Abstract
Oceanobacillus iheyensis HTE831 is an alkaliphilic and extremely halotolerant Bacillus-related species isolated from deep-sea sediment. We present here the complete genome sequence of HTE831 along with analyses of genes required for adaptation to highly alkaline and saline environments. The genome consists of 3.6 Mb, encoding many proteins potentially associated with roles in regulation of intracellular osmotic pressure and pH homeostasis. The candidate genes involved in alkaliphily were determined based on comparative analysis with three Bacillus species and two other Gram-positive species. Comparison with the genomes of other major Gram-positive bacterial species suggests that the backbone of the genus Bacillus is composed of approximately 350 genes. This second genome sequence of an alkaliphilic Bacillus-related species will be useful in understanding life in highly alkaline environments and microbial diversity within the ubiquitous bacilli.
INTRODUCTION
The nature of the chemical and physical environment, whether under stable or stressful conditions, influences the growth and physiology of microbial populations. Although the deep sea is principally an oligotrophic, cold, dark and high pressure environment, the bottom of the deep sea is not devoid of organisms. Numerous microorganisms have been isolated from deep-sea sediments collected at depths ranging from 1050 to 10897 m located near the southern part of Japan (1,2). Although some of these isolates showed piezophilic and/or psychrophilic phenotypes, most of those isolates were piezotolerant and/or psychrotolerant. Piezophiles are defined as bacteria that have optimal growth at a pressure higher than 0.1 MPa or by a requirement of increased pressure for growth. Psychrophiles are bacteria that have an optimal temperature for growth lower than 15°C, a maximal temperature for growth lower than 20°C and a minimal temperature for growth at 0°C or lower. Among these deep-sea isolates, many extremophilic bacteria, such as alkaliphiles, halophiles and thermophiles, have been found that would be expected to thrive in an extreme environment different from the in situ conditions of high hydrostatic pressure and low temperature in the deep sea sampling sites (2). The number of piezophilic and psychrophilic isolates recovered from deep-sea sediments showed a tendency to be smaller than those of alkaliphiles and thermophiles.
Bacillus species are distributed nearly ubiquitously in nature. These organisms have often been isolated from various terrestrial soils and deep-sea sediments. Some of these Bacillus species possess adaptations to extreme environments, including high and low temperature, high and low pH and high salinity (3,4). Oceanobacillus iheyensis HTE831 (JCM 11309, DSM 14371), which was isolated from a deep-sea sediment collected at a depth of 1050 m on the Iheya Ridge, and was recently reclassified from the genus Bacillus (formerly Bacillus sp. HTE831), shows extremely halotolerant and facultative alkaliphilic properties (5).
The genome sequences of two terrestrial Bacillus spp., Bacillus halodurans (6) and Bacillus subtilis (7), have previously been published. Here we report the complete nucleotide sequence of the genome of O.iheyensis. We provide comparative analysis of this genome with those of the alkaliphilic and moderately halotolerant B.halodurans and the neutrophilic and moderately halotolerant B.subtilis along with two Gram-positive species belonging to the genera Staphylococcus (8) and Clostridium (9). Special emphasis is placed on the mechanisms of adaptation to alkaline and highly saline environments.
MATERIALS AND METHODS
Sequencing and assembly
The genome of O.iheyensis HTE831 was primarily sequenced by the whole genome random sequencing method used in a previous work (6). DNA sequences determined by means of the MegaBace1000 DNA sequencer were assembled into contigs using Phrap (http://bozeman.mbt.washington.edu/phrap.docs/phrap.html) with default parameters. 2000 sequences were obtained from the reverse end of shotgun clones and both ends of genomic libraries containing inserts ranging from 4 to 6 kb. These sequences were assembled with consensus sequences derived from the contigs of random phase sequences using Phrap, resulting in a reduction in the number of contigs to 105. 1500 sequences from both ends of 20 kb inserts in λ phage clones were assembled to bridge the remaining contigs. The final gaps were closed by direct sequencing of the products amplified by long accurate PCR as described previously (6).
Gene prediction and annotation
The predicted protein coding regions were initially defined by searching for open reading frames longer than 100 codons using the Genome Gambler system (10; http://www.xanagen.com/index-e.html). An analysis of coding potential for the entire genome was performed with the GeneHacker Plus program (11) using hidden Markov models (http://elmo.ims.u-tokyo.ac.jp/GH/) trained with a set of O.iheyensis ORFs longer than 300 nt. Searches of the protein databases for amino acid similarities were performed using BLAST2 sequence analysis tools (12) with subsequent comparison of the putative proteins showing significant homology (>10–5 significance) performed using the Lipman–Pearson algorithm (13). The functional assignment of annotated protein coding sequences (CDSs) identified in the O.iheyensis genome was performed in the same manner as for B.subtilis (SubtiList, http://www.pasteur.fr/Bio/SubtiList.html).
Comparative analysis
The BLAST2 search program was used to generate a similarity matrix among the proteins deduced from all CDSs identified in the five compared genomes as the first step of comparative analysis. The alignment results of BLAST2 were recalculated by the dynamic programming method (14) with the JTT-PAM250 scoring matrix (15) when the e-value was <1e–3 and the Z-score was >80. The clustering based on the unweighted pair-group method (16) using the arithmetic mean was carried out by using all-against-all similarity search results, including both similarity scores and matched regions. Assignment of the proteins deduced from predicted CDSs to clusters of orthologous groups (COGs) was based on the results through this clustering procedure (http://mbgd.genome.ad.jp). The functional assignments embedded in the COG database (http://www.ncbi.nlm.nih.gov/COG/) were used for reconstruction of metabolic pathways and other functional systems in O.iheyensis with the KEGG database (http://www.genome.ad.jp/kegg/kegg2.html).
