Abstract
We have identified a novel component, Helicase89B, that is required for the inducible antimicrobial response in Drosophila larvae by means of a P-element insertional genetic screen. Helicase89B belongs to the Mot1p/BTAF1 subfamily of SNF2-like ATPases. This subfamily can interact with TATA-binding proteins, but whether the interaction leads to gene activation or repression is being debated. We found that Helicase89B is required for the inducible expression of antimicrobial peptide genes but not for the inducible expression of heat-shock genes. The antimicrobial peptide genes are activated by the Toll and immune deficiency (IMD) signalling pathways. Genetic experiments show that Helicase89B acts downstream of DIF and Relish, the two nuclear factor-κB (NF-κB)-related transcription factors that mediate Toll- and IMD-stimulated antimicrobial response. Thus, Helicase89B positively regulates gene expression during innate immune response and may act as a link between NF-κB-related transcription factors and the basal transcription machinery.
Keywords: DIF, Helicase89B, innate immunity, Mot1p, Toll
Introduction
The innate immune response serves as the first line of defence against microbial infection in insects and mammals (Medzhitov & Biron, 2003). In Drosophila, microbial infection induces the expression of more than 20 antimicrobial peptides, which are essential for the insect to survive infection. The induction of these peptides depends on the Toll and immune deficiency (IMD) signalling pathways (Hoffmann, 2003; Brennan & Anderson, 2004).
Toll is a transmembrane receptor that relays the signals of fungal and Gram-positive bacterial infections to the cytoplasm (Hoffmann, 2003; Brennan & Anderson, 2004). Through the adaptor proteins MyD88 and Tube and the kinase Pelle, Toll signalling triggers the nuclear transport of DIF and Dorsal, two nuclear factor-κB (NF-κB)-related transcription factors. The IMD pathway mediates the response to Gram-negative bacterial infection (Hoffmann, 2003; Brennan & Anderson, 2004). Recognition of Gram-negative bacterial components by peptidoglycan-recognition proteins, such as PGRP-LC, stimulates the adaptor protein IMD. IMD then relays the signal to the kinase TAK-1 and the IκB kinase-like complex to activate the third NF-κB-related protein Relish. The components of the Drosophila Toll and IMD pathways are highly homologous to those of the mammalian Toll-like receptor (TLR), interleukin-1 receptor (IL-1R) and tumour necrosis factor receptor signalling pathways.
In this study, we show that Helicase89B is a novel component that is required for the inducible expression of antimicrobial peptide genes. Helicase89B codes for a homologue of Mot1p and BTAF1, which are ATPases/helicases that interact with TATA-binding protein (TBP) and can activate or repress transcription (Pereira et al, 2003). Our results indicate that Helicase89B acts downstream of NF-κB-related proteins to mediate the activation of antimicrobial peptide genes.
Results
P-element screen for immunity gene expression mutants
To identify genes that are essential for the innate immune response, we carried out a P-element insertional genetic screen in Drosophila larvae. Approximately 750 P-element lines were set up and approximately 200 lines gave sufficient homozygous larvae for assaying the antimicrobial peptide gene expression induced by septic injury. Among the strains assayed, P1732 (BL11732), obtained from the Drosophila Stock Center, showed consistent defects in the induction of five representative genes: Diptericin, Defensin, Cecropin, Attacin and Drosomycin. In wild-type larvae, all of these genes can be induced to express higher levels of messenger RNA in 3–6 h after septic injury (Fig 1). In contrast, the P1732 strain showed much lower expression levels of the antimicrobial peptide genes after septic injury. Among the five genes tested, Drosomycin induction seemed to have a less significant defect in the mutant. Overall, our results indicate that we have obtained a mutant strain with defects in the inducible expression of five antimicrobial peptide genes. The induction of Drosomycin is primarily under the control of the Toll pathway and the induction of the other antimicrobial peptide genes is regulated primarily by the IMD pathway (Hoffmann, 2003; Brennan & Anderson, 2004). Thus, the normal function of the gene mutated in P1732 may be involved in both the Toll and the IMD pathways.
Figure 1.
