screen.
Met-puromycin dipeptide assays
Met-puromycin dipeptide assays were performed essentially as previously described (Lorsch and Herschlag 1999). Briefly, reactions included 60 nM ribosomes, 2 nM [35S]methionyl tRNA, 400 μM puromycin, and 1 μM model mRNA, in a buffer containing 500 μM GTP, 1.75 mM magnesium acetate, and other components as previously described (Lorsch and Herschlag 1999). 80S complexes were preformed prior to addition of puromycin and inhibitor. Reactions proceeded for 12 min at 26°C, with aliquots removed periodically and quenched in 3 M sodium acetate (pH 5.1). Quenched samples were resolved by cation-exchange TLC and 35S-Met-puromycin product quantitated. The observed rate constant at several concentrations of inhibitor was measured and normalized to a positive control reaction performed in the absence of inhibitor.
eEF-1 and eEF-2 dependent assays
Phe-tRNAPhe was prepared as previously described (Odom et al. 1990) except that yeast S100 was used as a source of tRNA synthetase. The tRNA was further purified by passage through a Sephadex G-50 column. Both enzymatic and nonenzymatic aminoacyl-tRNA binding experiments were performed following the procedure outlined (SirDeshpande and Toogood 1995). Reaction mixture containing HEPES buffer (20 mM HEPES-KOH at pH 7.5, 10 mM MgCl2, 100 mM KCl, 1.0 mM DTT), 89 pmoles of 80S ribosome, and inhibitor incubated for 10 min at 37°C; 0.15 mM GMPPNP, 0.2 μg of poly(U), 2.3 μg of eEF1, and 10 pmoles of [14C]Phe-tRNAPhe were then added and incubated at 37°C for another 30 min. To determine the amount of ribosome bound [14C]Phe-tRNAPhe, 2 aliquots (10%) of the assay mixture were removed, diluted with 0.8 mL of HEPES buffer, and filtered through a Millipore Type HA nitrocellulose filter. The filter was rinsed with the same buffer (5 × 2 mL), dried, and counted.
To measure nonenzymatic binding of [14C]Phe-tRNAPhe to ribosome, 177 pmoles of 80S ribosome were preincubated with inhibitor in the presence of Tris-HCl buffer (50 mM Tris-HCl at pH 7.5, 60 mM KCl) and 20 mM MgCl2 at 37°C for 10 min poly(U) (40 μg) and [14C]Phe-tRNAPhe were incubated for 30 min at 37°C. Two aliquots constituting 10% of the reaction mixture were removed and diluted into 0.8 mL of Tris-HCl buffer containing 20 mM MgCl2 and filtered through a Type HA nitrocellulose filter. The filter was rinsed with the same buffer (2 × 5 mL), dried, and counted to measure the amount of ribosome bound [14C]Phe-tRNAPhe.
eEF1-dependent [14C]Phe-tRNAPhe binding to 89 pmoles of ribosome was performed as described above. The amount of P site bound [14C]Phe-tRNAPhe was determined for an identical sample using the puromycin reaction (Wurmbach and Nierhaus 1979). To determine the inhibition of translocation, inhibitors were added into the rest of the 80% reaction mixture and incubated for 10 min at 37° C. Then, eEF2 (0.4 μg), HEPES buffer to compensate with the reaction volume, and 1 mM puromycin were added followed by 1 mM GTP to initiate translocation. Following incubation for an additional 30 min at 37° C, 2 aliquots (10%) of reaction mixture were quenched in 30 μL of 1 mM sodium acetate (pH 5.1) and the amount of P site bound [14C]Phe-tRNAPhe was determined by extraction of [14C]Phe-puromycin into ethyl acetate (1 mL). The amount of radioactivity was measured by scintillation counting of 900-μL aliquots of the organic layer.
To assay translocation following nonenzymatic binding of [14C]Phe-tRNAPhe to ribosome, 10× Tris-HCl buffer (pH 7.5) was added to compensate the final reaction volume, followed by addition of 1 mM GTP and 1 mM MgCl2 into the rest of the 80% assay mixture from the nonenzymatic tRNA-binding reaction. Following incubation for 15 min at 37° C, two 10% aliquots of reaction mixture were diluted into 0.8 mL of Tris-HCl buffer containing 10 mM MgCl2 and precipitated onto nitrocellulose as previously described to determine the amount of [14C]Phe-tRNAPhe bound to ribosomes immediately prior to translocation. Nonenzymatic translocation was measured by adding 10× Tris-HCl buffer containing 100 mM MgCl2 and 0.5 mM puromycin following incubation for 30 min at 37°C. The reaction (2 aliquots of 10%) was quenched with 60 μL NH4HCO3 and extracted with 1 mL ethyl acetate. The amount of [14C]Phe-puromycin formed was measured as previously described.