RESULTS AND DISCUSSION
General features of the genome
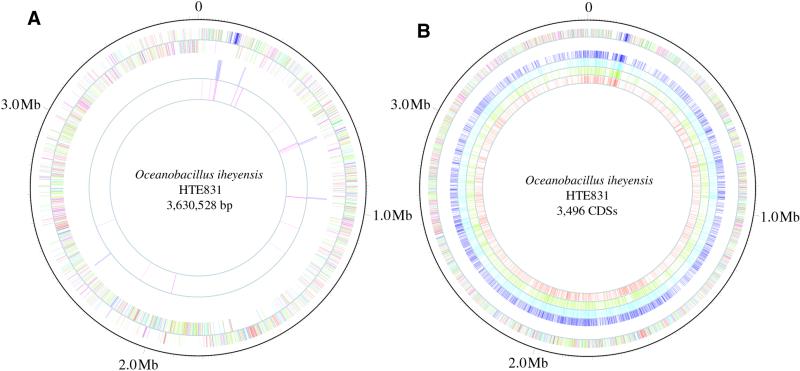
The genome of O.iheyensis is a single circular chromosome consisting of 3 630 528 bp with an average G+C content of 35.7% (Fig. 1A and Table 1). The G+C content of DNA in the coding region and non-coding region is 36.1 and 31.8%, respectively. On the basis of analysis of the G+C ratio and G-C skew (G-C/G+C), we estimated that the site of termination of replication (terC) is nearly 1.77–1.78 Mb (176°) from the replication origin. We identified 3496 CDSs, on average 883 nt in size, using a coding region analysis program (10,11). Coding sequences cover 85% of the chromosome. We found that 79.5% of the genes started with ATG, 7.8% with GTG and 12.7% with TTG. These values are quite similar to those of B.subtilis and B.halodurans, whose whole genomic sequences have been completely defined previously (Table 1). The average size of the predicted proteins in O.iheyensis is 32.804 kDa, ranging from 2.714 to 268.876 kDa. Predicted protein sequences were compared with sequences in non-redundant protein databases (nr-aa, SwissProt and PIR) and putative biological roles were assigned to 1972 (56.4%) of them. In this database search 1069 proteins deduced from CDSs (30.6%) were identified as conserved proteins of unknown function in comparison with proteins from other organisms and for 456 (13%) there was no database match. Sixty-nine tRNA species, organized into 10 clusters involving 63 genes plus 6 single genes, were identified (Fig. 1A and Table 1). Of the 10 tRNA clusters, 5 were organized in association with rRNA operons. Seven rRNA operons are present in the HTE831 genome and in two cases their organization is the same as that of B.subtilis and B.halodurans (6) (16S-23-S-5S, 16S-23S-5S-tRNA and tRNA-16S-23S-5S), but another gene order is also present (16S-tRNA-23S-5S-tRNA).
Figure 1.
Circular representation of the alkaliphilic and extremely halotolerant O.iheyensis genome. (A) General view of the O.iheyensis HTE831 chromosome. The distribution of CDSs is depicted by colored boxes according to functional categories and direction of transcription (outer circle, plus strand; inner circle, minus strand; red, cell wall, sensors, motility and chemotaxis, protein secretion, cell division and transformation/competence; magenta, transport/binding proteins and lipoptoteins and membrane bioenergetics; gold, sporulation and germination; yellow/green, intermediary metabolism; gray, DNA replication, DNA restriction/modification and repair, DNA recombination and DNA packaging and segregation; pink, RNA synthesis; blue, protein synthesis; forest green, miscellaneous functions; sky blue, conserved CDSs with unknown function; coral, non-conserved proteins). The third and fourth circles indicate the distribution of rRNA and tRNA in the genome, respectively. (B) Distribution of orthologs identified among five genomes of major Gram-positive bacterial species based on the proteins deduced from the CDSs identified in the O.iheyensis genome. The distribution of orthologs is depicted by colored boxes (outermost circle, O.iheyensis; second circle, B.halodurans; third circle, B.subtilis; fourth circle, S.aureus; innermost circle, C.acetobutylicum). The blank spaces on the outermost circle indicate the location of RNA elements with no ortholog compared with the HTE831 genome in the case of the other circles. A linear map of the genes, with color codes for functional categories, is available at http://www.jamstec.go.jp/jamstec-e/bio/jp/topj.html.
Table 1. General features of the O.iheyensis genome and its comparison with other Bacillus genomes.