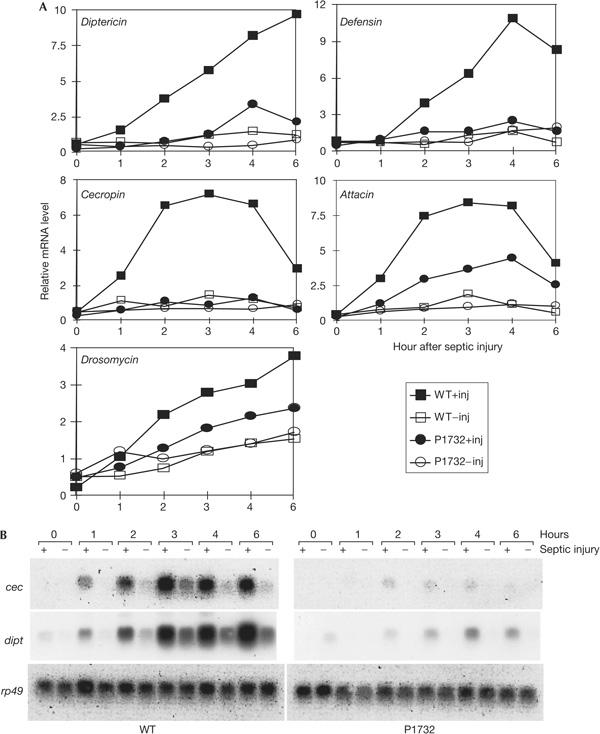
Immune defects in a P-insertion strain. (A) Plots of expression of five antimicrobial peptide genes after septic injury (+inj) of larvae. The signals on the northern blots were quantified and then normalized according to the signal of the ribosomal protein 49 (rp49) RNA. The results were plotted as relative messenger RNA levels using the lowest expressing sample set as 1. Expression was measured at the indicated time after septic injury. Uninjected animals were used for the time zero samples. (B) A set of representative autoradiographs of the northern blots for the Cecropin (cec), Diptericin (dipt) and rp49 genes in the wild-type (WT) and P1732 strain.
Helicase89B is required for innate immune response
The P element of the P1732 strain was mapped near or in the gene Helicase89B by the Drosophila genome project (http://flybase.bio.indiana.edu). The genomic organization of the Helicase89B region was previously reported, but the 5′ end of the gene remained ambiguous (Crosby et al, 1999). Thus, we mapped the 5′ end by the rapid amplification of cloned ends (RACE) method using mRNA isolated from wild-type Drosophila. On the basis of the sequences of the RACE clone and two other partial complementary DNA clones, we reconstructed the transcription unit, as shown in Fig 2A. The Helicase89B transcription unit is approximately 330 base pairs (bp) from the neighbouring gene moira. The two genes are transcribed divergently. By comparing the reconstructed transcription unit and the mapping data, we can place the P element of the P1732 strain in the first intron of Helicase89B, not in the promoter region as described in the database.
Figure 2.
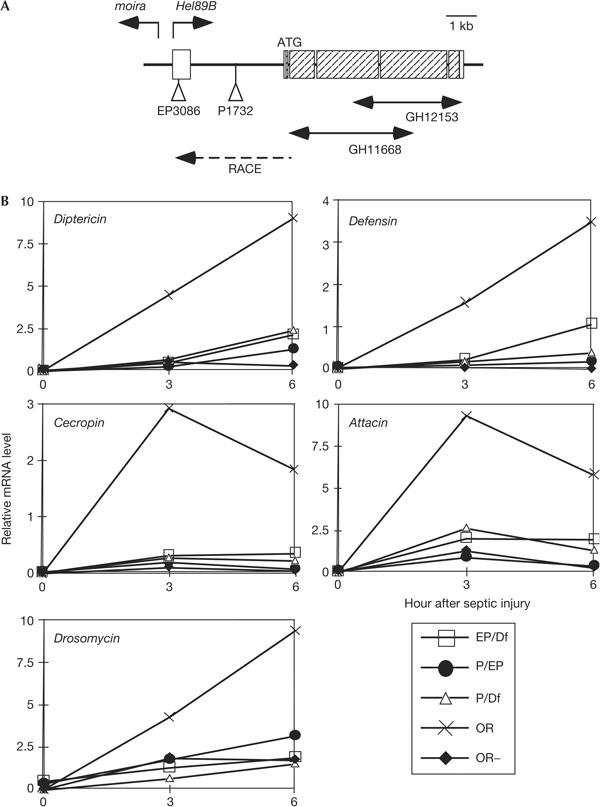
Analysis of immunity gene expression in Helicase89B mutant strains. (A) Genomic organization around the Helicase89B (Hel89B) locus. The boxes represent exons and the hatched regions represent protein-coding regions. The three complementary DNA clones that were used to regenerate the whole coding region are indicated below the gene locus. The two P-element insertion sites are indicated as EP3086 and P1732. Helicase89B and the neighbouring gene moira are transcribed divergently. RACE, rapid amplification of cloned ends. (B) Northern blot analysis of transheterozygotes. Hybridization signals on northern blots were quantified and normalized with ribosomal protein 49 (rp49) expression and are plotted as shown. P represents P1732, EP represents EP3086 and Df represents Df(3R)sbd105. The plots represent the results from larvae that had septic injury for 3 h, except the OR− samples, which were wild-type larvae that were not injected.
