Abstract
Tandem repeats of the telomeric DNA sequence d(T4G4) of Oxytricha nova are capable of forming unusually stable secondary structures incorporating Hoogsteen hydrogen bonding interactions. The biological significance of such DNA structures is supported by evidence of specific recognition of telomere end-binding proteins in the crystal state. To further characterize structural polymorphism of Oxytricha telomeric DNAs, we have obtained and interpreted Raman, ultraviolet resonance Raman (UVRR) and circular dichroism (CD) spectra of the tandem repeats d(G4T4G4) (Oxy1.5), d(T4G4)2 (Oxy2) and dT6(T4G4)2 (T6Oxy2) and related non-telomeric isomers in aqueous salt solutions. Raman markers of Oxy1.5 identify both C2′-endo/anti and C2′-endo/syn conformations of the deoxyguanosine residues and Hoogsteen hydrogen bonded guanine quartets, consistent with the quadruplex fold determined previously by solution NMR spectroscopy. Raman, UVRR and CD signatures and Raman dynamic measurements, to monitor imino NH→ND exchanges, show that the Oxy1.5 antiparallel quadruplex fold is distinct from the hairpin structures of Oxy2 and T6Oxy2, single-stranded structures of d(TG)8 and dT6(TG)8 and previously reported quadruplex structures of d(T4G4)4 (Oxy4) and dG12. Spectral markers of the telomeric and telomere-related DNA structures are tabulated and novel Raman and UVRR indicators of thymidine and deoxyguanosine conformations are identified. The results will be useful for probing structures of Oxytricha telomeric repeats in complexes with telomere end-binding proteins.
INTRODUCTION
Telomeres, the protein–DNA complexes that protect the ends of eukaryotic chromosomes from degradation and fusion, are essential for a variety of cellular functions (1). The telomeric DNA strand that is oriented in the 5′→3′ direction is distinguished by multiple repeats of the sequence d(TnAm G3–4), where n and m are small integers. The guanine-rich telomeric segment may range in size from several base pairs to tens of kilobase pairs. A single-stranded overhang at the terminus of the guanine-rich strand is a feature that is conserved among protozoa (2), yeast (3) and mammals (4). In the ciliated protozoan Oxytricha nova this telomeric 3′ overhang consists of the 16 base sequence d(T4G4)2, also referred to as Oxy2. Recent studies suggest that molecular mechanisms of telomere maintenance may also be conserved among different eukaryotic species (5,6).
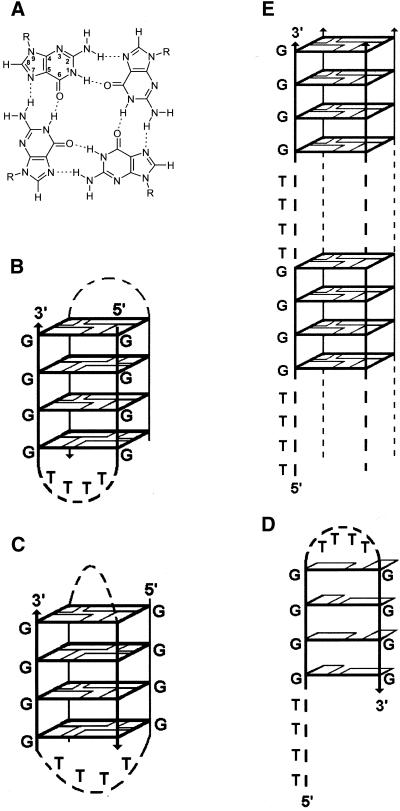
Telomeric DNA sequences are remarkable for their ability to form highly stable quadruplex structures in vitro (7). An intermolecular quadruplex structure of the Oxytricha telomeric sequence d(G4T4G4) (Oxy1.5), in which Hoogsteen hydrogen bonded guanine quartets (Fig. 1A) tether four strands in parallel (Fig. 1E), has been reported to accompany thermal denaturation of the Watson–Crick duplex d(G4T4G4)· d(C4A4C4) in the presence of high concentrations of alkali metal ions (8). On the other hand, the tandem repeat d(T4G4)4 (Oxy4) readily forms an intramolecular quadruplex distinguished by antiparallel folding of the DNA backbone and guanine quartets (8). Alkali metal ions facilitate conversion of this ‘foldback’ structure to the intermolecular parallel quadruplex (9,10). Variants of the foldback secondary structure have also been demonstrated for dimers of Oxy1.5 in the crystal (Fig. 1B) (11) and solution (Fig. 1C) states (12). In the case of Oxy2, a solution hairpin fold stabilized by Hoogsteen G·G pairs has been proposed (Fig. 1D; 10).
Figure 1.
(A) Hoogsteen hydrogen bonded guanine quartet. (B) Crystal structure of d(G4T4G4) (Oxy1.5) incorporating guanine quartets (11). (C) Solution structure of Oxy1.5 incorporating guanine quartets (12). (D) Hairpin structure of d(T4G4)2 (Oxy2) incorporating G·G pairs (10). (E) Parallel-stranded quadruplex structure of Oxy2 (9).
Folded secondary structures of Oxytricha telomeric sequences are characterized by thymidine loops (dT4) and Hoogsteen hydrogen bonded dG residues in both the C2′-endo/syn and C2′-endo/anti conformations. Because Raman marker bands have been established for both the dT4 loop and for syn and anti dG moieties (8,10,13–15), Raman spectroscopy provides a convenient probe of these structural features. In addition, Raman monitoring of hydrogen isotope exchange dynamics differentiates between guanines involved in the formation of Hoogsteen pairs and quartets (10,15). The capability of Raman spectroscopy to probe equilibrium and dynamic properties of telomeric DNA structures can be extended to dilute DNA solutions (∼10 µM) through the technique of UV resonance Raman spectroscopy (UVRR) using 257 nm excitation (16–19).
Here, we report Raman and UVRR signatures of the secondary structures formed by the Oxytricha telomeric DNA sequences Oxy1.5, Oxy2 and T6Oxy2 in aqueous solutions containing either Na+ or both Na+ and K+. We also compute difference spectra diagnostic of structural transitions between folded and unfolded states of these sequences. The T6Oxy2 oligomer differs from Oxy2 by the addition of six dT residues at the 5′ terminus. Such a 5′ leader may be required for recognition of telomeric DNA by cellular telomere-binding factors in vivo (20–22). The Raman and UVRR spectra are complemented by corresponding circular dichroism (CD) spectra, which are compared with previously reported CD profiles of telomeric DNA (23). The results allow identification of tertiary folds common to different repeats of the Oxytricha telomeric sequence and provide a reference library for future characterization of structure transformations induced by binding of telomere-specific factors. The present results extend previous Raman investigations of Oxy2 and Oxy4 (8–10,15) and X-ray and NMR studies of Oxy1.5 (11,12,24–26).
MATERIALS AND METHODS
Sample preparation
Oligonucleotides d(G4T4G4) (Oxy1.5), d(T4G4)2 (Oxy2), dT6(T4G4)2 (T6Oxy2), d(TG)8 and dT6(TG)8 were synthesized by standard methods and purified on an ISCO model 2350 HPLC system (ISCO, Lincoln, NE) using a Hamilton PRP-1 reversed-phase column. The column was heated to 90°C to ensure denaturation of DNA secondary structure during purification. For HPLC, samples were dissolved in 0.1 M triethylamine acetate, pH 8. Optima grade acetonitrile (Fisher Scientific, Pittsburgh, PA) was used for elution. The first chromatographic step separated detritylated failure sequences from the correct sequence containing the trityl group. Peak fractions were pooled, lyophilized and detritylated in 80% acetic acid. The pure oligonucleotide was obtained after a second chromatographic step. The samples were desalted by dialysis (1000 molecular weight cut-off) against water. DNA concentrations were determined using an average nucleotide molar extinction at 260 nm of 9850 M–1·cm–1. Purified DNA was dissolved to 15 mg/ml in water and adjusted to pH 7.2 ± 0.2, using either dilute HCl or NaOH, and lyophilized. The lyophilizate was dissolved to ∼30 mg/ml in 50 mM TEN sample buffer (10 mM Tris pH 7.2, 1 mM EDTA, 50 mM NaCl).
The samples of Oxy1.5, Oxy2 and T6Oxy2 examined by spectroscopic methods were also characterized by PAGE under non-denaturing conditions (data not shown). Oxy1.5 migrated as a single, highly folded species, presumed to be a quadruplex. Both Oxy2 and T6Oxy2 migrated more slowly than Oxy1.5 but more rapidly than the single-stranded models d(TG)8 and dT6(TG)8. This is consistent with the formation of similarly folded secondary structures distinct from the quadruplex (10).