| General features | Oceanobacillus iheyensis | Bacillus haloduransa | Bacillus subtilisb |
|---|---|---|---|
| Size (bp) | 3 630 528 | 4 202 352 | 4 214 630 |
| G+C content (mol%) | |||
| Total genome | 35.7 | 43.7 | 43.5 |
| Coding region | 36.1 | 44.4 | 43.4 |
| Non-coding region | 31.8 | 39.8 | 43.6 |
| Protein coding sequences | |||
| Predicted no. | 3496 | 4066 | 4106 |
| Average length (bp) | 883 | 879 | 896 |
| Percent of coding region | 85 | 85 | 87 |
| Initiation codon | |||
| Percent AUG | 79.5 | 78 | 78 |
| Percent GUG | 7.8 | 12 | 9 |
| Percent UUG | 12.7 | 10 | 13 |
| RNA elements | |||
| Percent stable RNA | 1.04 | 1.02 | 1.27 |
| No. of rrn operons | 7 | 8 | 10 |
| 16S-23S-5S | 2 | 2 | 3 |
| 16S-23S-5S-tRNA | 3 | 1 | 5 |
| tRNA-16S-23S-5S | 1 | 2 | 0 |
| 16S-tRNA-23S-5S | 0 | 3 | 2 |
| 16S-tRNA-23S-5S-tRNA | 1 | 0 | 0 |
| Nos of tRNA | 69 | 78 | 86 |
| Insertion elements | |||
| Phage-associated genes | |||
| PBSX | 8 | 2 | 37 |
| skin element | 0 | 0 | 3 |
| Spβ | 7 | 10 | 185c |
| Others | 12 | 30 | 43 |
| Transposase gene of IS | 14 | 93 | 0 |
| ISL3 family | 10 | 43 | 0 |
| IS200/IS605 family | 2 | 8 | 0 |
| IS30 family | 1 | 3 | 0 |
| IS1272/IS660 family | 1 | 5 | 0 |
| Recombinase of group II intron | 5 | 5 | 0 |
The O.iheyensis genome possesses 21 genes encoding putative transposases or recombinases with similarity to sequences present in the genomes of B.halodurans, Marinococcus halophilus or Enterococcus faecium. However, the variety and number of these are much less than the 112 that were categorized into 27 groups found in the B.halodurans genome (5). In a series of analyses of the genes encoding transposases or recombinases, we defined a gene divided into several pieces as one gene because the transposable elements are often mutagenized by deletions, insertions and substitutions of nucleotides. Fourteen of those genes showed significant similarity to transposases of IS elements present in B.halodurans, categorized into the ISL3, IS200/IS605, IS30 (17) and IS1272/IS660 families (18), and five were also similar to recombinases of the group II intron found in B.halodurans (Table 1). Although these putative genes encoding transposases and recombinases present in the O.iheyensis genome were not identified in B.subtilis 168, the transposase genes of IS elements belonging to the IS200/IS605 and IS1272/IS660 families and, especially, the recombinase gene of the group II intron were widely spread among other species of Bacillus and related genera as judged by Southern hybridization. The genome also contains 27 putative phage-associated genes, which are similar to those of B.subtilis, Staphylococcus aureus, Lactococcus lactis, Clostridium acetobutylicum and Streptococcus species, although the O.iheyensis genome contains no intact prophage such as Spβ, PBSX or skin found in the B.subtilis genome (7).
Ortholog analysis
The genome of O.iheyensis provides us with a unique opportunity to investigate the genes that underlie the capability to adapt to an alkaline or hypersaline environment. The first issue was addressed by comparing the orthologous relationships among the proteins deduced from all CDSs identified in the five genomes of Gram-positive bacteria (Figs 1B and 2). Out of 3496 proteins identified in the O.iheyensis genome, 838 putative proteins (24.0%) have no orthologous relationship to proteins encoded in the four other genomes (Fig. 2). 793 proteins (22.7%) were orthologs identified among five Gram-positive bacterial species and 354 (10.1%) were identified as common proteins only among Bacillus-related species. 243 putative proteins (7.0%) were shared only between the two alkaliphiles, O.iheyensis and B.halodurans. As shown in Figure 2, the trend of orthologous relationships was almost the same in the case of analyses based on each Bacillus-related species although the genome size of O.iheyensis is 600 kb smaller than those of the other two Bacillus genomes. In addition, orthologous relationships emerged in the comparison with all combinations among the five genomes used in this study (Fig. 2). The putative proteins characterized on the basis of orthologous relationships were assigned to the functional categories used for B.subtilis.
Figure 2.
Summary of orthologous relationships among the proteins deduced from all CDSs identified in the genomes of bacilli and other major Gram-positive species. (A) Orthologous relationships based on the O.iheyensis genome. (B) Orthologous relationships based on the B.halodurans genome. (C) Orthologous relationships based on the B.subtilis genome. OB, O.iheyensis; BH, B.halodurans; BS, B.subtilis; SA, S.aureus; CA, C.acetobutylicum.
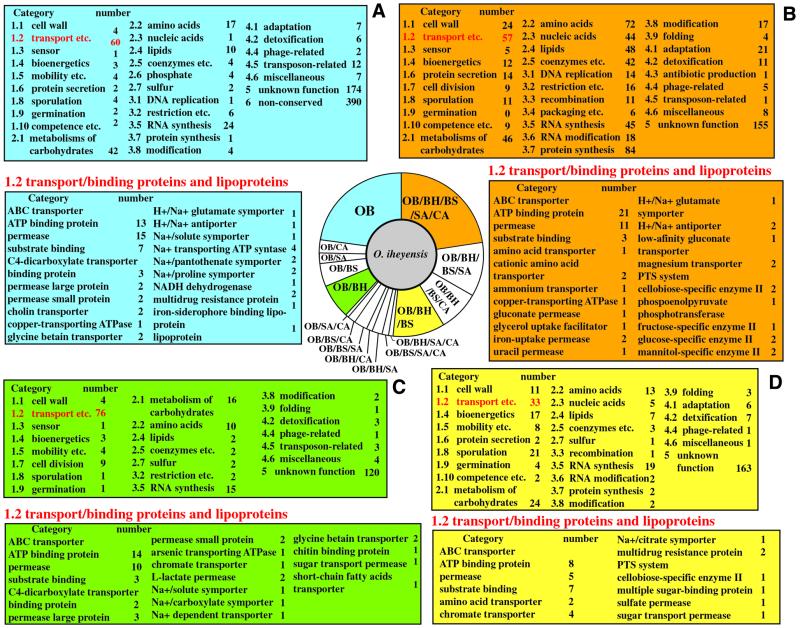
Out of 838 putative proteins without orthologous relationship to four other Gram-positive species, 390 were orphans showing no significant similarity of amino acid sequence to any other protein and 448 were conserved proteins in other organisms. 174 were conserved proteins of unknown function among 448 conserved proteins. Sixty proteins were grouped into transport/binding proteins and lipoproteins (category 1.2), which was numerically the most abundant category. Nearly half of these proteins are ABC transporter-related proteins (Fig. 3). Many of the orthologs which were shared between two to four species in the comparisons were also grouped into this category, indicating that some of these transport-related proteins are distinguishing characteristics for subsets of Gram-positive bacteria.