The deficiency strain Df(3R)sbd105 uncovers the 89B region, and the EP3086 strain is an independent P-element strain from the Drosophila Stock Center. On the basis of our reconstructed map, the P element of EP3086 is inserted in the first exon approximately 100 bp downstream of the 5′ end of the Helicase89B transcription unit. We examined larvae that were transheterozygous for P1732 and the genetic mutants Df(3R)sbd105 and EP3086 (Fig 2B). Northern blot analysis of the transheterozygotes showed that the induction of all five antimicrobial peptide genes after septic injury was significantly lower in the mutant than in wild-type larvae.
Helicase89B and the neighbouring gene moira, which encodes an SWI3 homologue of chromatin remodelling protein (Crosby et al, 1999), share a common and relatively short 5′ flanking sequence. Thus, we examined the expression of these two genes by northern blot analysis of the insertional mutant. The sizes of the transcripts are 7.5 and 4.5 kb for Helicase89B and moira, respectively (Crosby et al, 1999). The expression of Helicase89B in the P1732 homozygous larvae was reduced to an undetectable level (Fig 3A, left panel), whereas the expression of moira in the homozygous larvae seemed to be slightly higher (Fig 3A, right panel), possibly owing to the influence of an exogenous DNA insertion in the neighbourhood. Nonetheless, the result clearly shows that the P-element insertion reduces Helicase89B mRNA but not moira mRNA expression.
Figure 3.
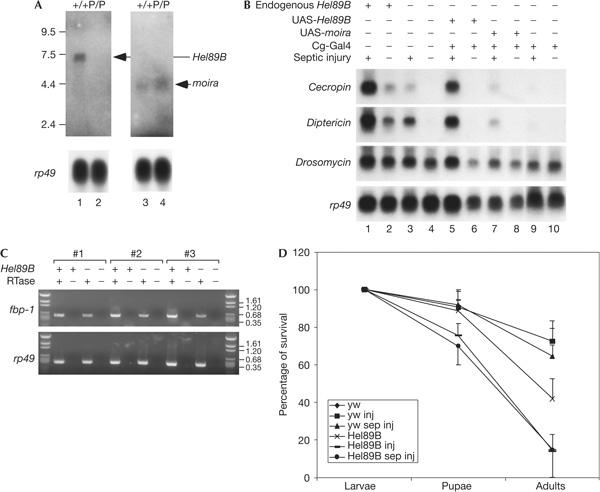
Loss of Helicase89B causes the immune deficiency phenotype. (A) Messenger RNA expression of Helicase89B (Hel89B) and moira in wild-type (+/+) and homozygous P1732 (P/P) larvae. The arrows indicate the hybridization signals corresponding to the expected sizes of the mRNA of the two genes. The ribosomal protein 49 (rp49) blots shown had a much shorter time of exposure than those shown for Helicase89B and moira. (B) Expression of antimicrobial peptide genes in the P1732 mutant and the transgenic rescued strains. Lanes 1 and 2 are wild-type animals, and the endogenous Helicase89B gene is indicated as +. All other lanes are homozygous P1732, and the endogenous Helicase89B gene is indicated as −. Each strain was tested by septic injury for 3 h, with uninduced larvae assayed in parallel for comparison. (C) Expression of fat body protein-1 (fbp-1) in larvae. Reverse transcription–PCR was carried out using total RNA as template, and the ethidium bromide-stained gels are shown. Three independent RNA preparations (#1–3) were used. Reverse transcriptase (RTase) was added or omitted (+ or −) in the complementary DNA synthesis step, as indicated. (D) Control larvae (y w) and P1732 homozygous mutant larvae were subjected to injury or septic injury for 3 h and were assayed for survival. Three independent experiments were carried out and the average was plotted, with standard deviation shown as error bars. The total number of animals scored in the three experiments was 116, 105, 130,144, 137 and 126, respectively, for the six samples listed in the label box.