In vivo metabolic labeling studies
For metabolic labeling studies, 7 × 104 HeLa cells were seeded into 2-cm2 plates in duplicate the day prior to the experiment. Cells were incubated in the presence of compound or vehicle alone for the indicated amounts of time. 35S-methionine (150–225 μCi/mL) was added to cells 5 min before harvesting, whereas 3H-uridine (24 μCi/mL) or 3H-thymidine (48 μCi/mL; Perkin Elmer Life Sciences) was added to cells 10 min before harvesting. For protein labeling, 35S-methionine was added in methionine-free media supplemented with 10% dialyzed FCS. For 3H-uridine and 3H-thymidine labeling, the isotopes were added in DMEM supplemented with 10% dialyzed FCS. Cells were washed once with phosphate-buffered saline, trypsinized, and harvested by centrifugation. The supernatant was removed by aspiration and the cell pellet was lysed in RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). Forty percent of the lysate was used for TCA precipitation. For detection of radiolabeled protein, the lysate was spotted onto Whatman 3 MM paper (that had been preblocked with 0.1% methionine), dried, and placed in cold 10% TCA for 20 min. Filters were transferred to 5% TCA, boiled for 15 min, washed once with 5% TCA and once with 95% ethanol, and dried. Radioactivity was determined by scintillation counting. The obtained counts were normalized to protein concentration in each sample, which had been determined using a modified Lowry assay (DC Protein Assay; Bio-Rad).
For detection of radiolabeled nucleic acid, 5% TCA was added to 50% of the lysate supplemented with 9 μg of yeast tRNA. This was incubated for 20 min at 4°C, after which the solution was filtered through a GF/C glass fiber filter (Whatmann) that had been preblocked with 0.1 M sodium pyrophosphate in 5% TCA, washed twice with 5 mL of 1% TCA, and rinsed with 95% ethanol. After drying, radioactivity was determined by scintillation counting. The obtained counts were normalized to the protein concentration in each sample, which had been determined using a modified Lowry assay (DC Protein Assay; Bio-Rad).
Polysome analysis in HeLa cells
The day prior to the experiment, HeLa cells were seeded at 4 × 105 cells/10 cm petri dish. Compound (10 μM final concentration) or DMSO (0.1% final concentration) was added to cells for 20 min, after which the cells were washed twice with PBS, harvested with a rubber policeman, and collected by brief centrifugation. The cell pellet was resuspended in lysis buffer (5 mM Tris-HCl at pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 0.5% Triton X-100, 0.5% sodium deoxycholate, 2 mM DTT), vortexed, and centrifuged for 2 min at 14,000g. The supernatants were loaded onto 10%–50% sucrose gradients prepared in 20 mM HEPES (pH 7.6), 100 mM KCl, 5 mM MgCl2 and centrifuged in an SW40 at 35,000 rpm for 2 h. Gradients were analyzed by piercing the tube with a Brandel tube piercer, passing 60% sucrose through the bottom of the tube, and monitoring the absorbance of the material eluting from the tube using an ISCO UA-6 UV Detector.
Acknowledgments
We thank Dr. Nahum Sonenberg for critical reading of the manuscript. We are immensely grateful to the NIH/NCI Developmental Therapeutics Program for their generous supply of phyllanthoside, nagilactone C, and other compounds for testing that were identified by the COMPARE algorithm. We are grateful to Dr. Jon Lorsch and Drew Applefield for their help in troubleshooting the peptidyl transferase assay. J.P. is a Canadian Institutes of Health Research (CIHR) senior investigator. This work was supported by grants from the National Cancer Institute of Canada (#011040, #012385) to J.P.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5200204.