Figure 3.
Functional assignment of CDSs based on the orthologous relationships. (A) Orphans which have no orthologous relationship to four other Gram-positive species. (B) Orthologs identified among the genomes of five Gram-positive bacterial species. (D) Orthologs identified among the genomes of three Bacillus-related species. (C) Orthologs identified between O.iheyensis and B.halodurans, both alkaliphiles. The functional categories used at the B.subtilis genome database (SubtiList, http://www.pasteur.fr/Bio/SubtiList.html) were applied to this study. (C) Orthologs identified between O.iheyensis and B.halodurans, both alkaliphiles. (D) Orthologs identified among the genomes of three Bacillus-related species.
Comparative analysis of the genome structure
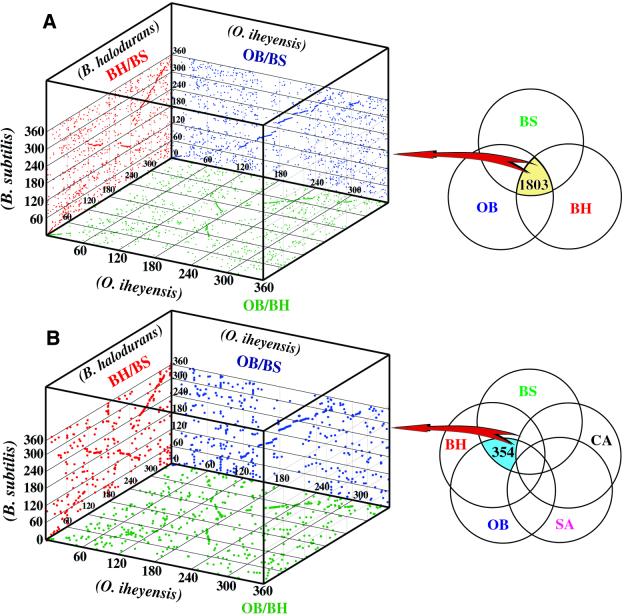
Although O.iheyensis and the other sequenced Bacillus-related species belong to the same phylogenetic clade (3,5), each is adapted to a different environment. Thus, the questions arise as to how these adaptive capabilities were represented in their genomes and what was the original genome structure in the ancestral progenitor Bacillus species. 1803 putative proteins were identified as orthologs by comparative analyses with the three genomes of Bacillus-related species (see subset in Fig. 4A). These orthologs represent ∼44.0–51.2% of each genome. About 980 orthologs are located at a similar position on each genome (Fig. 4A). The physical distribution of common genes between O.iheyensis and B.halodurans is largely collinear, but the direction of collinearity changes at ∼30–40° from terC in both directions (Fig. 4A and B). A similar result has previously been documented in comparisons between B.subtilis and B.halodurans. It has been reported that the B.halodurans genome has inversions between the regions around 112–153° and 212–240°, due to the action of IS elements (6,18).
Figure 4.
Comparison of ortholog organization among the genomes of three Bacillus-related species. (A) In the case of 1803 orthologs identified in comparison with only three Bacillus-related species. (B) In the case of 354 orthologs identified in comparison with all five Gram-positive species (OB/BH/BS in Fig. 2). The dots are plotted at the positions where the gene for an orthologous protein exists when comparing two genomes. The scale of 360°, beginning with 0 at the dnaA locus (oriC), is based on the B.halodurans and B.subtilis maps. OB, O.iheyensis; BH, B.halodurans; BS, B.subtilis, SA, S.aureus; CA, C.acetobutylicum.
354 orthologs were identified only among the three Bacillus-related species when the proteins deduced from all CDSs of the five genomes were compared (see subset in Fig. 4B), indicating that these orthologs may reflect a basic gene set for constructing an ancestral Bacillus species genome backbone. 158 (44.6%) of these orthologs were located at similar positions on each genome (Fig. 4B). Most of the collinear orthologs among the three Bacillus-related species were assigned to eight major functional categories: membrane bioenergetics, motility and chemotaxis, sporulation, germination, transformation and competence, RNA modification, aminoacyl-tRNA synthetases and adaptation to atypical conditions.