A genetic rescue experiment was carried out to examine whether expression of Helicase89B is sufficient to restore the immune defect. Transgenic flies containing Helicase89B and moira cDNA in the pUAST vector were crossed with the P1732 mutant. Expression of the transgenes was driven by Cg-Gal4, which directs the expression in the larval fat body and in the haemocytes, which are the main immune tissues (Bettencourt et al, 2004). The inducible expression of Cecropin, Diptericin and Drosomycin was then assayed in these larvae (Fig 3B). P1732 mutant larvae with the Cg-Gal4 and UAS-Helicase89B transgenes showed increased levels of expression of all three genes after septic injury (Fig 3B, lane 5). Other control strains, such as Cg-Gal4/UAS-moira (Fig 3B, lane 7) or Cg-Gal4 alone (Fig 3B, lane 9), did not show a comparable level of rescue. These results indicate that the antimicrobial peptide genes become inducible again in the mutant larvae when transgenic Helicase89B is expressed. Thus, the results from the phenotypic analysis of the transheterozygotes, mRNA expression in the P1732 mutant strain and the genetic rescue indicate that Helicase89B participates in the inducible expression of antimicrobial peptide genes during innate immune response.
Drosophila larvae undergo moulting and metamorphosis in a relatively short time. The developmental stage may affect immune competence (Tingvall et al, 2001; Ligoxygakis et al, 2002) and for this reason, the third-instar larvae for our experiments were selected on the basis of size and observable maturity, not on the basis of the hour after the egg was laid. To further assess maturity, we used reverse transcription–PCR to assay for the expression of the fat body protein-1 (fbp-1) gene, which is a target for the master regulatory hormone, ecdysone. Although there was an approximately twofold reduction, the substantial expression of fbp-1 could consistently be detected in the Helicase89B mutant larvae (Fig 3C). This phenotype is different from that of the previously characterized 18wheeler mutant larvae, which have low or no detectable fbp-1-lacZ expression in the larval fat body (Ligoxygakis et al, 2002). Similarly to other mutant analyses, we cannot completely separate fat body developmental defect from immune response defect. Nonetheless, the substantial expression of fbp-1 mRNA supports the idea that the mutant larvae that we used should be competent to respond to microbial challenge.
A survival assay was carried out to further characterize the Helicase89B mutant. Homozygous P1732 larvae or control larvae were subject to injury or septic injury. Wild-type larvae showed strong viability (more than 90%) throughout the larval and pupal stages after septic injury. Uninjected P1732 larvae showed a slightly reduced viability before pupariation but only 42% of the mutant larvae hatched as adults, which suggests a semi-lethal phenotype. After injury or septic injury, the survival rate of mutant larvae dropped from 89% to 73% before pupariation and dropped from 42% to 14% before adulthood. The results indicate that the Helicase89B mutant has increased susceptibility to injury and septic injury, but we have not been able to link susceptibility directly to infection.
Helicase89B acts downstream of DIF and Relish
Helicase89B has homology to Mot1p and BTAF1, which act as TAFs for general or inducible gene expression, including heat-shock response in yeast (Geisberg & Struhl, 2004). However, the heatshock-induced expression of hsp83, hsp70b and hsp26 was normal in mutant Helicase89B larvae (Fig 4A). The heatshock genes were also inducible in the fat bodies of Helicase89B mutant larvae (Fig 4B), which indicates that the mutant larvae are able to respond to at least one stress signal. We also examined whether the antimicrobial peptide gene expression is affected in adult flies. The three antimicrobial peptide genes tested, that is, Cecropin, Diptericin and Drosomycin, showed substantial induction after septic injury (Fig 4C). These results show that Helicase89B is important for the induction of antimicrobial peptide genes in larvae, but not for the induction of these genes in adults and for the induction of heatshock genes in larvae.
Figure 4.