Raman spectroscopy
Aliquots (6 µl) of samples were sealed in glass capillary tubes (Kimax no. 34507) for Raman spectroscopy. Spectra were excited with the 532 nm line of a solid state Nd:YVO4 laser (Verdi model V; Coherent, Santa Clara, CA) using 200 mW of radiant power at the sample. Rayleigh scattered light was separated by a notch filter. Raman scattered light was collected with a single spectrograph (500M; SPEX Industries, Metuchen, NJ) equipped with a liquid nitrogen-cooled, charge-coupled device detector. The spectrometer was calibrated with indene and CCl4 as wavenumber standards. Reported Raman wavenumbers are accurate to ±1 cm–1 for sharp bands and to within ±2 cm–1 for broad or overlapping bands. Sample temperature was maintained at 10°C during data collections by use of a thermostat designed for the 90° scattering geometry. In a typical data collection protocol, four 10 min exposures were accumulated and averaged. Solvent compensations and other spectral manipulations were accomplished using standard software routines (Galactic Industries Corp., Salem, NH).
Time-resolved Raman spectroscopy to monitor deuterium exchange of Oxy1.5 in D2O solution was carried out using protocols similar to those described previously for Oxy4 and Oxy2 (10,15). Briefly, the DNA was lyophilized from 50 mM TEN sample buffer and redissolved to the same volume in D2O. The sample was sealed in a glass capillary and thermostated at 20°C for collection of data as H→D exchange progressed.
UV resonance Raman spectroscopy
Samples of dT6(TG)8, dT6(T4G4)2, d(TG)8, d(T4G4)2 and d(G4T4G4) were prepared in low salt (25 mM Na2SO4) and high salt (25 mM Na2SO4, 150 mM KCl) buffers at DNA concentrations of 10, 10, 14, 14 and 19 µM, respectively, by dilution of 30 mg/ml stock solutions. An aliquot (120 µl) of each sample was transferred to a cylindrical quartz cell mounted in an air-driven thermostated sample rotator (∼300 r.p.m.). UVRR spectra were excited with the 257 nm line of a continuous wave, frequency-doubled argon ion laser (model FreD; Coherent, Santa Clara, CA) using ∼1 mW of radiant power at the sample. Further details of this instrumentation have been described (27,28). No sample photodecomposition could be detected for exposure times up to ∼6 min. Wavenumbers of reported UVRR bands are accurate to ±2 cm–1.
Circular dichroism spectroscopy
CD spectra in the region 240–330 nm were recorded on a Jasco-720 spectropolarimeter (Jasco, Easton, MD) using cells of 1 cm optical path and an instrument scan speed of 20 nm/min with a response time of 1 s. For CD spectroscopy, DNA solutions were prepared at 3 µM in 10 mM sodium phosphate, pH 7.5.
RESULTS AND INTERPRETATION
Characterization of telomeric and non-telomeric DNA sequences
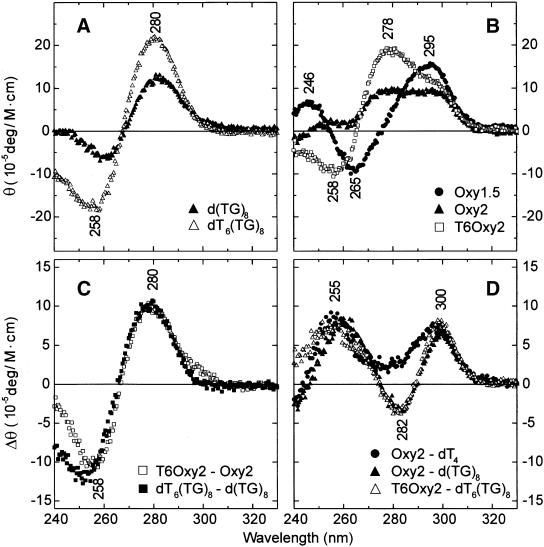
Circular dichroism spectroscopy. CD spectra (240–330 nm) of non-telomeric sequence isomers d(TG)8 and dT6(TG)8 (Fig. 2A) exhibit a single sharp ellipticity maximum at 280 nm and a minimum of lesser amplitude near 258–260 nm, consistent with single-stranded helical conformations stabilized by the stacking of bases (29,30). CD spectra of Oxy1.5, Oxy2 and T6Oxy2 (Fig. 2B) are more complex. Oxy1.5, two molecules of which form a quadruplex containing only interstrand hydrogen bonding (Fig. 1C; 12), exhibits a trough at 265 nm and peaks at 246 and 295 nm diagnostic of guanine quartets (23). Figure 2B shows that this signature changes as the d(G4T4G4) sequence is extended by 5′-thymidylate leaders of four (Oxy2) and 10 residues (T6Oxy2). Thus, Oxy2 exhibits substantially lower amplitudes at 246, 265 and 295 nm and a distinct positive band at 278 nm not apparent in the Oxy1.5 CD spectrum. For T6Oxy2, the attenuations of positive ellipticities at wavelengths below 260 nm and above 290 nm are even more pronounced and the spectrum is strongly dominated by a maximum at 278 nm and minimum at 258 nm.
Figure 2.
Circular dichroism spectra (240–330 nm). (A) d(TG)8 (filled triangle) and dT6(TG)8 (open triangle). (B) Oxy1.5 (filled circle), Oxy2 (filled triangle) and T6Oxy2 (open square). (C) Differences: T6Oxy2 – Oxy2 (open square) and dT6(TG)8 – d(TG)8 (filled square). (D) Differences: Oxy2 – dT4 (filled circle), Oxy2 – d(TG)8 (filled triangle) and, T6Oxy2 – dT6(TG)8 (open triangle). Data were obtained on a Jasco-720 spectropolarimeter (20 nm/min scan speed; 1 s response time; 1 cm path) from 3 µM DNA samples in 10 mM sodium phosphate, pH 7.5, at 20°C.
Computed T6Oxy2 – Oxy2 and dT6(TG)8 – d(TG)8 difference spectra are virtually superimposable (Fig. 2C), indicating similar folds. Also, the difference signatures of Figure 2C closely approximate the spectra of Figure 2A, implying a single-stranded base stack for the dT6 leader. Likewise, the Oxy2 – d(TG)8 and T6Oxy2 – dT6(TG)8 differences of Figure 2D are nearly congruent, consistent with a common G4T4G4 fold. CD signatures of these sequences are thus well explained as sums of modules contributed by constituent folds.
A further manipulation of the CD data is instructive. Assuming that the CD contribution from the single-stranded 5′-dT4 leader of Oxy2 can be represented by 2/3 of the CD contribution from the dT6 leader of T6Oxy2 (as generated in Fig. 2C), then the difference Oxy2 – dT4 (Fig. 2D, filled circles) should approximate the CD contribution of the G4T4G4 fold of Oxy2. We propose this CD profile, characterized by ellipticity maxima near 255 and 300 nm, as diagnostic of the secondary structure of the folded G4T4G4 domain of Oxy2. This fold consists of a hairpin stabilized by Hoogsteen G·G pairs (10) and the proposed CD signature differs significantly from that of the Oxy1.5 quadruplex (Fig. 2B). The data of Figure 2D are also consistent with a hairpin secondary structure for the folded G4T4G4 domain of T6Oxy2. In summary, the CD data distinguish the antiparallel quadruplex fold of the Oxy1.5 dimer (Fig. 1C) from the antiparallel hairpin folds of Oxy2 and T6Oxy2 monomers (Fig. 1D). Further distinction is provided by the Raman and UVRR spectra discussed below.
The telomeric DNA samples yielding the CD spectra of Figure 2B were also examined by PAGE under non-denaturing conditions (data not shown). The gels confirm that T6Oxy2 and Oxy2 migrate as hairpins and that the preparations are free of appreciable quantities of quadruplexes or higher order aggregates.