Candidate genes involved in the alkaliphilic phenotype
Generally, alkaliphilic Bacillus strains cannot grow or grow poorly under neutral pH conditions, but grow well at a pH higher than 9.5 (19). Oceanobacillus iheyensis grows up to pH 10 with a broad pH optimum for growth ranging from 7.0 to 9.5 (5). Over the past three decades alkaliphilic microorganisms have been studied to elucidate their mechanisms of adaptation to alkaline environments. As shown in Figure 2A and B, 243 orthologs were identified between only the two alkaliphiles (O.iheyensis and B.halodurans) and 76 genes among these orthologs were grouped into category 1.2. These included various ABC transporters, transporters associated with C4-dicarboxylate, organic osmotic solute transport and Na+ uptake (Fig. 3). The ABC transporters could be categorized into three groups based on substrate specificity. All five genes encoding ABC transporters for branched chain amino acids identified in the O.iheyensis genome show orthologuous relationships only to B.halodurans among the five Gram-positive species compared. These particular transporters could be important for alkaliphily. Branched chain amino acids such as leucine, isoleucine and valine are believed to be converted to l-glutamate in the presence of 2-oxoglutarate and pyridoxal 5-phosphate by a branched chain amino acid aminotransferase (20,21). This protein is widely distributed in various organisms, including O.iheyensis. Since l-glutamate should be negatively charged at pH values higher than its pKa (3.9 or 4.3), the converted l-glutamate and its accompanying proton could contribute to the acidification of the alkaliphilic Bacillus cytoplasm, the pH of which is maintained ∼8–8.5, despite a pH outside the cell is ∼10.5 (19,22). Twenty-one genes encoding oligopeptide ABC transporters were identified in the O.iheyensis genome, as well as 28 in the case of B.halodurans. This is in contrast to only 11 such genes in B.subtilis, although C.acetobutylicum has 29 (9). However, only six of these genes identified in O.iheyensis showed orthologous relationships between the two alkaliphiles. Similarly to the case of ABC transporters for branched chain amino acids, these transport systems presumably work in the uptake of oligopeptides during growth under alkaline pH conditions. Thus, they can contribute to the acidification of the cytoplasm if the acidic amino acids, such as glutamate and aspartate, are released by digestion of the incorporated oligopeptide by a peptidase or are converted to free amino acids from other digests by an aminotransferase. These acidic amino acids are an important source of protons and a resource for acidic polymers in the cell wall.
The cell wall is crucial in alkaliphilic Bacillus species because their protoplasts lose stability in alkaline environments (19). Alkaliphilic Bacillus species contain certain acidic polymers containing galacturonic acid, gluconic acid, glutamate, asparatate and phosphoric acid. It is known that the amount of acidic polymers increases during growth under alkaline pH conditions. The Donnan equilibrium theory of electrolytes between an anionic polymer layer and the bulk aqueous phase was applied to cell wall systems and a pH reduction at the cell wall–membrane boundary has been shown (23). According to this calculation, the pH values estimated to exist inside a polymer layer (cell wall) are more acidic than those of the surrounding environment by 1–1.5 U. Teichuronopeptide (TUP), which is a co-polymer of polyglutamate and polyglucuronic acid, is one of the important components in the cell wall of the alkaliphilic B.halodurans (19) and actually contributes to the regulation of pH homeostasis in the cytoplasm. The putative protein (OB2920) showing significant similarity to the tupA gene product involved in TUP biosynthesis in B.halodurans (6) is only shared between the two alkaliphiles (Fig. 5). No homolog of tupA has been identified in any other organism, except the two alkaliphiles, thus far.
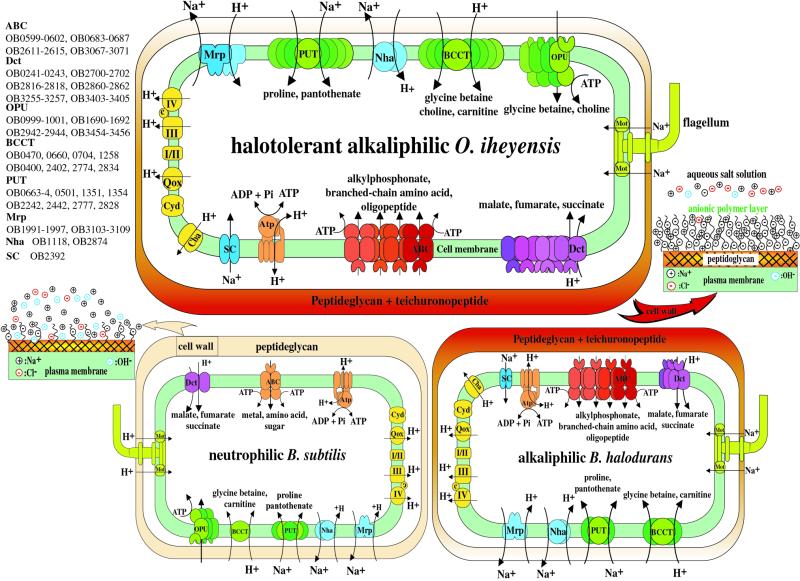
Figure 5.
Overview of putative major transport systems that govern alkaliphily and extreme halotolerance in O.iheyensis. Overview of the cell wall system and the enzymes involved in the respiratory chain (yellow) are also shown. Transporters are grouped by their category: sodium cycle (blue), organic osmotic solutes such as glycine betaine, choline, carnitine, proline and pantothenate (green), carboxylic acids such as malate, fumarate and succinate (purple) and peptides and amino acids (red). ABC transport systems are shown as composite figures of oval, circular and sickle shape. The gene number of O.iheyensis corresponding to each component of the transporter is described below or above each figure. Atp, F1F0 ATP synthase; ABC, ABC transporter; BCCT, glycine betaine, carnitine/H+ symporter; Cba, cytochrome c oxidase (bo3-type); Cyd, cytochrome bd oxidase; I, NADH dehydrogenase; II, succinate dehydrogenase; III, menaquinol cytochrome c oxidoreductase; IV, cytochrome caa3 oxidase; Dct, C4-dicarboxylate transport system; Mrp, Na+/H+ antiporter; Mot, channel for energization of motility; Nha, Na+/H+ antiporter; OPU, glycine betaine ABC transporter; PUT, proline/Na+ symporter and pantothenate/Na+ symporter; and Qox, cytochrome aa3 quinol oxidase; Sc, voltage-gated sodium channel.