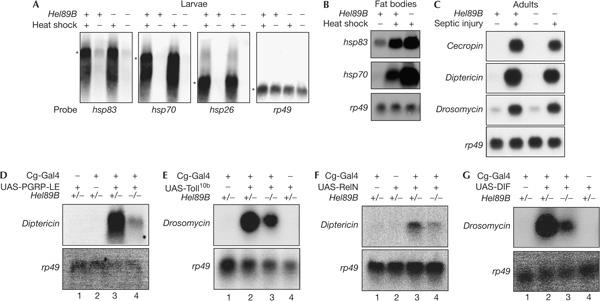
Requirement of Helicase89B in the Toll and immune deficiency pathways. (A) Autoradiographs of northern blots for the heat-shock genes in whole larvae. The messenger RNA sizes of the three heat-shock genes and ribosomal protein 49 (rp49) gene are 2.6, 2.5, 1.1 and 0.6 kb, respectively, as indicated by the asterisk. The P1732 homozygous mutant was used. (B) Autoradiographs of northern blots for heat-shock genes in the larval fat body. (C) Autoradiographs of northern blots for Cecropin, Diptericin and Drosomycin expression in wild-type and P1732 mutant adult flies after septic injury for 3 h. (D–G) Genetic interaction of Helicase89B with the Toll and immune deficiency (IMD) pathways. Overexpression of PGRP-LE (a peptidoglycan-recognition protein), Toll10b, RelN and DIF in larvae was achieved by the Cg-Gal4-UAS system. The IMD pathway components PGRP-LE and RelN can induce Diptericin expression without bacterial challenge, but the effect is suppressed in the Helicase89B (P1732) mutant. The expression of Drosomycin mRNA is induced by the expression of Toll10b and DIF, but in the Helicase89B mutant background, the expression is lower.
To gain a further insight into the mechanism of Helicase89B function, we carried out a series of genetic suppression experiments. Expression of the peptidoglycan-recognition proteins, PGRP-LC or PGRP-LE, can stimulate the IMD pathway and increase the expression of antimicrobial peptide genes in the absence of septic injury (Choe et al, 2002; Gottar et al, 2002; Takehana et al, 2002). Thus, we crossed the Cg-Gal4 and UAS-PGRP-LE strains with the Helicase89B mutant background. PGRP-LE overexpression induced Diptericin expression, but the loss of Helicase89B suppressed this effect (Fig 4D). This result shows that Helicase89B is a downstream component of the IMD pathway.
The well-characterized Toll10b is a ligand-independent, constitutively active allele of the receptor Toll. The expression of Toll10b can increase the expression of Drosomycin in adult flies in the absence of infection (Hu et al, 2004). Thus, we used the Gal4-UAS system to express Toll10b in wild-type and Helicase89B mutant larvae. The expression of Toll10b in wild-type larvae also led to highly increased expression of Drosomycin (Fig 4D). This Toll10b-induced Drosomycin expression was partly suppressed in the Helicase89B mutant background (Fig 4D). This result shows that Helicase89B is also a downstream component of the Toll pathway.
Because of the homology to BTAF1/Mot1p, Helicase89B may mediate the activation of antimicrobial peptide genes by the NF-κB-related transcriptional activators in the Toll and IMD pathways. Thus, we generated a truncated Relish construct (RelN) that resembles the naturally cleaved Relish during immune response. Transgenic expression of RelN in the larval fat body could activate Diptericin (Fig 4E), but not Drosomycin (data not shown), in the absence of infection. The level of Diptericin activation by RelN is weaker than that by septic injury (compare lanes 3 of panels F and D of Fig 4, respectively). Nonetheless, this constitutive activation of Diptericin by RelN was suppressed in the Helicase89B mutant (Fig 4E), which suggests that Helicase89B functions downstream of Relish in the IMD pathway.
DIF is a key transcription factor of the Toll pathway. Overexpression of DIF by the Gal4-UAS system led to constitutive activation of Drosomycin (Fig 4F). The effect of DIF on Drosomycin expression was reduced in Helicase89B mutant larvae (Fig 4F), which suggests that Helicase89B acts downstream of DIF.
