Abstract
Mutations in mitochondrial DNA (mtDNA), particularly those in the 12S rRNA gene, have been shown to be associated with sensorineural hearing loss. Recently, a systematic and extended mutation screening of the mitochondrial 12S rRNA gene has been initiated in the large clinical population of the Otology Clinic at the Chinese PLA General Hospital with the aim of identifying mtDNA mutations associated with hearing loss. Here we report the clinical and molecular characterization of a Chinese patient with auditory neuropathy. Sequence analysis of mtDNA in this patient identified a T-to-C transition at position 1095 (T1095C) in the 12S rRNA gene and other nucleotide changes. The T1095C mutation is expected to disrupt an evolutionarily conserved A-to-U base-pair, which is at the highly conserved P-site of 12SrRNA. TheT1095C mutation has also been found to be associated with hearing loss in several unrelated families. Among other nucleotide changes, two novel variants: the I175V mutation in the CO2 and the V112M mutation in the ND6 localize at highly evolutionarily conserved residues from different organisms. Furthermore, the absence of mutation in the otoferlin related to auditory neuropathy showed that otoferlin may not be involved in the phenotypic expression of T1095C mutation in this subject. These data suggest that the T1095C mutation may be associated with auditory neuropathy in this subject, and two novel variants I175V and V112M may play a role in the phenotypic expression of the T1095C mutation.
Keywords: hearing loss, inherited, mitochondria 12S RNA, mutation, auditory neuropathy
INTRODUCTION
Mitochondrial DNA (mtDNA) mutations are likely to be one of the important causes of sensorineural hearing loss, especially nonsyndromic forms [Fischel-Ghodsian, 1999; Guan, 2004]. In particular, the 12S rRNA gene has been shown to be a hot spot for aminoglycoside induced and nonsyndromic hearing loss. Several deafness-associated mtDNA mutations have been identified in this gene. Of those, an A to G transition at position 1555 (A1555G) in the highly conserved decoding site of the 12S rRNA has been associated with both aminoglycoside induced and nonsyndromic hearing loss in many families of different ethnic origins [Prezant et al., 1993; Matthijs et al., 1996; Pandya et al., 1997; Estivill et al., 1998; del Castillo et al., 2003; Li et al., 2004b]. Recently, the C1494T mutation in the same gene has been associated with both aminoglycoside induced and nonsyndromic hearing loss in a large Chinese family [Zhao et al., 2004]. In addition, a C-insertion or deletion at position 961 of the 12S rRNA gene has been shown to be associated only with aminoglycoside-induced deafness [Bacino et al., 1995; Casano et al., 1999], while the novel T961G mutation has been implicated to be responsible for the nonsyndromic hearing loss in five Caucasian pediatric subjects [Li et al., 2004a]. Furthermore, the T1095C mutation has also been shown to be associated with hearing impairment [Thyagarajan et al., 2000; Tessa et al., 2001].
With the aim of identifying mtDNA mutations associated with hearing loss, a systematic and extended mutation screening of the mitochondrial 12S rRNA gene has been initiated in the large clinical population of Otology Clinic at the Chinese PLA General Hospital. As a consequence of this study, 34 pedigrees with a maternally inherited pattern of nonsyndromic and aminoglycoside-induced hearing loss have been identified, including 15 pedigrees carrying the A1555G mutation and one pedigree carrying the C1494T mutation in the 12S rRNA gene [Zhao et al., 2004]. In the present study, we have performed a clinical and molecular analysis of a Chinese patient with auditory neuropathy associated with the T1095C mutation in the 12S rRNA gene.
SUBJECTS AND METHODS
As the part of genetic screening program for the hearing impairment, a 27-year-old female was ascertained at the Otology Clinic at PLA General Hospital. A comprehensive history and physical examination were performed to identify any syndromic findings or genetic factors related to the hearing loss. An age-appropriate audiological examination was performed, and this examination included pure-tone audiometry (PTA) and/or auditory brainstem response (ABR), immittance testing, and distortion product otoacoustic emissions (DPOAE). The PTA was calculated from the sum of the audiometric thresholds at 500, 1,000 and 2,000, 4,000, and 8,000 Hz. The severity of hearing impairment was classified into five grades: normal <26 dB; mild = 26–40 dB; moderate = 41–70 dB; severe = 71–90 dB; and profound >90 dB. Informed consent was obtained from the participant prior to their participation in the study, in accordance with the Cincinnati Children’s Hospital Medical Center Institutional Review Board and Ethnic Committee of Chinese PLA General Hospital.