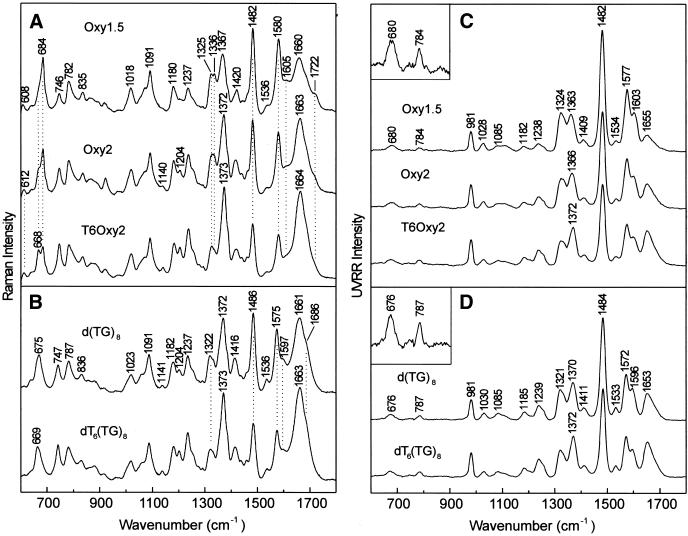
Raman spectroscopy. The Raman spectrum of Oxy1.5 (Fig. 3A, top trace) exhibits prominent phosphodiester markers at 835 and 1091 cm–1, indicating g–/g– O–P conformations of the C5′–O5′–P–O3′–C3′ network. This conformation also occurs in double-helical B-DNA and telomeric quadruplexes (8,31). Raman markers at 1336 and 684 cm–1 identify the C2′-endo/anti dG conformation, whereas the marker at 1325 cm–1 and the partially resolved shoulder near 670 cm–1 identify C2′-endo/syn dG (8,32,33). Thymidines of the Oxy1.5 loop produce the weak band near 610 ± 2 cm–1 and also contribute to the Raman intensity near 668 cm–1 (15,30). Markers near 610 and 668 cm–1 are assigned to thymidines of the dT4 loop (15,34). The data indicate comparable populations of C2′-endo/anti and C2′-endo/syn dG conformers and predominantly C2′-endo/anti dT conformers in Oxy1.5. Raman markers at 1482 and 1722 cm–1, which are sensitive to guanine hydrogen bonding (8,15,35,36), indicate that both N7 and O6 participate in interbase hydrogen bonding (Table 1). All of the key Raman markers of Oxy1.5 are therefore consistent with the structure of Figure 1C.
Figure 3.
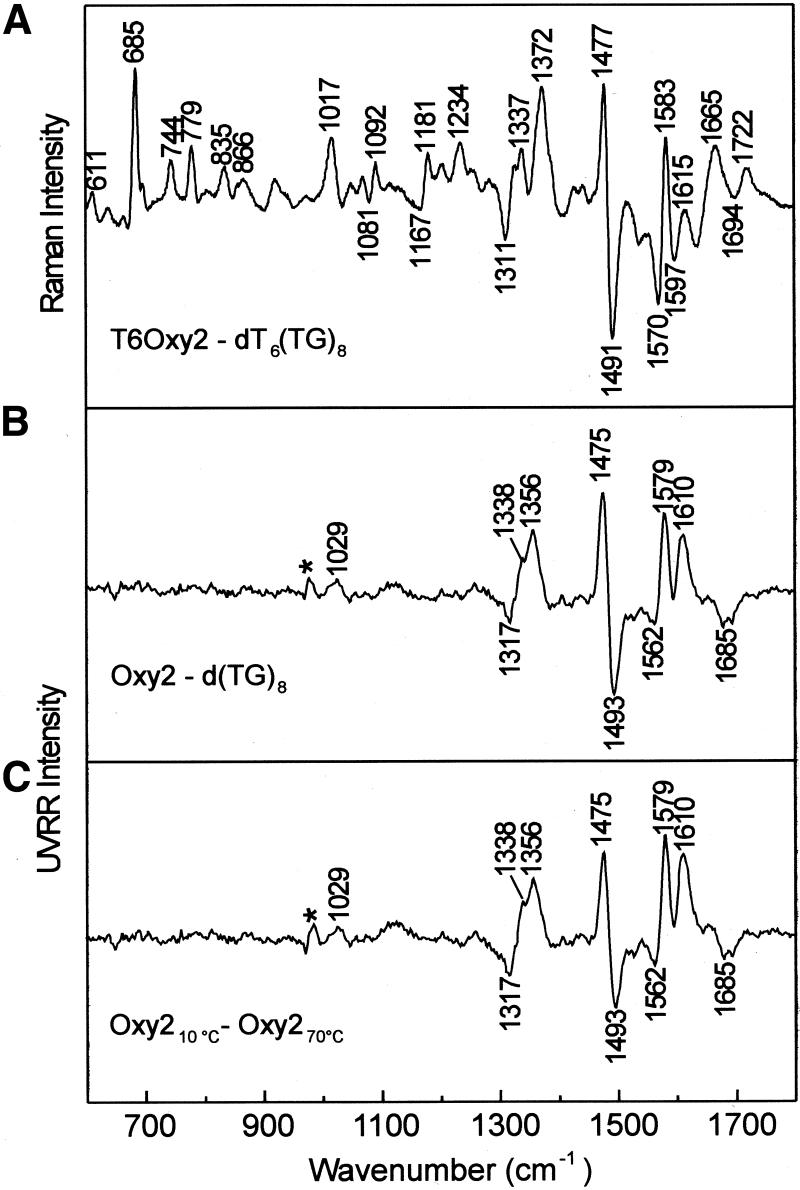
(A) Raman spectra of Oxy1.5, Oxy2 and T6Oxy2. (B) Raman spectra of d(TG)8 and dT6(TG)8. In (A) and (B) solutions contained DNA at 30 mg/ml in 10 mM Tris pH 7.2, 1 mM EDTA and 50 mM NaCl and were maintained at 10°C. Dashed lines in (A) and (B) indicate bands discussed extensively in the text. (C) UVRR spectra of Oxy1.5, Oxy2 and T6Oxy2. (D) UVRR spectra of d(TG)8 and dT6(TG)8. In (C) and (D) the DNA solutions (10– 20 µM, see text) also contained 25 mM Na2SO4 for use of the 981 cm–1 band of SO42– as a reference intensity and wavenumber standard. The inset at the upper left in (C) and (D) shows a 5-fold ordinate amplification of the 600–900 cm–1 interval.
Table 1. Raman and UV resonance Raman bands of Oxytricha telomeric DNA and related non-telomeric DNA sequences in H2O and D2O solutionsa.
| Telomeric repeatb | Non-telomeric isomerc | Assignmentd | Commente | ||
|---|---|---|---|---|---|
| Ramanf | UVRRg | Ramanf | UVRRg | ||
| 608 (603) | dT | T4 diagonal loop | |||
| 612 | T4 hairpin loop | ||||
| 668 (651) | 668 (655) | dT | C2′-endo/anti | ||
| 675 (679) | 680h (677) | 675 (675) | 676 (677) | dG | C2′-endo/syn |
| 684 (683) | 680h (677) | dG | C2′-endo/anti | ||
| 746 (734) | 740 (732) | 747 (736) | 737 (733) | dT | T(N3)i |
| 782 (782) | 784 (785) | 787 (786) | 787 (784) | bk OPO; dT | g–/g– |
| 835 (832) | 836 (831) | bk OPO | g–/g– | ||
| (980) | dG ND def | G(N1, N2)j | |||
| 1018 | 1023 (1018) | dG NH def; dT | G(N2) | ||
| 1028 (1030) | 1030 (1034) | dG | |||
| 1085 (1085) | 1085 (1085) | dG | |||
| 1091 (1091) | 1091 (1090) | bk PO2– | intensity standard | ||
| 1140 | 1141 | dT | |||
| 1180 (1155) | 1182 (1157) | 1182 (1157) | 1185 (1157) | unpaired dT | T(N3) |
| 1237 (1242) | 1238 (1242) | 1237 (1237) | 1239 (1244) | dT | T(N3) |
| (1268) | (1267) | (1264) | dT ND def | T(N3) | |
| 1325 (1325) | 1324 (1320) | 1322 (1318) | 1321 (1319) | dG | C2′-endo/syn |
| 1336 (1336) | u u | u | u u | dG | C2′-endo/anti |
| 1363k (1357) | 1363l(1360) | u (1359) | u (1356) | dG ND2 def | G(N2), C2′-endo/anti |
| 1372 (1372) | ul (1373) | 1372 (1374) | 1371 (1374) | dT; dG | dG C2′-endo/syn |
| (1403) | (1403) | dG ND,ND2 def | (N1, N2)j | ||
| 1412 (1410s) | (1407) | 1411 (1410s) | dG; dT | ||
| 1420 (1420) | 1416 (1415) | C5′H2 def | |||
| 1482 (1478) | 1482 (1478) | 1486 (1476) | 1484 (1477) | dG | N7 H-bond |
| 1536 (1535) | 1534 (1535) | 1536 (1536) | 1533 (1534) | dG | |
| 1580 (1580) | 1577 (1578) | 1575 (1574) | 1572 (1574) | dG | N7 H-bond |
| 1605 | 1603 | 1597 (1603) | 1596 | dG NH def | N1H H-bond |
| 1662 (1662) | 1655 (1659) | 1661 (1659) | 1653 (1658) | dT C=O str | O4 H-bond |
| 1722 (1689) | 1686 (1685) | dG C=O str | O6 H-bond |
aRaman and UVRR bands are given in cm–1 units (no data indicates no band observed; u indicates unresolved wavenumber; D2O solution data are in parentheses). Relative Raman and UVRR intensities (not listed) vary among sequences in accordance with T:G content and conformation (see text). Data are compiled from this work and previous results (8,10,15,30,32,34,37,45).