Discussion
Our genetic experiments suggest that Helicase89B mediates the activation of gene expression in both the Toll and IMD pathways. We speculate that Helicase89B links the three NF-κB-related activators in both pathways to the basal transcription complex. Conversely, the GATA factor Serpent in Drosophila has been shown to cooperate with NF-κB factors to activate antimicrobial peptide genes (Petersen et al, 1999; Senger et al, 2004). It is also possible that Helicase89B acts downstream of the GATA/NF-κB activation module. Either way, our results are consistent with the idea that Helicase89B has a functional homology to the Mot1p/BTAF1 family of ATPases. Mot1p forms a distinct complex with TBP that is separable from the TFIID complex (Auble et al, 1994; Poon et al, 1994). Mot1p and BTAF1 may use ATP hydrolysis to extract TBP from promoters, resulting in transcriptional repression (Wade & Jaehning, 1996; Auble et al, 1997; Gumbs et al, 2003). However, Mot1p has also been shown to activate transcription by regulating the distribution of TBP, by interacting with an active form of the TBP complex or with the Spt3 chromatin remodelling complex (Muldrow et al, 1999; Dasgupta et al, 2002; Geisberg & Struhl, 2004; Topalidou et al, 2004). Thus, Mot1p/BTAF1 may exist in multiple complexes in vivo and perform various functions. Helicase89B has been reported to bind to TBP using bacterially expressed proteins (Adamkewicz et al, 2001). We also detected Helicase89B and TBP interaction by co-immunoprecipitation after transient transfection in S2 cells (data not shown). However, we have not been able to detect complex formation between Helicase89B and the Drosophila NF-κB proteins. The manner in which Helicase89B links the upstream activators with TBP requires further investigation.
Whereas Mot1p regulates many inducible processes, including heat-shock response, in yeast (Geisberg & Struhl, 2004), Helicase89B is required for immune response but not heatshock response in Drosophila larvae. Moreover, the induction of the same antimicrobial peptide genes in adult flies does not require Helicase89B. Our genetic analysis also shows that the Helicase89B mutants are semi-lethal, with approximately 70% viability. These observations suggest that Helicase89B may be involved in other processes but not general transcription of all genes. Further study may resolve whether Helicase89B regulates other processes in Drosophila.
Methods
Fly strains and septic injury. P-element lines on the third chromosome were balanced with TM6B and those on the second chromosome were balanced with T(2:3)Cy;TM6B. Homozygous larvae were selected by the absence of the Tubby marker-associated TM6B balancer. Transgenic lines that contain rescue constructs on the second chromosome were crossed with the P1732 line and stable lines were established over the T(2:3)Cy;TM6B balancer chromosomes. The Cg-Gal4 driver chromosome (Bettencourt et al, 2004) was also established in the P1732 background. The mating of the two lines resulted in flies that had one copy of Gal4 and one copy of the pUAST transgene in the homozygous P1732 background. The PGRP-LE overexpression line (GS1068) was a GS vector insertion line into the PGRP-LE gene (Takehana et al, 2002).
Septic injury was carried out by puncturing larvae or adults with a needle that was previously dipped in a concentrated bacterial solution of Enterobacter cloacae, Micrococcus luteus and Erwinia carotovora. After septic injury, the larvae were placed on a wet filter paper inside a moist chamber and the adults were placed in food vials for the duration indicated in the figures.
Molecular cloning and northern analysis. 5′ RACE was carried out using a kit from Roche Diagnostic (Indianapolis, IN, USA) and mRNA from CantonS flies as the template. To regenerate the Helicase89B cDNA, partially digested EcoRI fragments from GH12153 and GH11668 (two partial cDNA clones from Research Genetics, Invitrogen, Carlsbad, CA, USA; Fig 2A) were sequentially cloned into the same site in pBluescript and then an EcoRI fragment from the 5′ RACE clone was inserted at the 5′ end. To generate the rescue construct, the full-length Helicase89B cDNA fragment obtained from BamHI and partial XhoI digestion of the pBluescript clone was ligated into a BglII/XhoI-digested pUAST vector. The moira cDNA (RE46850) was digested with KpnI and NotI and cloned into the same sites of pUAST. The DIF cDNA fragment (from 40 bp upstream of ATG to 10 bp downstream of the stop codon) was PCR amplified and cloned into the KpnI and XbaI sites of pUAST. The RelN cDNA fragment (codes for amino acids 1–545 of Relish) was cloned into the KpnI and XbaI sites of pUAST. For northern blots, total RNA from frozen animals was isolated using the TRIZOL (MRC Inc., Cincinnati, OH, USA) or the RNeasy kit (Qiagen, Valencia, CA, USA). For the expression of Helicase89B and moira, mRNA that was purified with the Oligotex mRNA Midi Kit (Qiagen) was used. For the analysis of antimicrobial peptide genes, total RNA was used. Formaldehyde agarose gel (1%) and GeneScreen Plus membrane (DuPont) were used for northern blots. Fragments from various cDNA clones were used as templates for labelling in the presence of radioactive or digoxygenin nucleotides. Hybridization signals were quantified using a PhosphoImager and normalized to those of the ribosomal protein 49 (rp49) gene.