REFERENCES
- Abid, M.R., Li, Y., Anthony, C., and DeBenedetti, A. 1999. Translational regulation of ribonucletide reductase by eukaryotic initiation factor 4E links protein synthesis to control of DNA replication. J. Biol. Chem. 274: 35991–35998. [DOI] [PubMed] [Google Scholar]
- Adachi, H., Nishimura, Y., and Takeuchi T. 2002. Synthesis and activities of bactobolin derivatives having new functionality at C-3. J. Antibiot. 55: 92–98. [DOI] [PubMed] [Google Scholar]
- Ahuja, D., Vera, M.D., SirDeshpande, B.V., Morimoto, H., Williams, P.G., Joullie, M.M., and Toogood, P.L. 2000. Inhibition of protein synthesis by didemnin B: How EF-1α mediates inhibition of translocation. Biochemistry 39: 4339–4346. [DOI] [PubMed] [Google Scholar]
- Anand, N., Murthy, S., Amann, G., Wernick, M., Porter, L.A., Cukier, I.H., Collins, C., Gray, J.W., Diebold, J., Demetrick, D.J., et al. 2002. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 31: 301–305. [DOI] [PubMed] [Google Scholar]
- Ayuso, M.S. and Heredia, C.F. 1968. Guanosine trisphosphate dependent enzymic binding of aminoacyl transfer ribonucleic acid to yeast ribosomes. Eur. J. Biochem. 7: 111–118. [DOI] [PubMed] [Google Scholar]
- Birkmayer, G.D. and Balda, B.R. 1970. Proflavine inhibition of protein synthesis in malignant hamster melanoma. FEBS Lett. 11: 221–223. [DOI] [PubMed] [Google Scholar]
- Burres, N.S. and Clement, J.J. 1989. Antitumor activity and mechanism of action of the novel marine natural products mycalamide-A and -B and onnamide. Cancer Res. 49: 2935–2940. [PubMed] [Google Scholar]
- Carrasco, L., Fernandez-Puentes, C., and Vazquez, D. 1975. Effects of ricin on the ribosomal sites involved in the interaction of the elongation factors. Eur. J. Biochem. 54: 499–503. [DOI] [PubMed] [Google Scholar]
- Carriere, M., Vijayabaskar, V., Applefield, D., Harvey, I., Philippe, G., Lorsch, J.R., Lapidot, A., and Pelletier, J. 2002. Inhibition of protein synthesis by aminoglycoside-arginine conjugates. RNA 8: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh, A.H., Hempel, W.M., Taylor, L.J., Rogalsky, V., Todorov, G., and Rothblum, L.I. 1995. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374: 177–180. [DOI] [PubMed] [Google Scholar]
- Chapman, D.E., Moore, D.J., Melder, D.C., Breau, A., and Powls, G. 1989. Isolation, identification and biological activity of a phyllanthoside metabolite produced in vitro by mouse plasma. Canc. Chemother. Pharmacol. 25: 184–188. [DOI] [PubMed] [Google Scholar]
- Cleaveland, E.S., Monks, A., Vaigro-Wolff, A., Zaharevitz, D.W., Paull, K., Ardalan, K., Cooney, D.A., and Ford Jr., H. 1995. Site of action of two novel pyrimidine biosynthesis inhibitors accurately predicted by the COMPARE program. Biochem. Pharmacol. 49: 947–954. [DOI] [PubMed] [Google Scholar]
- Cleaveland, E.S., Zaharevitz, D.W., Kelley, J.J., Paull, K., Cooney, D.A., Ford Jr., H. 1996. Identification of a novel inhibitor (NSC 665564) of dihydroorotate dehydrogenase with a potency equivalent to brequinar. Biochem. Biophys. Res. Commun. 223: 654–659. [DOI] [PubMed] [Google Scholar]
- Craig, N. and Kostura, M. 1983. Inhibition of protein synthesis in CHO cells by actinomycin D: Lesion occurs after 40S Initiation complex formation. Biochemistry 22: 6064–6071. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia, N., Gustavsson, P., Andersson, B., Pettersson, M., Willig, T.N., Dianzani, I., Ball, S., Tchernia, G., Klar, J., Matsson, H., et al. 1999. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21: 169–175. [DOI] [PubMed] [Google Scholar]
- Dyer, B.W., Ferrer, F.A., Klinedinst, D.K., and Rodriguez, R. 2000. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 282: 158–161. [DOI] [PubMed] [Google Scholar]
- Ennis, H.L. 1966. Synthesis of ribonucleic acid in L cells during inhibition of protein synthesis by cycloheximide. Mol. Pharmacol. 2: 543–557. [PubMed] [Google Scholar]
- Fresno, M., Jimenez, A., and Vazquez, D. 1977. Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72: 323–330. [DOI] [PubMed] [Google Scholar]