Likewise, the alkylphosphonate ABC transporters shared only between the two alkaliphiles are not found in the other three Gram-positive species. Four genes encoding alkylphosphonate ABC transporters (two permeases and one ATP-binding and one phosphonate-binding protein) are shared between O.iheyensis and B.halodurans. Escherichia coli has been known for some time to cleave carbon–phosphorus (C-P) bonds in unactivated alkylphosphonates (24). When an alkylphosphonate, such as methylphosphonate, is degraded by bacterial C-P lyase (CH3PO32– → CH4 + HPO42–), the resultant monohydrogen phosphate ion is utilized as a source of phosphorus for growth (25). On the other hand, in the case of mitochondria, monohydrogen phosphate ion is an important substrate to exchange itself for dicarboxylates such as malate and succinate, by means of the dicarboxylate carrier (26). It is unclear whether these alkylphosphonate ABC transporters contribute to the regulation of pH homeostasis of the bacterial cytoplasm because the C-P lyase gene has not been identified in the genome of either alkaliphile.
Oceanobacillus iheyensis possesses 18 genes in total encoding six sets of C4-dicarboxylate carriers, which are all grouped into the DctA family (27). Of these, seven genes, encoding two sets of C4-dicarboxylate carriers and one permease large protein, are shared only between O.iheyensis and B.halodurans (Fig. 5). The other 11 genes, encoding three C4-dicarboxylate binding proteins, two permease large proteins and two permease small proteins, are unique among the five Gram-positive species. Although the seven shared proteins showed highest similarity among the alkaliphiles, the other 11 unique proteins showed significant similarity to those of Gram-negative strains such as Sinorizobium meiloti, Salmonella typhimurium and Pseudomonas aeruginosa. In aerobic bacteria, dicarboxylate transport carriers catalyze uptake of C4-dicarboxylates by a H+ or Na+ symport (27). This transport system may play a role in the regulation of pH homeostasis and the sodium cycle of the alkaliphilic bacterial cytoplasm.
The Na+ cycle plays an important role in the remarkable capacity of aerobic alkaliphilic Bacillus species (22) for pH homeostasis. The capacity for pH homeostasis directly determines the upper pH limit of growth. The first major actor in the alkaliphile Na+ cycle is the Na+/H+ antiporter, which achieves net H+ accumulation coupled to Na+ efflux. The Na+/H+ antiport activity is also an important mechanism for maintaining low intracellular sodium concentrations in halophilic or halotolerant aerobic bacteria (28). The major antiporter for pH homeostasis is thought to be the mrp encoded antiporter, first identified in B.halodurans. Mrp could function as part of a complex with six other gene products. The O.iheyensis genome possesses 14 genes encoding two sets of Mrp-related proteins and all 14 genes showed various orthologous relationships among four of the five Gram-positive species, but not C.acetobutylicum. The genome contains two genes for additional antiporters, including NhaC, a property that is shared among all five Gram-positive species. These antiporters presumably play fundamental roles in the pH homeostasis of the bacterial cell.
The second major actor of the Na+ cycle is Na+ re-entry via the Na+/solute symporter and presumably the ion channel associated with the Na+-dependent flagella motor (22). Two genes encoding a Na+/solute symporter and a Na+-dependent transporter are shared only between O.iheyensis and B.halodurans. These alkaliphile-specific transporters could also be important for Na+ re-entry, although there is no experimental evidence for such a function under alkaline pH conditions. Recently, the first prokaryotic voltage-gated sodium channel was discovered in the alkaliphilic B.halodurans, based initially on the sequences of transmembrane domains (29). It was confirmed experimentally that the channel is activated by voltage and is selective for sodium, although it is blocked by calcium channel blockers. A putative protein (OB2392) identified in O.iheyensis shows significant similarity to this sodium channel. Because this sequence has not been detected in any other prokaryotic genome except for the two alkaliphiles, voltage-gated channels could be a common property of alkaliphilic Bacillus-related species. This channel likely provides a transient supply of Na+ for the sodium cycle under both alkaline and neutral pH conditions.
In total, 32 putative proteins associated with the flagella were identified in the O.iheyensis genome and all of these proteins, except for one, were well conserved among the three Bacillus-related species. Alkaliphile motility is Na+ coupled and is observed only in cells growing at the high alkaline end of the pH range for growth (22). In fact, O.iheyensis is extremely motile under alkaline pH conditions. Therefore, these flagella-associated proteins identified in the O.iheyensis genome are likely to work as Na+ channels for Na+ entry at high pH to ensure an adequate supply of Na+ for the increased antiport coupled to H+ accumulation (Fig. 5). This is in contrast to the H+-driven flagella motors present in the neutrophilic B.subtilis (30). On the other hand, there are no major differences in the proteins associated with the terminal oxidase of the respiratory chain between the neutrophilic B.subtilis and the other two alkaliphilic bacilli, although B.subtilis is missing cytochrome bd oxidase and this enzyme is conserved in both alkaliphiles as well as the thermophile Geobacillus stearothermophilus (http://www.genome.ou.edu/bstearo.html).