Genomic DNA was isolated from whole blood of participants using the Puregene DNA Isolation Kits (Gentra Systems, Minneapolis, MN). First, the subject’s DNA fragments spanning the entire mitochondrial 12S rRNA gene or tRNASer(UCN) gene were amplified by PCR using oligodeoxynucleotides corresponding to the mitochondrial genome at positions 618–635 and 1988–2007 [Guan et al., 1996; Zhao et al., 2004] and 7148–7167 and 8076–8095 [Guan et al., 1998; Li et al., 2004c], respectively. Each fragment was purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using the Big Dye Terminator Cycle sequencing reaction kit. mtDNA sequence alignments were carried out using seqweb program GAP (GCG).
The entire mitochondrial genomes of the subject carrying the T1095C mutation were PCR amplified in 24 overlapping fragments by use of sets of the light-strand and the heavy strand oligonucleotide primers, as described elsewhere [Rieder et al., 1998]. Each fragment was purified and subsequently submitted for sequence analysis as described above. The resultant sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_001807) [Anderson et al., 1981].
The DNA fragments spanning the entire coding region of the otoferlin (OTOF) gene were PCR-amplified in this subject, and subsequent sequencing analysis was performed as detailed elsewhere [Varga et al., 2003]. The results were compared with the wild-type OTOF sequence (GenBank accession number: AF183185) to identify the mutations.
RESULTS AND DISCUSSION
A female Chinese subject came to the otology clinic at the Chinese PLA General Hospital at the age of 27 years old. She had suffered from speech discrimination difficulties for 6 months before visiting this clinic. Clinical characterization showed that her speech discrimination disorder was out of proportion to her hearing level. As shown in Figure 1, the audiological evaluation, including pure-tone audiomery, revealed a reverse “cookie bite” pattern (worse in the low and high frequencies) sensorineural hearing loss in right ear, and a primary low and mid frequency sensorineural hearing loss in the left ear. Auditory brainstem responds (ABR) were absent in both sides. DPOAE was normal on the left side with a decrease in amplitude at 4,000 Hz. It was not inhibited by noise from opposite side. She had no history of exposure of aminoglycosides, including gentamicin, streptomycin, and kanamycin. The results of other clinical and biochemical examinations, such as blood biochemistry tests, magnetic resonance imaging (MRI), electrophoresis of anti-membranous labyrinth protein, were normal. Comprehensive medical examinations of this individual showed no other neurological disorders. Thus, this subject showed a typical phenotype of auditory neuropathy.
Fig. 1.
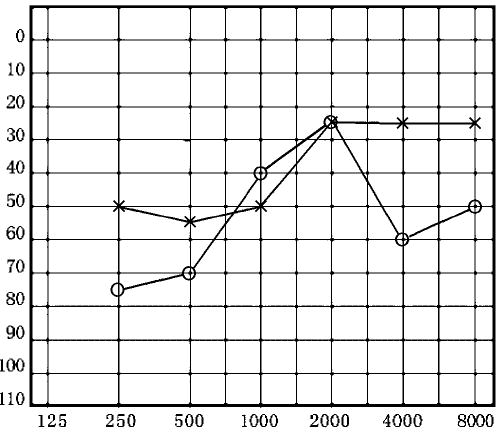
Air conduction audiogram of the affected subject with the T0195C mutation. Symbols: X-left, O-right ear.