Acknowledgments
We thank L. Zhou and X. Hu for technical assistance. We acknowledge the Bloomington Stock Center and the Drosophila genome project for the stocks and mapping information. We thank Dr S. Kurata for the generous gift of PGRP-LE strain. This work was supported by a National Institutes of Health grant (GM53269). Core resources supported by the Diabetes Endocrinology Research Center grant DK32520 were also used.
References
- Adamkewicz JI, Hansen KE, Prud'homme WA, Davis JL, Thorner J (2001) High affinity interaction of yeast transcriptional regulator, Mot1, with TATA box-binding protein (TBP). J Biol Chem 276: 11883–11894 [DOI] [PubMed] [Google Scholar]
- Auble DT, Hansen KE, Mueller CG, Lane WS, Thorner J, Hahn S (1994) Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev 8: 1920–1934 [DOI] [PubMed] [Google Scholar]
- Auble DT, Wang D, Post KW, Hahn S (1997) Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol Cell Biol 17: 4842–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt R, Asha H, Dearolf C, Ip YT (2004) Hemolymph-dependent and -independent response to microbial substances in Drosophila immune tissues. J Cell Biochem 92: 849–863 [DOI] [PubMed] [Google Scholar]
- Brennan CA, Anderson KV (2004) Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol 22: 457–483 [DOI] [PubMed] [Google Scholar]
- Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV (2002) Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296: 359–362 [DOI] [PubMed] [Google Scholar]
- Crosby MA, Miller C, Alon T, Watson KL, Verrijzer CP, Goldman-Levi R, Zak NB (1999) The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol Cell Biol 19: 1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Darst RP, Martin KJ, Afshari CA, Auble DT (2002) Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci USA 99: 2666–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K (2004) Cellular stress alters the transcriptional properties of promoter-bound Mot1–TBP complexes. Mol Cell 14: 479–489 [DOI] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J (2002) The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416: 640–644 [DOI] [PubMed] [Google Scholar]
- Gumbs OH, Campbell AM, Weil PA (2003) High-affinity DNA binding by a Mot1p–TBP complex: implications for TAF-independent transcription. EMBO J 22: 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA (2003) The immune response of Drosophila. Nature 426: 33–38 [DOI] [PubMed] [Google Scholar]
- Hu X, Yagi Y, Tanji T, Zhou S, Ip YT (2004) Multimerization and interaction of Toll and Spatzle in Drosophila. Proc Natl Acad Sci USA 101: 9369–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis P, Bulet P, Reichhart JM (2002) Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defense of Drosophila. EMBO Rep 3: 666–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Biron CA (2003) Innate immunity. Curr Opin Immunol 15: 2–4 [Google Scholar]
- Muldrow TA, Campbell AM, Weil PA, Auble DT (1999) MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol Cell Biol 19: 2835–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LA, Klejman MP, Timmers HT (2003) Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription. Gene 315: 1–13 [DOI] [PubMed] [Google Scholar]
- Petersen UM, Kadalayil L, Rehorn KP, Hoshizaki DK, Reuter R, Engstrom Y (1999) Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. EMBO J 18: 4013–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon D, Campbell AM, Bai Y, Weil PA (1994) Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)–TBP-associated factor complex distinct from transcription factor IID. J Biol Chem 269: 23135–23140 [PubMed] [Google Scholar]
- Senger K, Armstrong GW, Rowell WJ, Kwan JM, Markstein M, Levine M (2004) Immunity regulatory DNAs share common organizational features in Drosophila. Mol Cell 13: 19–32 [DOI] [PubMed] [Google Scholar]
- Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S (2002) Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates IMD/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA 99: 13705–13710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingvall TO, Roos E, Engstrom Y (2001) The GATA factor Serpent is required for the onset of the humoral immune response in Drosophila embryos. Proc Natl Acad Sci USA 98: 3884–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, Papamichos-Chronakis M, Thireos G, Tzamarias D (2004) Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. EMBO J 23: 1943–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Jaehning JA (1996) Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol Cell Biol 16: 1641–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]