We have highlighted the genes involved in the alkaliphilic phenotype based on comparative analysis with three Bacillus species and two other Gram-positive species through this paper. However, out of 243 genes shared only between the two alkaliphiles (OB/BH), 120 genes are still functionally unknown, as shown in Figure 3. Therefore, it will be necessary to analyze the gene expression pattern under alkaline and neutral pH conditions to clarify the genes responsible for the alkaliphilic phenotype.
Candidate genes involved in the halotolerant phenotype
Bacteria subjected to sudden increases in osmolarity respond with an adaptive response that is characterized by two distinct phases. The key physiological role of compatible solute accumulation as an adaptive response of B.subtilis, which can grow in medium containing NaCl up to 7% (31), to high osmolarity environments has been firmly established. Bacillus subtilis responds to a sudden increase in external osmolarity by an initial rapid uptake of K+, followed by the accumulation of large amounts of the compatible solute proline through de novo synthesis. The genome of O.iheyensis, which can grow at salinities of 0–21% (3.6 M) at pH 7.5 and 0–18% (3.1 M) at pH 9.5 (5), contains two putative proteins showing significant similarity to both Na+ and K+ transporters from various bacterial species (32). These two proteins could play a major role in the initial rapid uptake of K+. Bacillus subtilis uses a high affinity, substrate-specific transport system, OpuE (osmoprotectant uptake), to transport proline efficiently for osmoprotective purposes (32). The OpuE transporter consists of a single component and is a member of the sodium/solute symporter family (SSF). The O.iheyensis genome possesses five genes encoding Na+/proline symporters (there are four in B.subtilis) as well as three more genes for Na+/pantothenate symporters (there is only one in B.subtilis), which is another member of the SSF. Bacillus subtilis employs transporters to scavenge glycine betaine from the environment up to cellular levels that surpass 1 M in osmotically stressed cultures (32). Glycine betaine can also be synthesized when the precursor choline is available to the cell. The glycine betaine and choline ABC transport systems are composed of an ATP-binding protein, permease and substrate-binding protein, while a secondary glycine betaine transporter is composed of a single component. There is not a great difference in the glycine betaine and choline ABC transport systems between the moderately halotolerant B.subtilis and extremely halotolerant O.iheyensis, although O.iheyensis has one more set of glycine betaine ABC transport systems (Fig. 5). The principal difference is found in the glycine betaine transporters as well as in the choline/H+ and carnitine/H+ symporters between the moderate and extremely halotolerant bacilli. In contrast to only one glycine betaine transporter present in B.subtilis, O.iheyensis possesses six genes encoding glycine betaine transporters and one each for choline/H+ and carnitine/H+ symporters. Oceanobacillus iheyensis must fully employ a large number of osmoprotectant transporters (Fig. 5) in order to survive in neutral or alkaline hypersaline environments that surpass 3 M NaCl.
CONCLUSION
The genomic sequence of O.iheyensis should facilitate the study of the model transport systems in alkaliphiles and extremely halotolerant bacteria. Comparative genomics among several species showing different adaptive capabilities in species of the genus Bacillus and other closely related genera will provide a better understanding of the features of the ancestral Bacillus species prior to its diversification. The genome sequence may also provide important clues to the understanding of the mechanisms of adaptation to extreme environments and the stress response at different levels through the analyses of metabolic and regulatory networks. Finally, O.iheyensis could reveal some properties unique to members of the benthic microbial community in the oligotrophic deep-sea sediment because cell growth of this strain occurs at pressures of up to 30 MPa, corresponding to the pressure at a depth of ∼3000 m. A new database specifically established for the O.iheyensis sequences, ExtremoBase, will be accessible through the World Wide Web server at http://www.jamstec.go.jp/jamstec-e/bio/jp/topj.htm. The sequence has been deposited in DDBJ/EMBL/GenBank with the accession nos AP004593–AP004605.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to M. Tsudome, H. Oida, S. Shimamura, H. Suzuki, S. Nishi, Y. Shen, S. Goda, Y. Hidaka and A. Matsuki for their technical assistance. We thank Dr G. C. Chee and S. Matsui for computing assistance. Thanks are also due to Drs H. Hirakawa and S. Kuhara of Kyushu University for their help with drawing the genome maps. Finally we thank Drs K. Horikoshi of JAMSTEC and D. Bartlett and D. Valentine of the University of California, San Diego for their useful suggestions and critical reading of the manuscript.
DDBJ/EMBL/GenBank accession nos AP004593–AP004605
REFERENCES
- 1.Takami H., Inoue,A., Fuji,F. and Horikoshi,K. (1997) Microbial flora in the deepest sea mud of Mariana Trench. FEMS Microbiol. Lett., 152, 279–285. [DOI] [PubMed] [Google Scholar]
- 2.Takami H., Kobata,K., Nagahama,T., Kobayashi,H., Inoue,A. and Horikoshi,K. (1999) Biodiversity in deep-sea sites near the south part of Japan. Extremophiles, 3, 97–102. [DOI] [PubMed] [Google Scholar]
- 3.Priest F.G. (1993) Systematics and ecology of Bacillus. In Sonenshein,A.L., Hoch,J.A. and Losick,R. (eds), Bacillus subtilis and Other Gram-Positive Bacteria. ASM Press, Washington, DC, pp. 3–16.