To elucidate the molecular basis of the auditory neuropathy, we performed a mutation analysis of the mitochondrial genome in this subject. Firstly, DNA fragments spanning mitochondrial 12S rRNA and tRNASer(UCN) genes, which are the hot spots for deafness-associated mutations [Guan, 2004], were PCR amplified and each fragment was purified and subsequently analyzed by DNA sequencing. We failed to detect the presence of the A7445G, T7510C, 7472insC, T7511C mutations in the tRNASer(UCN) gene and the A1555G, C1494T, and961 mutations in this 12S rRNA gene [Guan, 2004]. However, the T1095C mutation in the 12S rRNA gene was found in this subject apparently in the homoplasmic form (Fig. 2). This mutation was absent in 364 Chinese control [Zhao et al., 2004]. This mutation has been found in an Italian family with auditory neuropathy, aminoglycoside-induced hearing loss and Parkinsonism [Thyagarajan et al., 2000], and other Italian family with maternally inherited deafness but in the absence of auditory neuropathy phenotype [Tessa et al., 2001]. This T-to-C transition disrupted an evolutionarily conserved base-pair at stem loop of the helix 25 of 12S rRNA [Neefs et al., 1991]. This nucleotide is also located at the P-site of ribosome, suggesting an important role in the initiation of mitochondrial protein synthesis [Thyagarajan et al., 2000]. The alteration of the tertiary or quaternary structure of this rRNA by the T1095C mutation may lead to impairing mitochondrial protein synthesis, thereby causing the mitochondrial dysfunction associated with hearing impairment. In fact, the occurrence of the T1095C mutation in these several genetically unrelated pedigrees affected by hearing impairment strongly indicates that this mutation is involved in the pathogenesis of hearing impairment, especially in auditory neuropathy.
Fig. 2.
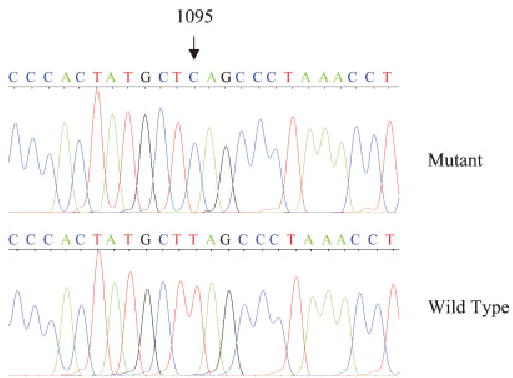
Identification of T1095C mutation in the mitochondrial 12S rRNA gene. Partial sequence chromatograms of the 12S rRNA gene from control and auditory neuropathy subjects.
To understand the role of mitochondrial haplotype in the phenotypic expression of the T1095C mutation, we also performed a PCR-amplification of fragments spanning entire mitochondrial genome and subsequent DNA sequence analysis in this subject. The comparison of the resultant sequence with the Cambridge consensus sequence [Anderson et al., 1981] identified a number of nucleotide changes as shown in Table I. Apart from T1095C mutation, there are two variants A750G and A1438G in the 12S rRNA gene and variant A2706G in the 16S rRNA gene, which are previously reported in control population [Kogelnik et al., 1998]. Of other nucleotide changes in this mitochondrial genome, 10 variants in the D-loop and 18 variants in the protein encoding genes were previously found in the Chinese control populations [Kogelnik et al., 1998]. Furthermore, the following mutations are novel missense polymorphisms in the Chinese population: A8108G (I175V) mutation in the CO2, G11969A (A406T) in the ND4, and C14340T (V112M) in the ND6. These variants were further evaluated by phylogenetic analysis of these mtDNA variants and mtDNAs from other organisms. The A406T mutation in the ND4 was not highly conserved, whereas the I175V mutation in the CO2 and the V112M mutation in the ND6 are localized at sites that are highly conserved in human [Anderson et al., 1981], mouse [Bibb et al., 1981], bovine [Gadaleta et al., 1989], and Xenopus [Roe et al., 1985]. There is an increasing evidence showing that the background sequences (haplotype) of the mtDNA modulate the severity and penetrance of the phenotypic expression of pathogenic mtDNA mutation(s) associated with some clinical abnormalities including hearing loss [Guan et al., 1998; Li et al., 2004c] and blindness [Torroni et al., 1997]. Here, the I175V mutation in the CO2 and the V112M mutation in the ND6, showing high evolutionary conservation, may contribute to the phenotypic expression of the T1095C mutation in this patient.