- Gharehbaghi, K., Paull, K., Kelley, J.J., Barchi Jr., J.J., Marquez, V.E., Cooney, D.A., Monks, A., Scudiero, D., Kohn, K., and Jayaram, H.N. 1994. Cytotoxicity and characterization of an active metabolite of benzamide ribosode: A novel inhibitor of IMP dehydrogenase. J. Cancer 56: 892–899. [DOI] [PubMed] [Google Scholar]
- Graff, J.R. and Zimmer, S.G. 2003. Translational control and metastatic progression: Enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin. Exp. Met. 20: 265–273. [DOI] [PubMed] [Google Scholar]
- Grollman, A.P. 1967. Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J. Biol. Chem. 242: 3226–3233. [PubMed] [Google Scholar]
- ———. 1968. Inhibitors of protein biosynthesis. V. Effects of emetine on protein and nucleic acid biosynthesis in HeLa cells. J. Biol. Chem. 243: 4089–4094. [PubMed] [Google Scholar]
- Harvey, I., Garneau, P., and Pelletier, J. 2002. Forced engagement of a RNA/protein complex by a chemical inducer of dimerization to modulate gene expression. Proc. Natl. Acad. Sci. 99: 1882–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, Y., Sakan, T., Sakurai, Y., and Tashiro, T. 1975. Antitumor activity of angilactones. Jpn. J. Cancer Res. 66: 587–588. [PubMed] [Google Scholar]
- Hayashi, Y., Kim, Y., and Hayashi, Y. 1992. Nagilactones as an anti-feedent from Podocarpus nagi for herbivorous mammals. Biosci. Biotech. Biochem. 56: 1302–1303. [Google Scholar]
- Hershey, J.W.B. and Miyamoto S. 2000. Translational control of cancer. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 637–654. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Holcik, M.N., Sonenberg, N., and Korneluk, R.G. 2000. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16: 469–473. [DOI] [PubMed] [Google Scholar]
- Huang, M.T. 1975. Harringtonine, an inhibitor of initiation of protien biosynthesis. Mol. Pharmacol. 11: 511–519. [PubMed] [Google Scholar]
- Huang, M.T. and Grollman, A.P. 1972. Mode of action of tylocrebrine: Effects on protein and nucleic acid synthesis. Mol. Pharmacol. 8: 538–550. [PubMed] [Google Scholar]
- Jansen, R.P. 1999. RNA-cytoskeleton associations. FASEB J. 13: 455–466. [PubMed] [Google Scholar]
- Kantarjian, H.M., Talpaz, M., Santini, V., Murgo, A., Cheson, B., and O’Brien, S.M. 2001. Homoharringtonine: History, current research, and future direction. Cancer 92: 1591–1605. [DOI] [PubMed] [Google Scholar]
- Kohlhagen, G., Paull, K., Cushman, M., Nagafuji, P., and Pommier, Y. 1998. Protein-linked DNA strand breaks induced by NSC 314622: A novel non-camptothecin topoisomerase I poison. Mol. Pharmacol. 54: 50–58. [DOI] [PubMed] [Google Scholar]
- Kubo, I., Muroi, H., and Himejima, M. 1993. Combination effects of antifungal nagilactones against Candida albicans and two other fungi with phenylpropanoids. J. Nat. Prod. 56: 220–226. [DOI] [PubMed] [Google Scholar]
- Kupchan, S.M., LaVoie, E.J., Branfman, A.R., Fei, B.Y., Bright, W.M., and Bryan, R.F. 1977. Phyllanthocin, a novel bisabolane aglycone from the antileukemic glycoside, phyllanthoside. J. Am. Chem. Soc. 99: 3199–3201. [DOI] [PubMed] [Google Scholar]
- Lieberman, I., Ove, P., Abrams, R., and Hunt, N. 1963. Levels of enzyme activity and deoxyribonucleic acid synthesis in mammalian cells cultured from animal J. Biol. Chem. 238: 3955–3962. [PubMed] [Google Scholar]
- Liljas, A. 1996. Protein synthesis: Imprinting through molecular mimicry. Curr. Biol. 6: 247–249. [DOI] [PubMed] [Google Scholar]
- Lorsch, J.R. and Herschlag, D. 1999. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J. 18: 6705–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric, F. and Hunt, K.K. 2003. Translation initiation in cancer: A novel target for therapy. Mol. Canc. Ther. 1: 971–979. [PubMed] [Google Scholar]
- Momparler, R.L., Karon, M., Siegel, S.E., and Avila, F. 1976. Effect of adriamycin on DNA, RNA, and protein synthesis in cell-free systems and intact cells. Cancer Res. 36: 2891–2895. [PubMed] [Google Scholar]
- Moore, D.J. and Powls, G. 1986. Disposition and metabolism of the antitumor glycoside phyllanthoside in mouse and beagle dog. Canc. Chemother. Pharmacol. 16: 218–222. [DOI] [PubMed] [Google Scholar]