- 4.Takami H. (1999) Isolation and characterization of microorganisms from deep-sea mud. In Horikoshi,K. and Tsujii,K. (eds), Extremophiles in Deep-Sea Environments. Springer-Verlag, Tokyo, Japan, pp. 3–26.
- 5.Lu J., Nogi,Y. and Takami,H. (2001) Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol. Lett., 205, 291–297. [DOI] [PubMed] [Google Scholar]
- 6.Takami H., Nakasone,K., Takaki,Y., Maeno,G., Sasaki,R., Masui,N., Fuji,F., Hirama,C., Nakamura,Y., Ogasawara,N. et al. (2000) Complete genome sequence of alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with B.subtilis. Nucleic Acids Res., 28, 4317–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunst F., Ogasawara,N., Mozer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S. et al. (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 8.Kuroda M., Ohta,T., Uchiyama,I., Baba,T., Yuzawa,H., Kobayashi,I., Cui,L., Oguchi,A., Aoki,K., Nagai,Y. et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet, 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 9.Nölling J., Breton,G., Omelchenko,M.V., Makarova,K.S., Zeng,Q., Gibson,R., Lee,H.M., Dubois,J., Qiu,D., Hitti,J. et al. (2001) Complete sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol., 183, 4823–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakiyama T., Takami,H., Ogasawara,N., Kuhara,S., Kozuki,T., Doga,K., Ohyama,A. and Horikoshi,K. (2000) An automated system for genome analysis to support microbial whole-genome shotgun sequencing. Biosci. Biotech. Biochem., 64, 670–673. [DOI] [PubMed] [Google Scholar]
- 11.Yada T., Totoki,Y., Takagi,T. and Nakai,K. (2001) A novel bacterial gene-finding system with top-class accuracy in locating start codons. DNA Res., 8, 97–106. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 15.Jones D.T., Taylor,W.R. and Thornton,J.M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci., 8, 275–283. [DOI] [PubMed] [Google Scholar]
- 16.Sokal R. and Sneath,P.H.A. (1963) Principles of Numerical Taxonomy. W.H. Freeman and Co., San Francisco, CA.
- 17.Mahillon J. and Chandler,M. (1998) Insertion sequence. Microbiol. Mol. Biol. Rev., 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takami H., Han,C.G., Takaki,Y. and Ohtsubo,E. (2001) Identification and distribution of new insertion sequences in the genome of alkaliphilic Bacillus halodurans C-125. J. Bacteriol., 183, 4345–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horikoshi K. (1999) Alkaliphiles. Haward Academic, Amsterdam, The Netherlands.
- 20.Schadewaldt P., Hummel,W., Wendel,U. and Adelmeyer,F. (1995) Enzymatic method for determination of branched-chain amino acid aminotransferase activity. Anal. Biochem., 230, 199–204. [DOI] [PubMed] [Google Scholar]
- 21.Kagamiyama H. and Hayashi,H. (2000) Branched-chain amino acid aminotransferase of E. coli. Methods Enzymol., 324, 103–113. [DOI] [PubMed] [Google Scholar]
- 22.Krulwich T.A., Ito,M. and Guffanti,A.A. (2001) The Na+-dependence of alkaliphily in Bacillus. Biochim. Biophys. Acta, 1505, 156–168. [DOI] [PubMed] [Google Scholar]
- 23.Tsujii K. (2002) Donnan equilibria in microbial cell walls: a pH-homeostatic mechanism in alkaliphiles. Colloids Surf. Biointerfaces, 24, 247–251. [Google Scholar]
- 24.Chen C.M., Ye,Q.Z., Zhu,Z., Wanner,B.L. and Walsh,C.T. (1990) Molecular biology of carbon-phosphorus bond cleavage. J. Biol. Chem., 265, 4461–4471. [PubMed] [Google Scholar]
- 25.Murata K., Higaki,N. and Kimura,A. (1988) Detection of carbon-phosphorus lyase activity in cell free extracts of Enterobacter aerogenes. Biochem. Biophys. Res. Commun., 57, 190–195. [DOI] [PubMed] [Google Scholar]
- 26.Krämer R. and Palmieri,F. (1992) Metabolite carriers in mitochondria. Molecular. In Ernster,L. (ed.), New Comprehensive Biochemistry: Mechanisms in Bioenergetics. Elsevier Science B.V., Amsterdam, The Netherlands, Vol. 23, pp. 359–384.
- 27.Janausch I.G., Zientz,E., Tran,Q.H., Kröger,A. and Unden,G. (2002) C4-dicarboxlylate carriers and sensors in bacteria. Biochim. Biophys. Acta, 1553, 39–56. [DOI] [PubMed] [Google Scholar]
- 28.Ventosa A., Nieto,J. and Oren,A. (1998) Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev., 62, 504–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren D., Navarro,B., Xu,H., Yue,L., Shi,Q. and Clapham,D.E. (2001) A prokaryotic voltage-gated sodium channel. Science, 294, 2372–2375. [DOI] [PubMed] [Google Scholar]
- 30.Shioi J.I., Matsuura,S. and Imae,Y. (1980) Quantitative measurements of proton motive force and motility in Bacillus subtilis. J. Bacteriol., 144, 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sneath P.H.A., Mair,N.S., Sharp,M.E. and Holt,J.G. (1986) Bergey’s Manual of Systematic Bacteriology, Vol. 2. Wiliams and Wikins, Baltimore, MD.
- 32.Kempf B. and Bremer,E. (1998) Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol., 170, 319–330. [DOI] [PubMed] [Google Scholar]