bData apply to Oxy1.5, Oxy2 and T6Oxy2 unless otherwise noted.
cData apply to d(TG)8 and dT6(TG)8.
dAbbreviations: bk, backbone; def, deformation; str, stretch. Standard notation is used for deoxynucleosides and chemical groups.
eParentheses indicate sites of deuteration. Standard notation is used for deoxynucleotide conformations.
fOff-resonance excitation (532 or 514.5 nm).
gUV resonance excitation (257 nm).
hComposite of C2′-endo/syn dG (∼675 cm–1) and C2′-endo/anti dG (∼684 cm–1) markers.
iVery weak in UVRR spectra.
jThe band shifts to ∼1392 cm–1 in d(G12) upon N2 amino deuteration and to ∼1400 cm–1 when the N1 imino site is additionally deuterated.
kShoulder to the more intense band of dT at 1372 cm–1.
lOverlap of dT and dG markers.
The Oxy2 and T6Oxy2 Raman signatures are distinguished from Oxy1.5 by the intensities of thymidine marker bands (Fig. 3A). Oxy2 and T6Oxy2 also exhibit electrophoretic mobilities and protium/deuterium (H/D) exchange kinetics characteristic of a hairpin structure containing Hoogsteen G·G pairs (Fig. 3A) (10). As in the case of Oxy1.5, these secondary structures contain equal populations of C2′-endo/syn and C2′-endo/anti dG conformers and primarily C2′-endo/anti dT conformers in their dT4 loops.
In contrast to the telomeric DNA sequences of Figure 3A, for which the dominant peak of the 660–690 cm–1 interval occurs at 684 cm–1, the non-telomeric sequences of Figure 3B exhibit their dominant peak at significantly lower wavenumber. Thus, the bands centered at 675 cm–1 in d(TG)8 and at 669 cm–1 in dT6(TG)8 consist mainly of overlapping contributions from the dT conformer (C2′-endo/anti dT at ∼668 cm–1) and the principal dG conformer (C2′-endo/syn dG at 675 cm–1). The greater dT composition in dT6(TG)8 versus d(TG)8 has the effect of displacing the band to a slightly lower wavenumber value (Table 1). Interestingly, the 1300– 1350 cm–1 profile of each non-telomeric sequence also appears to consist of two overlapping components, a dominant peak at 1322 cm–1 (C2′-endo/syn dG) and a weaker shoulder at 1336 cm–1 (C2′-endo/anti dG). The minor population of the latter dG conformer presumably accounts for the broad wing on the high wavenumber side of the 675 cm–1 peak in d(TG)8 and the 669 cm–1 peak in dT6(TG)8. The non-telomeric sequences of Figure 3B lack a dT loop and therefore give no marker near 610 ± 2 cm–1. Likewise, Raman bands characteristic of solvent-exposed guanine N7 and O6 sites (1486 and 1686 cm–1) replace the markers (1482 and 1722 cm–1) diagnostic of Hoogsteen N7 and C6=O hydrogen bonding interactions of the G·G pairs. Also, the guanine Raman marker at 1575 cm–1, which is sensitive to solvation of the exocyclic C2–NH2 group, differs from the analogous band (1580 cm–1) in telomeric DNA (15). Thus, Raman markers diagnostic of either hairpin or quadruplex secondary structures are absent from Raman spectra of d(TG)8 and dT6(TG)8. On the other hand, the prominent DNA backbone markers at 836 and 1091 cm–1 in Figure 3B indicate the same local phosphodiester conformation (g–/g) as in telomeric sequences. The secondary structure for both of the non-telomeric sequences is apparently a base-stacked single strand.
UVRR spectroscopy. The strucutral significance of the UVRR signatures of Figure 3C and D can be understood by reference to UVRR studies of model structures (17,28,36,37) and comparison with the off-resonance data of Figure 3A and B. For telomeric structures, the guanine UVRR markers at 1324 and 1482 cm–1 (Fig. 3C) coincide with their off-resonance counterparts and indicate Hoogsteen hydrogen bonding and C2′-endo/syn dG, respectively (Table 1). Conversely, for the non-telomeric structures, the guanine UVRR markers at 1321 and 1484 cm–1 identify C2′-endo/syn dG without Hoogsteen hydrogen bonding. Unique to the telomeric UVRR signatures of Figure 3C is a well-resolved shoulder (∼1603 cm–1) to the intense guanine marker at 1577 cm–1. The corresponding shoulder in non-telomeric structures, which appears at much lower wavenumber (∼1596 cm–1), has been assigned to a guanine ring vibration involving in-plane bending of the C2–N1–H linkages (36). We ascribe the observed shift to the presence of Hoogsteen hydrogen bonding (N1–H···O6) in telomeric sequences in lieu of solvent hydrogen bonding (N1–H···OH2) in non-telomeric sequences.
In contrast to the distinctive UVRR markers of dG conformers, UVRR markers of dT are rather similar for telomeric and non-telomeric DNAs. Bands at 784 and 1182 cm–1 in Figure 3C shift slightly to 787 and 1185 cm–1 in Figure 3D, consistent with the observations in Figure 3A and B. In addition, no UVRR band is observed near 610 cm–1 for the dT4 loop. The shift of the 1363 cm–1 dT marker of Oxy1.5 to 1366 cm–1 in Oxy2 and 1372 cm–1 in T6Oxy2 (cf. Fig. 3C and D) indicates an underlying conformation-sensitive (syn versus anti) contribution from dG rather than a change of dT conformation. For d(TG)8 and dT6(TG)8 the sharp peak observed near 1370–1372 cm–1 reflects the virtual coincidence of a C2′-endo/syn dG marker (∼1371 cm–1) with the C2′-endo/anti dT marker (∼1371 cm–1). On the other hand, in Oxy1.5 the population of C2′-endo/anti dG conformers contributes at a wavenumber lower than 1371 cm–1, which results in the appearance of the composite band peak near 1363 cm–1. Similar overlapping effects explain the data observed for Oxy2 and T6Oxy2. The thymine UVRR marker of highest wavenumber (∼1653 cm–1) is quite distinct from the Raman marker (∼1662 cm–1), consistent with experimental and theoretical studies (34) showing that the UVRR and Raman bands are due to coupled double bond stretching vibrations (involving both C4=O and C5=C6 internal coordinates) rather than to a pure C4=O stretching mode.