Table I.
mtDNA Variants in the Chinese Patient With Auditory Neuropathy
| Gene | Position | Replacement | Conservationa H/B/M/X | Previously reportedb |
|---|---|---|---|---|
| D-Loop | 73 | A to G | Yes | |
| 146 | T to C | Yes | ||
| 215 | A toG | Yes | ||
| 263 | A to G | Yes | ||
| 311 | C insertion | Yes | ||
| 318 | T to C | Yes | ||
| 326 | A to G | Yes | ||
| 489 | T to C | Yes | ||
| 16172 | C to T | Yes | ||
| 16223 | C to T | Yes | ||
| 12S rRNA | 750 | A to G | A/A/G/- | Yes |
| 1095c | T to C | T/T/T/T | Yes | |
| 1438 | A to G | A/A/A/G | Yes | |
| 16S rRNA | 2706 | A to G | A/G/AA | Yes |
| ND2 | 4769 | A to G | Yes | |
| CO1 | 7028 | C to T | Yes | |
| CO2 | 7642 | G to A | Yes | |
| 8108 | A to G (Ile to Val) | I/I/I/I | No | |
| A6 | 8701 | A to G (Thr to Ala) | T/S/L/Q | Yes |
| 8860 | A to G (Thr to Ala) | T/A/A/T | Yes | |
| CO3 | 9383 | C to T | No | |
| 9540 | T to C | Yes | ||
| 9667 | A to G (Asn to Ser) | N/G/G/G | Yes | |
| 9950 | T to C | Yes | ||
| ND3 | 10398 | A to G (Thr to Ala) | T/T/T/A | Yes |
| 10400 | C to T | Yes | ||
| ND4 | 10873 | T to C | Yes | |
| 11719 | G to A | Yes | ||
| 11969 | G to A (Ala to Thr) | A/A/G/A | No | |
| ND5 | 12705 | A to G | Yes | |
| 13074 | A to G | No | ||
| ND6 | 14180 | T to C (Tyr to Cys) | Y/V/F/Y | Yes |
| 14340 | C to T(Val to Met) | V/V/V/V | No | |
| Cytochromeb | 14783 | C to T | Yes | |
| 15043 | G to A | Yes | ||
| 15301 | G to A | Yes | ||
| 15326 | A to G (Thr to Ala) | T.M.I. I. | Yes |
Conservation of amino acid for polypeptides or nucleotide for RNAs, in human (H), bovine (B), mouse (M), and Xenopus laevis (X).
Conserved nucleotide and residues are boldfaced.
Furthermore, the mutations in the otoferlin have been shown to be associated with auditory neuropathy [Varga et al., 2003]. To examine the role of the otoferlin in the phenotypic expression of the T1095C mutation, we performed the mutational screening of the otoferlin in this subject. Sequence analysis revealed the absence of mutation in the otoferlin gene in this subject, ruling out the possible involvement of otoferlin in the phenotypic expression of T1095C mutation.
In summary, our data indicated that the T1095C mutation in the 12S rRNA gene may be associated with auditory neuropathy in this patient. Two novel variants, the I175V mutation in the CO2 and the V112M mutation in the ND6, may play a role in the pathogenesis of this disorder. Furthermore, it seems that the otoferlin may not be involved in the phenotypic expression of T1095C mutation in this subject.
Acknowledgments
This work was supported by the National Institutes of Health grant DC04958 and DC05230 from the National Institute on Deafness and Other Communication Disorders and a Research Grant Award from the Deafness Research Foundation (to M.X.G).
Footnotes
Qiuju Wang and Roughua Li equally contributed to this work.