- Mueller, G.C., Kajiwara, K., and Stubblefield, E. 1962. Molecular events in reproduction of animal cells. 1. Effects of puromycin on duplication of DNA. Cancer Res. 22: 1084–1090. [PubMed] [Google Scholar]
- Nahvi, A., Sudarsan, N., Ebert, M.S., Zou, X., Brown, K.L., and Breaker, R.R. 2002. Genetic control by a metabolite binding mRNA. Chem. Biol. 9: 1043–1049. [DOI] [PubMed] [Google Scholar]
- Odom, O.W., Picking, W.D., and Hardesty, B. 1990. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry 29: 10734–10744. [DOI] [PubMed] [Google Scholar]
- Ottenheijm, H.C. and van den Broek, L.A. 1988. The development of sparsomycin as an anti-tumour drug. Anticancer Drug Des. 2: 333–337. [PubMed] [Google Scholar]
- Paull, K.D., Shoemaker, R.H., Hodes, L., Monks, A., Scudiero, D.A., Rubinstein, L., Plowman, J., and Boyd, M.R. 1989. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: Development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 81: 1088–1092. [DOI] [PubMed] [Google Scholar]
- Paull, K., Lin, C.M., Malspeis, L., and Hamel, E. 1992. Identification of novel antimitotic agents acting at the tubulin level by computer-assisted evaluation of differential cytotoxicity data. Cancer Res. 52: 3892–3900. [PubMed] [Google Scholar]
- Pestka, S. 1977. Inhibitors of protein synthesis. Academic Press, New York.
- Raught, B., Gingras, A.-C., and Sonenberg, N. 2000. Regulation of ribosome recruitment in eukaryotes. In Translational control of gene expression (eds. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews), pp. 245–293. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rinehart, K.L. 2000. Antitumor compounds from tunicates. Med. Res. Rev. 20: 1–27. [DOI] [PubMed] [Google Scholar]
- Ruggero, D. and Pandolfi, P.P. 2003. Does the ribosome translate cancer? Nat. Rev. 3: 179–192. [DOI] [PubMed] [Google Scholar]
- Shrestha, K., Banskota, A.H., Kodata, S., Shrivastava, S.P., Strobel, G., and Gewali, M.B. 2001. An antiproliferative norditerpene dilactone, Nagilactone C, from Podocarpus neriifolius. Phytomedicine 8: 489–491. [DOI] [PubMed] [Google Scholar]
- Sing, P., Fennemore, P.G., and Dugdale, J.S. 1978. The insecticidal activity of foliage from New Zealand conifers. Biochem. System Ecol. 6: 103–106. [Google Scholar]
- SirDeshpande, B.V. and Toogood, P.L. 1995. Mechanism of protein synthesis inhibition by didemnin B in vitro. Biochemistry 34: 9177–9184. [DOI] [PubMed] [Google Scholar]
- Szittner, R. and Meighen, E. 1990. Nucleotide sequence, expression, and properties of luciferase coded by lux genes from a terrestrial bacterium. J. Biol. Chem. 265: 16581–16587. [PubMed] [Google Scholar]
- Vazquez, D. 1979. Inhibitors of protein biosynthesis. Mol. Biol. Biochem. Biophys. 30: 1–312. [DOI] [PubMed] [Google Scholar]
- Watkins, S.J. and Norbury, C.J. 2002. Translation initiation and its deregulation during tumorigenesis. Br. J. Cancer 86: 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, I.B. and Finkelstein, I.H. 1967. Proflavine inhibition of protein synthesis. J. Biol. Chem. 242: 3757–3762. [PubMed] [Google Scholar]
- Wurmbach, P. and Nierhaus, K.H. 1979. Codon–anticodon interaction at the ribosomal P (peptidyl-tRNA) site. Proc. Natl. Acad. Sci. 76: 2143–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, C.W. 1966. Inhibitory effects of acetoxycycloheximide, puromycin, and pactamycin upon synthesis of protein and DNA in asynchronous populations of HeLa cells. Mol. Pharmacol. 2: 50–55. [PubMed] [Google Scholar]
- Zaharevitz, D.W., Gussio, R., Loest, M., Senderowicz, A.M., Lahusen, T., Kunick, C., Meijer, L., and Sausville, E.A. 1999. Discovery and initial characterization of the paullones: A novel class of small-molecule inhibitors of cyclin-dependent kinases. Cancer Res. 59: 2566–2569. [PubMed] [Google Scholar]
- Zaharevitz, D.W., Holbeck, S.L., Bowerman, C., and Svetlik, P.A. 2002. COMPARE: A web accessible tool for investigating mechanisms of cell growth inhibition. J. Mol. Graph. Mod. 20: 297–303. [DOI] [PubMed] [Google Scholar]