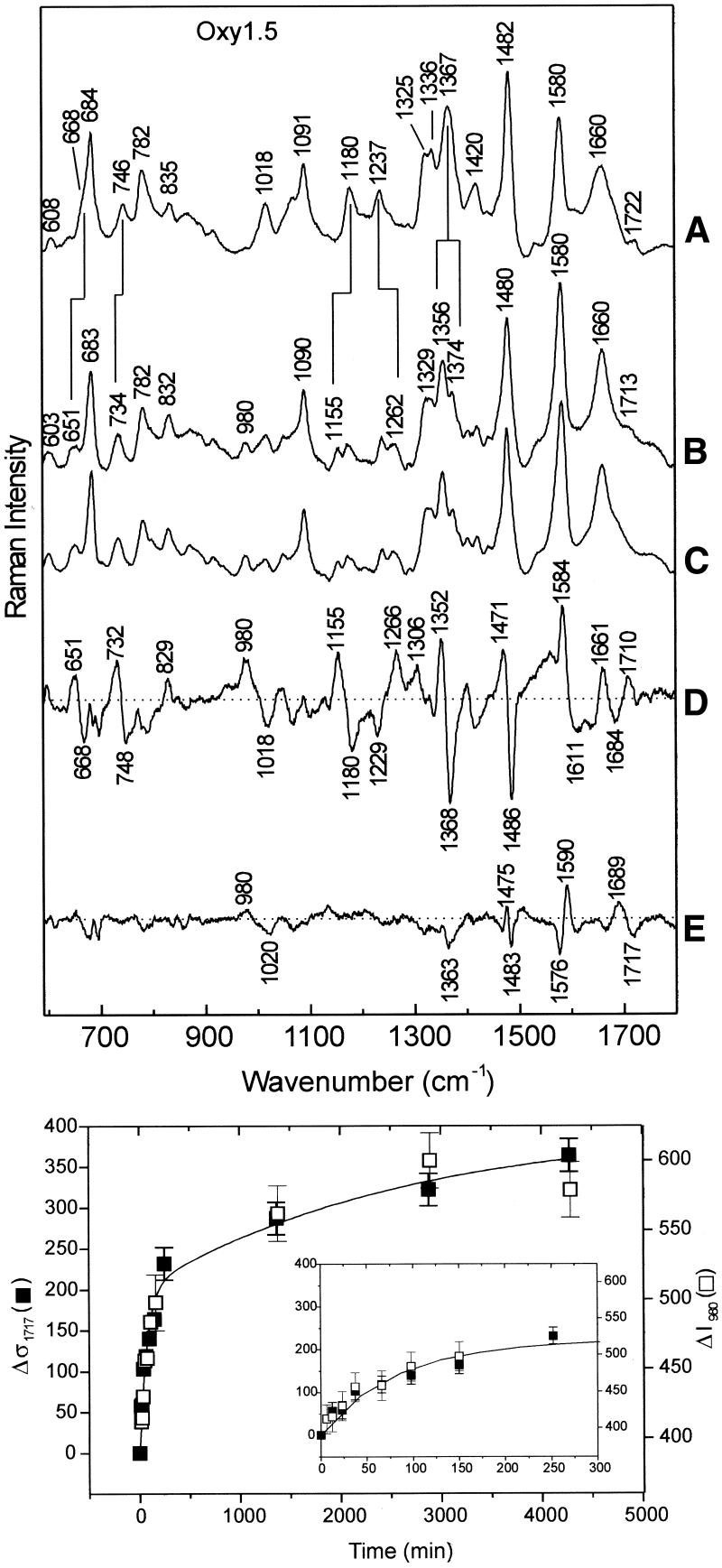
Hydrogen isotope exchange of guanine imino sites in Oxy1.5. Telomeric DNA secondary structures exhibit retardation of guanine amino (N2H2→N2D2) and imino (N1H→N1D) exchanges, which are conveniently measured by time-resolved Raman spectroscopy (15). Representative Oxy1.5 data (Fig. 4, top) illustrate resolution of amino (fast) and imino (slow) exchanges. Difference spectra for sequential exchange processes are shown in Figure 4 traces D (= B – A) and E (= C – B), respectively. Guanine H/D exchanges produce shifts of Raman peaks from ∼1018 to 980 cm–1, 1367 to 1356 cm–1, 1482 to 1480 cm–1 and 1722 to 1713 cm–1 (cf. traces A, B and C of Fig. 4) (15). In the computed difference spectra, the separate effects of amino and imino exchanges are more clearly differentiated. Note, for example, that the imino exchange process results in a peak/trough profile at 1689/1717 cm–1 which provides a basis for quantification of the N1H→N1D exchange rate. The traces in the upper panel of Figure 4 and related time-dependent spectra yield the exchange kinetics profiles shown in the bottom panel of Figure 4.
Figure 4.
(Top) Time-resolved Raman spectra (600–1800 cm–1, 532 nm excitation) of guanine exchanges in Oxy1.5 at 20°C. (A) H2O solution (pH 7.5) prior to the onset of exchange; (B) D2O solution (pD 7.5) following 10 min exchange; (C) D2O solution (pD 7.5) following 4320 min exchange; (D) difference (B) – (A); (E) difference (C) – (B). (Bottom) Guanine imino exchange (N1H→N1D) monitored by the downward shift of the 1717 cm–1 marker band, Δσ1717 (filled square), and intensity increase of the 980 cm–1 marker band, ΔI980 (open square), versus time of exchange (t). The results for both markers suggest at least two exchange regimes, each of which is approximated by an exponential fit (line) to the data points. The inset shows the data points over the period 0 < t < 300 min on an expanded scale (see text).
We interpret the results of Figure 4 (bottom) as indicative of at least two distinct phases of guanine imino exchange in Oxy1.5. The more rapidly exchanging fraction (phase 1), which is complete at time t < 100 min with apparent rate k ≈ 1.39 × 10–2 min–1, represents N1H sites that are only marginally protected by the quadruplex secondary structure. These are likely to reside in the peripheral G quartets (Fig. 1C). The more slowly exchanging fraction (phase 2), which exchanges in the period ∼100 < t < ∼4275 min with apparent rate k ≈ 3.86 × 10–4 min–1, represents more protected N1H sites, presumably those of the internal G quartets (10,15). The phase 2 protons of Oxy1.5 exchange at a rate that is more similar to those of hairpins (10) and extended quadruplexes (15). The guanine H/D exchange data in the bottom panel of Figure 4 may also be fitted to a single exponential function (not shown) with first order rate constant k ≈ 5.9 × 10–3 min–1, which is close to values reported for hairpins (10). The results of Figure 4 further imply that approximately equal populations of protons exchange in phases 1 and 2. We conclude that G quartets of the NMR determined Oxy1.5 quadruplex (12) are not as well protected from H/D exchange as those in the quadruplex structure of Oxy4 (15). This could reflect either the fundamental structural difference between the two quadruplexes (i.e. two interleaved chains in Oxy1.5 versus a single foldback chain in Oxy4) or different exchange mechanisms.
In Oxy1.5, as in previously investigated Oxytricha telomeric DNA structures (10,15), the thymine imino exchange (N3H→N3D) is not time-resolved by the experimental protocol employed here. Nevertheless, thymine imino deuteration is evident from the deuteration shifts of several thymine Raman bands, including 668→651, 746→734, 1180→1155 and 1237→1262 cm–1, as shown in traces A and B of the top panel of Figure 4.
Signatures of Oxytricha telomeric DNA structure transformations
TEBP subunits bind to the Oxytricha telomeric DNA repeat and induce conformational rearrangements (20–22,38). In the case of Oxy1.5, the fold induced in the DNA chain by the nucleotide-binding domains of the α and β subunits has been revealed in the X-ray crystal structure of a ternary complex (25,26). Because no other telomere complex has proven amenable to X-ray structure determination, alternative probes of TEBP-induced DNA structure transformations are required. Raman and UVRR difference signatures have the potential to reveal such DNA transformations (39,40) and provide a reference library for identification of tertiary folds common to other telomeric sequences.
Hairpin. A Raman difference spectrum diagnostic of hairpin formation in T6Oxy2 can be generated by subtracting the spectrum of the single-stranded sequence isomer dT6(TG)8 from the spectrum of T6Oxy2 (Fig. 5A). Because the dT and phosphodiester conformations are very similar in T6Oxy2 and dT6(TG)8, they make no major contribution to this difference spectrum. Figure 5A thus exhibits peaks diagnostic of the dT4 loop, G·G pairs and the mixed dG conformer (syn + anti) populations of T6Oxy2 and troughs diagnostic of the unfolded chain and uniform dG conformer (all syn) populations of dT6(TG)8.
Figure 5.
(A) Raman signature (10°C) of the hairpin fold of T6Oxy2 generated by the difference T6Oxy2 – dT6(TG)8. (B) UVRR signature (10°C) of the hairpin fold of Oxy2 generated by the difference Oxy2 – d(TG)8. (C) UVRR signature of the hairpin fold of Oxy2 generated by the difference Oxy210°C – Oxy270°C. Asterisks in traces (B) and (C) indicate uncompensated intensity of the reference intensity standard (981 cm–1 band of SO42–).
In Figure 5A difference peaks (+) and troughs (–) at 611(+), 744(+), 779(+), 1167(–), 1181(+), 1234(+), 1372(+) and 1665(+) cm–1 are due to dT and those at 685(+), 1311(–), 1337(+), 1477(+), 1491(–), 1570(–), 1583(+), 1597(–), 1615(+), 1694(–) and 1722(+) cm–1 are due to dG. The peaks at 685 and 1337 cm–1 identify the C2′-endo/anti dG conformation of the hairpin, which is only a minor component of single-stranded dT6(TG)8. The trough at 1311 cm–1 is assigned to C2′-endo/syn dG, which is the dominant conformation in the single strand. The C2′-endo/syn and C2′-endo/anti dG conformations alternate in the quadruplex and presumably also in the hairpin. The peak/trough pairs at 1477/1491 and 1583/1570 cm–1 reflect the N7 hydrogen bonds of G·G pairs in the hairpin, which are formed at the expense of solvated N7 sites in the single strand. Similarly, the 1722/ 1694 cm–1 pair reflects different O6 hydrogen bonding environments in the hairpin and single strand. The negative difference band at 1597 cm–1 and relative maximum at 1615 cm–1 may reflect altered hydrogen bonding of imino proton (N1H) sites. Other generally weak difference bands in the 800–1100 cm–1 region are assigned to the expectedly small conformational differences between the deoxyribosyl phosphate backbones of T6Oxy2 and dT6(TG)8. The prominent peak at 1017 cm–1, which originates in large part from a guanine mode involving the exocyclic C2–NH2 group (10), may also reflect different NH2 hydrogen bonding environments in hairpin and strand. It is interesting that the difference spectrum of Figure 5A exhibits many features in common with the previously computed difference spectrum of Oxy2 between 10°C (hairpin) and 90°C (thermally unfolded) (41), although the latter reflects additional spectral perturbations caused by base unstacking at the higher temperature. Subtraction of the Raman spectrum of d(TG)8 from that of Oxy2 yields a difference spectrum (not shown) that is very similar to Figure 5A.