References
- Anderson S, Bankier AT, Barrell BG, Debruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Rose BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young I. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bacino C, Prezant TR, Bu X, Fournier P, Fischel-Ghodsian N. Susceptibility mutations in the mitochondrial small ribosomal RNA gene in aminoglycoside induced deafness. Pharmacogenetics. 1995;5:165–172. doi: 10.1097/00008571-199506000-00005. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Casano RA, Johnson DF, Bykhovskaya Y, Torricelli F, Bigozzi M, Fischel-Ghodsian N. Inherited susceptibility to aminoglycoside ototoxicity: Genetic heterogeneity and clinical implications. Am J Otolaryngol. 1999;20:151–156. doi: 10.1016/s0196-0709(99)90062-5. [DOI] [PubMed] [Google Scholar]
- del Castillo FJ, Rodriguez-Ballesteros M, Martin Y, Arellano B, Gallo-Teran J, Morales-Angulo C, Ramirez-Camacho R, Cruz Tapia M, Solanellas J, Martinez-Conde A, Villamar M, Moreno-Pelayo MA, Moreno F, del Castillo I. Heteroplasmy for the 1555A >G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet. 2003;40:632–636. doi: 10.1136/jmg.40.8.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill X, Govea N, Barceló A, Perelló E, Badenas C, Romero E, Moral L, Scozzari R, D’Urbano L, Zeviani M, Torroni A. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am J Hum Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed. Hum Mut. 1999;13:261–270. doi: 10.1002/(SICI)1098-1004(1999)13:4<261::AID-HUMU1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: Cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- Guan MX. Molecular pathogenetic mechanism of maternally inherited deafness. Ann NY Acad Sci. 2004;1011:259–271. doi: 10.1007/978-3-662-41088-2_25. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet. 1996;5:963–971. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- Guan MX, Enriquez JA, Fischel-Ghodsian N, Puranam R, Lin CP, Marion MA, Attardi G. The deafness-associated mtDNA 7445 mutation, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase ND6 subunit gene expression. Mol Cell Biol. 1998;18:5868–5879. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP: A human mitochondrial genome database—1998 update. Nucleic Acids Res. 1998;26:112–115. doi: 10.1093/nar/26.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Greinwald JH, Yang L, Choo DI, Wenstrup RJ, Guan MX. Molecular analysis of mitochondrial 12S rRNA and tRNASer(UCN)genes in pediatric subjects with nonsyndromic hearing loss. J Med Genet. 2004a;41:615–620. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Xing G, Yan M, Cao X, Liu XZ, Bu X, Guan MX. Cosegregation of C-insertion at position 961 with A1555G mutation of mitochondrial 12S rRNA gene in a large Chinese family with maternally inherited hearing loss. Am J Med Genet. 2004b;124A:113–117. doi: 10.1002/ajmg.a.20305. [DOI] [PubMed] [Google Scholar]
- Li X, Fischel-Ghodsian N, Schwartz F, Yan Q, Friedman RA, Guan MX. Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids Res. 2004c;32:867–877. doi: 10.1093/nar/gkh226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijs G, Claes S, Longo-Bbenza B, Cassiman J-J. Non-syndromic deafness associated with a mutation and a polymorphism in the mitochondrial 12S ribosomal RNA gene in a large Zairean pedigree. Eur J Hum Genet. 1996;4:46–51. doi: 10.1159/000472169. [DOI] [PubMed] [Google Scholar]
- Neefs JM, Van de Peer Y, De Rijik P, Goris A, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1991;19(Suppl):1987–2018. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya A, Xia X, Radnaabazar J, Batsuuri J, Dangaansuren B, Fischel-Ghodsian N, Nance WE. Mutation in the mitochondrial 12S ribosomal-RNA gene in two families from Mongolia with matrilineal aminoglycoside ototoxicity. J Med Genet. 1997;34:169–172. doi: 10.1136/jmg.34.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezant TR, Agapian JV, Bohlman MC, Bu X, Öztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, Shohat M, Fischel-Ghodsian N. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: Analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe A, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- Tessa A, Giannotti A, Tieri L, Vilarinho L, Marotta G, Santorelli FM. Maternally inherited deafness associated with a T1095C mutation in the mDNA. Eur J Hum Genet. 2001;9:147–149. doi: 10.1038/sj.ejhg.5200601. [DOI] [PubMed] [Google Scholar]
- Thyagarajan D, Bressman S, Bruno C, Przedborski S, Shanske S, Lynch T, Fahn S, DiMauro S. A novel mitochondrial 12SrRNA point mutation in parkinsonism, deafness, and neuropathy. Ann Neurol. 2000;48:730–736. [PubMed] [Google Scholar]
- Torroni A, Petrozzi M, D’Urbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De Negri A, Scozzari R. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- Varga R, Kelley PM, Keats BJ, Starr A, Leal SM, Cohn E, Kimberling WJ. Non-syndromic recessive auditory neuropathy is the result of mutations in the otoferlin (OTOF) gene. J Med Genet. 2003;40:45–50. doi: 10.1136/jmg.40.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, Bai Y, Young WY, Guan MX. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]