Figure 5B shows the UVRR difference spectrum computed between Oxy2 and d(TG)8, both at 10°C. With the possible exception of a barely distinguishable peak near 1029 cm–1 due mainly to guanine, no peaks or troughs occur in the region 600–1300 cm–1. This is consistent with the low UVRR cross-sections for vibrational modes within this spectral interval (28). However, a rich pattern of difference bands occurs in the 1300–1700 cm–1 interval. The observed UVRR difference spectrum, with peaks and troughs at 1317(–), 1338(+), 1356(+), 1475(+), 1493(–), 1562(–), 1579(+), 1610(+) and 1685(–) cm–1, closely resembles the corresponding off- resonance difference profile (compare the 1300–1700 cm–1 intervals of Figure 5A and B), confirming similar folds. A simplifying advantage of the UVRR signature is the absence of Raman bands from the deoxyribosyl phosphate backbone.
Figure 5C shows the UVRR difference spectrum computed for Oxy2 between 10 and 70°C. This difference spectrum reflects thermal denaturation of the Oxy2 secondary structure. It is very similar to that of Figure 5B, which underscores the attribution of peaks to dG and dT residues of the hairpin fold and troughs to dG and dT in the single strand. This similarity further confirms that the d(TG)8 single strand is a reasonable model for the denatured hairpin of similar base composition. The UVRR difference profile, unlike the off-resonance Raman difference profile, is uncomplicated by Raman contributions from the DNA backbone.
We also computed the UVRR difference spectrum (not shown) for d(TG)8 between 10°C (minuend) and 70°C (subtrahend). As expected, this difference spectrum is feeble in comparison with that of Figure 5C. It consists only of troughs of low intensity in the region 1300–1700 cm–1, which reflect recovery of Raman hypochromism at the higher temperature upon disordering of the d(TG)8 single strand.
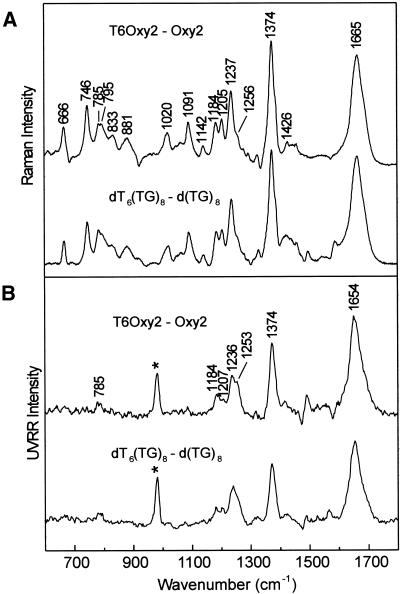
Thymidine leader. The upper trace of Figure 6A shows the Raman difference spectrum obtained by subtracting the spectra of Oxy2 and T6Oxy2, which represents the Raman spectrum of dT6 attached to a hairpin fold (Oxy2). Similarly, the lower trace of Figure 6A represents the Raman spectrum of dT6 attached to an alternating dT/dG single strand. The Raman signature of the thymidine leader is not significantly affected by the DNA fold to which it is attached. This signature is very similar to the Raman spectrum of poly(dT) (30). Figure 6B illustrates the same point for the UVRR spectrum.
Figure 6.
(A) Raman signatures (10°C) of the thymidine 5′ leader generated by the differences T6Oxy2 – Oxy2 (top trace) and dT6(TG)8 – d(TG)8 (bottom trace). (B) UVRR signatures (10°C) of the thymidine 5′ leader generated by the differences T6Oxy2 – Oxy2 (top trace) and dT6(TG)8 – d(TG)8 (bottom trace). Asterisks indicate uncompensated intensity of the reference intensity standard (981 cm–1 band of SO42–).
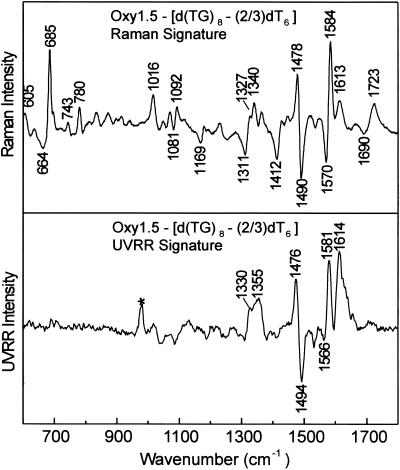
Quadruplex. The top panel of Figure 7 shows the difference spectrum obtained by compensating the Raman spectrum of the Oxy1.5 quadruplex for contributions of dG and dT residues in a single-stranded sequence isomer. The corresponding UVRR difference signature is shown in the bottom panel of Figure 7. Interestingly, the Raman and UVRR signatures of Figure 7 exhibit many similarities to their counterparts in Figure 5. Important distinctions, however, are the relative amplitudes of peaks and troughs in the 1450– 1750 cm–1 interval. For example, the band near 1723 cm–1, which is a specific marker of Hoogsteen O6 interaction, is more intense in Figure 7 than in Figure 5A, consistent with the greater number of Hoogsteen hydrogen bonding interactions per guanine residue of the quadruplex. Another interesting difference between Figure 5A and the top panel of Figure 7 is the wavenumber value of the thymidine loop marker (611 versus 605 cm–1). This may signify different loop geometries in the respective structures. We propose the difference spectra of Figure 7 as the Raman and UVRR signatures diagnostic of the Oxy1.5 quadruplex.
Figure 7.
Raman (top) and UVRR (bottom) signatures of the guanine quartet component of Oxy1.5 generated by compensating corresponding Oxy1.5 spectra for contributions from dT (see text).
DISCUSSION AND CONCLUSIONS
We have reported and interpreted Raman, UVRR and CD spectra of the Oxytricha telomeric DNA sequences Oxy1.5, Oxy2 and T6Oxy2 and of the non-telomeric sequence isomers d(TG)8 and dT6(TG)8. This work extends the database of DNA structures probed by the UVRR method and correlates the UVRR spectra (257 nm excitation) of Oxy1.5, Oxy2, T6Oxy2, d(TG)8 and dT6(TG)8 with corresponding off-resonance Raman spectra (532 nm excitation). The greater sensitivity of UVRR spectroscopy facilitates identification of telomeric DNA folds in solutions that are several orders of magnitude more dilute than those typically employed in off-resonance Raman studies. Raman and UVRR spectra are complemented by CD spectra, confirming and extending previously reported CD results and providing a correlation between CD and vibrational spectra. We find that Oxy1.5 exhibits the positive and negative CD bands near 295 and 265 nm, respectively, proposed as diagnostic of a quadruplex (23). Conversely, Oxy2 and T6Oxy2 contain only the positive bands near 300 and 255 nm, proposed as diagnostic of the hairpin fold (10).
In the 2.5 Å X-ray crystal structure of Oxy1.5, two strands of he dodecanucleotide form a quadruplex having antiparallel strand topology (Fig. 1B; 11). However, in the NMR solution structure of Oxy1.5, the quadruplex comprises two strands interleaved to combine both parallel and antiparallel strand orientations (Fig. 1C). The quadruplex structure of Figure 1C has also been discovered within the lattice of an O.nova TEBP/DNA co-crystal solved at 1.86 Å resolution (25). Because the buffer conditions employed in the present experiments more closely approximate those in the solution NMR and co-crystal X-ray studies, it is reasonable to assume that the Oxy1.5 structure examined here adopts the quadruplex strand topology of Figure 1C rather than that of Figure 1B. This is consistent with the Raman, UVRR and CD results and is also supported by the H/D exchange measurements.
Raman marker bands diagnostic of specific dG conformations and interactions in telomeric DNA structures have been reported previously (8,42). The present work shows that comparable dG markers occur within the 1300–1700 cm–1 interval of UVRR spectra. UVRR spectra of telomeric DNA sequences also exhibit a novel dG marker at 1600 ± 5 cm–1. No comparable band can be resolved in off-resonance Raman spectra, owing to an apparently low off-resonance Raman scattering cross-section and possible obscuration by intense neighboring bands of the guanine (1575 cm–1) and thymine rings (1660 cm–1). In accordance with previous work (8,15,36), we assign the 1600 cm–1 marker to a vibration involving the guanine N1H imino group, and note that it appears at a higher wavenumber value in natively folded structures (∼1605 cm–1, Fig. 3A) than in single-stranded non-telomeric DNA (∼1597 cm–1, Fig. 3B). We propose this UVRR marker as a means of distinguishing the N1H···O6 hydrogen bond between two guanines (e.g. in Hoogsteen G·G pairs or G quartets) from the N1H···OH2 hydrogen bond between guanine and solvent.
It is interesting to note that at DNA concentrations (10– 20 µM) employed for UVRR spectroscopy a parallel-stranded quadruplex is not induced by prolonged heating of either Oxy1.5, Oxy2 or T6Oxy2, irrespective of salt concentration. Conversely, such quadruplexes are readily formed at the much higher DNA concentrations (10–100 mM) usually employed in off-resonance Raman studies (9). Thus, UVRR provides a decisive advantage over off-resonance Raman spectroscopy for investigating antiparallel quadruplexes without the complication of parallel quadruplex formation.
The H/D exchange of guanine imino (N1H→N1D) sites in Oxy1.5 is biphasic, with rates of 1.39 × 10–2 and 3.86 × 10–4 min–1. The overall exchange process can also be described in terms of a single rate constant of 5.9 × 10–3 min–1, which is intermediate between values reported for Oxy2 (4.6 × 10–3) and T6Oxy2 (9.6 × 10–3 min–1) (10). Much slower rates (∼10–6 min–1) are exhibited by the foldback (antiparallel) and extended (parallel) quadruplex structures of Oxy4 (15). Therefore, although the Oxy1.5 quadruplex contains G quartets of high stability, it does not exhibit the same degree of H/D exchange protection observed for Oxy4 quadruplexes. One plausible explanation for this finding is that guanine exchange depends not simply on the nature of interbase hydrogen bonding but also on DNA strand topology. For example, in both the solution NMR and co-crystal X-ray structures of the Oxy1.5 quadruplex (12,25), the strand topology results in a very wide groove (10 Å) that is not present in the Oxy4 quadruplex structures (43,44). The wide groove of the Oxy1.5 structure may facilitate solvent penetration for more rapid guanine proton exchanges. This is analogous to the case of B-DNA. It is also possible that H/D exchanges may proceed by different mechanisms in Oxy1.5 and Oxy4 structures.
The distinctive structure and H/D exchange kinetics observed for Oxy1.5 vis-à-vis Oxy2 and T6Oxy2 demonstrate that the single-stranded 5′ leader sequence (the so-called 3′ overhang) plays an important role in telomeric DNA structure and dynamics. The single-stranded dT4 sequence of Oxy2 (or dT10 sequence of T6Oxy2) may function to stabilize hairpin formation in the d(G4T4G4) sequence to which it is linked. Quadruplex formation, whether by a mechanism of strand association in the case of Oxy1.5 or folding in the case of Oxy4, is presumably disfavored if a stable hairpin is formed initially. The single-stranded overhang may thus provide a natural barrier to quadruplex associations.
In Table 2 we list the key Raman and UVRR markers identified in this and previous work. This tabulation includes many new Raman markers not previously recognized as diagnostic of dG and dT conformations and of base hydrogen bonding interactions in telomeric DNA folds. The data of Table 2 are considered important for Raman and UVRR classifications of telomeric DNA structures, for the design of future experiments to probe structures of Oxytricha telomeric repeats in complexes with TEBP subunits and for the use of Raman and UVRR probes in investigating other protein–DNA interactions.
Table 2. Raman and UVRR markers of Oxytricha telomeric DNA and related foldsa.
| Marker (cm–1) | Assignment | Diagnostic telomeric DNA foldb |
|---|---|---|
| 610 ± 2 | dT4 loop | anti-|| quadruplex or hairpin |
| 674 ± 3 | C2′-endo/syn dG | anti-|| quadruplex or hairpin (50% dG) |
| 682 ± 2 | C2′-endo/anti dG | anti-|| quadruplex or hairpin (50% dG) |
| || quadruplex (100% dG) | ||
| 1324 ± 2 | C2′-endo/syn dG | anti-|| quadruplex or hairpin (50% dG) |
| 1333 ± 3 | C2′-endo/anti dG | anti-|| quadruplex or hairpin (50% dG) |
| || quadruplex (100% dG) | ||
| 1361 ± 2c | C2′-endo/anti dG | anti-|| quadruplex or hairpin (50% dG) |
| || quadruplex (100% dG) | ||
| 1481 ± 2 | dG N7 strong Hoogsteen H-bond | anti-|| quadruplex or || quadruplex |
| ∼1486 | dG N7 weak H-bond to H2O | |
| 1579 ± 1 | dG N2H interbase H-bond | anti-|| quadruplex, || quadruplex or hairpin |
| ∼1574 | dG N2H H-bond to H2O | |
| 1603 ± 2 | dG N1H interbase H-bond | anti-|| quadruplex, || quadruplex or hairpin |
| ∼1595 | dG N1H H-bond to H2O | |
| 1722 ± 2 | dG O6 interbase H-bond | anti-|| quadruplex, || quadruplex or hairpin |
| ∼1686 | dG O6 H-bond to H2O |
bData apply to Oxy1.5, Oxy2 and T6Oxy2. Symbols: ||, parallel; anti-||, antiparallel. Where appropriate, percentage residues affected are given in parentheses.
cBand center depends on G:T content due to overlap with dT marker, 1370 cm–1.
Acknowledgments
ACKNOWLEDGEMENTS
Part LXXX in the series Raman Spectral Studies of Nucleic Acids. Support of this research by grant GM54378 from the National Institutes of Health is gratefully acknowledged.
REFERENCES
- 1.McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Annu. Rev. Genet., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- 2.Zakian V.A. (1995) Telomeres: beginning to understand the end. Science, 270, 1601–1607. [DOI] [PubMed] [Google Scholar]
- 3.Zakian V.A. (1996) Structure, function and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet., 30, 141–172. [DOI] [PubMed] [Google Scholar]
- 4.McElligott R. and Wellinger,R.J. (1997) The terminal DNA structure of mammalian chromosomes. EMBO J., 16, 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann P. and Cech,T.R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science, 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- 6.de Lange T. (2001) Cell biology. Telomere capping–one strand fits all. Science, 292, 1075–1076. [DOI] [PubMed] [Google Scholar]
- 7.Williamson J.R. (1994) G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 8.Miura T. and Thomas,G.J.,Jr (1994) Structural polymorphism of telomere DNA: interquadruplex and duplex-quadruplex conversions probed by Raman spectroscopy. Biochemistry, 33, 7848–7856. [DOI] [PubMed] [Google Scholar]
- 9.Miura T., Benevides,J.M. and Thomas,G.J.,Jr (1995) A phase diagram for sodium and potassium ion control of polymorphism in telomeric DNA. J. Mol. Biol., 248, 233–238. [DOI] [PubMed] [Google Scholar]
- 10.Laporte L. and Thomas,G.J.,Jr (1998) A hairpin conformation for the 3′ overhang of Oxytricha nova telomeric DNA. J. Mol. Biol., 281, 261–270. [DOI] [PubMed] [Google Scholar]
- 11.Kang C., Zhang,X., Ratliff,R., Moyzis,R. and Rich,A. (1992) Crystal structure of four-stranded Oxytricha telomeric DNA. Nature, 356, 126–131. [DOI] [PubMed] [Google Scholar]
- 12.Smith F.W. and Feigon,J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature, 356, 164–168. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura Y., Tsuboi,M., Nakano,T., Higuchi,S., Sato,T., Shida,T., Uesugi,S., Ohtsuka,E. and Ikehara,M. (1983) Raman diagnosis of nucleic acid structure: sugar-puckering and glycosidic conformation in the guanosine moiety. Nucleic Acids Res., 11, 1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benevides J.M., Wang,A.H.J., van der Marel,G.A., van Boom,J.H. and Thomas,G.J.,Jr (1989) Effect of the GT mismatch on backbone and sugar conformations of Z-DNA and B-DNA: analysis by Raman spectroscopy of crystal and solution structures of d(CGCGTG) and d(CGCGCG). Biochemistry, 28, 304–310. [DOI] [PubMed] [Google Scholar]
- 15.Miura T. and Thomas,G.J.,Jr (1995) Structure and dynamics of interstrand guanine association in quadruplex telomeric DNA. Biochemistry, 34, 9645–9654. [DOI] [PubMed] [Google Scholar]
- 16.Wen Z.Q., Overman,S.A. and Thomas,G.J.,Jr (1997) Structure and interactions of the single-stranded DNA genome of filamentous virus fd: investigation by ultraviolet resonance Raman spectroscopy. Biochemistry, 36, 7810–7820. [DOI] [PubMed] [Google Scholar]
- 17.Wen Z.Q., Armstrong,A. and Thomas,G.J.,Jr (1999) Demonstration by ultraviolet resonance Raman spectroscopy of differences in DNA organization and interactions in filamentous viruses Pf1 and fd. Biochemistry, 38, 3148–3156. [DOI] [PubMed] [Google Scholar]
- 18.Wen Z.Q. and Thomas,G.J.,Jr (2000) Ultraviolet-resonance Raman spectroscopy of the filamentous virus Pf3: interactions of Trp 38 specific to the assembled virion subunit. Biochemistry, 39, 146–152. [DOI] [PubMed] [Google Scholar]
- 19.Wen Z.Q., Overman,S.A., Bondre,P. and Thomas,G.J.,Jr (2001) Structure and organization of bacteriophage Pf3 probed by Raman and ultraviolet resonance Raman spectroscopy. Biochemistry, 40, 449–458. [DOI] [PubMed] [Google Scholar]
- 20.Fang G. and Cech,T.R. (1993) The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell, 74, 875–885. [DOI] [PubMed] [Google Scholar]
- 21.Fang G. and Cech,T.R. (1993) Oxytricha telomere-binding protein: DNA-dependent dimerization of the alpha and beta subunits. Proc. Natl Acad. Sci. USA, 90, 6056–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang G., Gray,J.T. and Cech,T.R. (1993) Oxytricha telomere-binding protein: separable DNA-binding and dimerization domains of the alpha-subunit. Genes Dev., 7, 870–882. [DOI] [PubMed] [Google Scholar]
- 23.Balagurumoorthy P., Brahmachari,S.K., Mohanty,D., Bansal,M. and Sasisekharan,V. (1992) Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res., 20, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultze P., Hud,N.V., Smith,F.W. and Feigon,J. (1999) The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4)). Nucleic Acids Res., 27, 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath M.P. and Schultz,S.C. (2001) DNA G-quartets in a 1.86 Å resolution structure of an Oxytricha nova telomeric protein-DNA complex. J. Mol. Biol., 310, 367–377. [DOI] [PubMed] [Google Scholar]
- 26.Horvath M.P., Schweiker,V.L., Bevilacqua,J.M., Ruggles,J.A. and Schultz,S.C. (1998) Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- 27.Russell M.P., Vohník,S. and Thomas,G.J.,Jr (1995) Design and performance of an ultraviolet resonance Raman spectrometer for proteins and nucleic acids. Biophys. J., 68, 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen Z.Q. and Thomas,G.J.,Jr (1998) Ultraviolet resonance Raman spectroscopy of DNA and protein constituents of viruses: assignments and cross sections for excitations at 257, 244, 238 and 229 nm. Biopolymers, 45, 247–256. [DOI] [PubMed] [Google Scholar]
- 29.Gray D.M., Ratliff,R.L. and Vaughan,M.R. (1992) Circular dichroism spectroscopy of DNA. Methods Enzymol., 211, 389–406. [DOI] [PubMed] [Google Scholar]
- 30.Movileanu L., Benevides,J.M. and Thomas,G.J.,Jr (1999) Temperature dependence of the Raman spectrum of DNA. I. Raman signatures of premelting and melting transitions of poly(dA-dT)·poly(dA-dT). J. Raman Spectrosc., 30, 637–649. [DOI] [PubMed] [Google Scholar]
- 31.Erfurth S.C., Kiser,E.J. and Peticolas,W.L. (1972) Determination of the backbone structure of nucleic acids and nucleic acid oligomers by laser Raman scattering. Proc. Natl Acad. Sci. USA, 69, 938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benevides J.M., Wang,A.H.J., van der Marel,G.A., van Boom,J.H., Rich,A. and Thomas,G.J.,Jr (1984) The Raman spectra of left-handed DNA oligomers incorporating AT base pairs. Nucleic Acids Res., 12, 5913–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benevides J.M., Wang,A.H.J., Rich,A., Kyogoku,Y., van der Marel,G.A., van Boom,J.H. and Thomas,G.J.,Jr (1986) The Raman spectra of single crystals of r(GCG)d(CGC) and d(CCCCGGGG) as models for A-DNA, their structure transitions in aqueous solution and comparison with double helical poly(dG)·poly(dC). Biochemistry, 25, 41–50. [DOI] [PubMed] [Google Scholar]
- 34.Tsuboi M., Komatsu,K., Hoshi,J., Kawashima,E., Sekine,T., Ishido,Y., Russell,M.P., Benevides,J.M. and Thomas,G.J.,Jr (1997) Raman and infrared spectra of (2′S)-[2′-2H] thymidine: vibrational coupling between deoxyribosyl and thymine moieties and structural implications. J. Am. Chem. Soc., 119, 2025–2032. [Google Scholar]
- 35.Nishimura Y., Tsuboi,M., Sato,T. and Aoki,K. (1986) Conformation-sensitive Raman lines of mononucleotides and their use in a structure analysis of polynucleotides: guanine and cytosine nucleotides. J. Mol. Struct., 146, 123–153. [Google Scholar]
- 36.Toyama A., Hanada,N., Ono,J., Yoshimitsu,E. and Takeuchi,H. (1999) Assignments of guanosine UV resonance Raman bands on the basis of 13C, 15N and 18O substitution effects. J. Raman Spectrosc., 30, 623–630. [Google Scholar]
- 37.Thomas G.J. Jr and Tsuboi,M. (1993) Raman spectroscopy of nucleic acids and their complexes. In Bush,C.A. (ed.), Advances in Biophysical Chemistry, Vol. 3. JAI Press, Greenwich, CT, pp. 1–70.
- 38.Fang G. and Cech,T.R. (1993) Characterization of a G-quartet formation reaction promoted by the β-subunit of the Oxytricha telomere-binding protein. Biochemistry, 32, 11646–11657. [DOI] [PubMed] [Google Scholar]
- 39.Benevides J.M., Chan,G., Lu,X.J., Olson,W.K., Weiss,M.A. and Thomas,G.J.,Jr (2000) Protein-directed DNA structure. I. Raman spectroscopy of a high-mobility-group box with application to human sex reversal. Biochemistry, 39, 537–547. [DOI] [PubMed] [Google Scholar]
- 40.Benevides J.M., Li,T., Lu,X.J., Srinivasan,A.R., Olson,W.K., Weiss,M.A. and Thomas,G.J.,Jr (2000) Protein-directed DNA structure. II. Raman spectroscopy of a leucine zipper bZIP complex. Biochemistry, 39, 548–556. [DOI] [PubMed] [Google Scholar]
- 41.Laporte L., Benevides,J.M. and Thomas,G.J.,Jr (1999) Molecular mechanism of DNA recognition by the α subunit of the Oxytricha telomere binding protein. Biochemistry, 38, 582–588. [DOI] [PubMed] [Google Scholar]
- 42.Miura T. and Thomas,G.J.,Jr (1995) Optical and vibrational spectroscopic methods. In Glasel,J.A. and Deutscher,M.P. (eds), Introduction to Biophysical Methods for Protein and Nucleic Acid Research. Academic Press, New York, NY, pp. 261–315.
- 43.Laughlan G., Murchie,A.I., Norman,D.G., Moore,M.H., Moody,P.C., Lilley,D.M. and Luisi,B. (1994) The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science, 265, 520–524. [DOI] [PubMed] [Google Scholar]
- 44.Aboul-ela F., Murchie,A.I., Norman,D.G. and Lilley,D.M. (1994) Solution structure of a parallel-stranded tetraplex formed by d(TG4T) in the presence of sodium ions by nuclear magnetic resonance spectroscopy. J. Mol. Biol., 243, 458–471. [DOI] [PubMed] [Google Scholar]
- 45.Benevides J.M., Wang,A.H.J., van der Marel,G.A., van Boom,J.H. and Thomas,G.J.,Jr (1988) Crystal and solution structures of the B-DNA dodecamer d(CGCAAATTTGCG) probed by Raman spectroscopy: heterogeneity in the crystal structure does not persist in the solution structure. Biochemistry, 27, 931–938. [DOI] [PubMed] [Google Scholar]