Abstract
To understand the specific genetic instabilities associated with deficiencies in RecQ family helicases, we have studied the substrate preferences of two closely related members of this family, human BLM and Saccharomyces cerevisiae Sgs1p. Here we show that both BLM and Sgs1p preferentially unwind G4 DNA relative to Holliday junction substrates, and that substrate preference reflects binding affinity and maps to the conserved central helicase domain. We identify the porphyrin N-methyl mesoporphyrin IX (NMM) as a specific inhibitor of G4 DNA unwinding, and show that in the presence of NMM the helicase becomes trapped on the NMM–G4 DNA complex, consuming ATP but unable to unwind or dissociate. These results suggest that BLM and Sgs1p function proactively in replication to remove G4 DNA structures which would otherwise present obstacles to fork progression, rather than by promoting recombination to restart a fork that has stalled.
INTRODUCTION
The RecQ helicases comprise a small family of enzymes present in prokaryotes and eukaryotes. These helicases share a central domain approximately 400 amino acids in length, which includes the conserved helicase motifs bearing the DEXH signature sequence and a Walker A and B box which confer the ability to bind and hydrolyze ATP. Among the best-studied members of this helicase family are the eponymous RecQ from Escherichia coli, Sgs1p from Saccharomyces cerevisiae, Rqh1 of Schizosaccharomyces pombe, and human BLM, WRN and RECQL4. The RecQ family helicases that have been purified and characterized in vitro all unwind DNA with a 3′ to 5′ polarity in a reaction dependent on Mg2+ and ATP.
Eukaryotic helicases of the RecQ family are required for the maintenance of genomic stability, and helicase deficiency produces both characteristic pathology and characteristic genetic instability. In humans, the absence of BLM helicase activity results in Bloom’s syndrome, a genetic disease characterized by elevated levels of sister chromatid exchange and formation of quadriradials, and early onset of malignancy (1). Deficiency in WRN activity results in Werner’s syndrome, a disease characterized by chromosomal deletions and translocations, and premature aging (2,3). Deficiency in RECQL4 has been linked to Rothmund–Thomson syndrome (4), characterized by chromosomal mosaicism, development of osteosarcomas (usually a rare tumor), and other symptoms including telangiectasia. Saccharomyces cerevisiae strains carrying mutations in SGS1 display increased mitotic recombination, defective chromosome segregation and reduced proliferative capacity, a phenotype which may be thought of as analogous to premature aging in humans lacking WRN (5,6).
RecQ family members are unusual in that they can unwind not only B-form DNA but also alternatively structured DNA molecules, including G4 DNA, synthetic D-loops and Holliday junctions (HJ) (7–12). G4 DNA is a four-stranded structure in which the repeating unit is a G-quartet, a planar array of guanines stabilized by hydrogen bonds (Fig. 1). Even though G4 DNA is not a substrate for all helicases, both yeast Sgs1p and its close human homolog, BLM, unwind G4 DNA with at least 15-fold preference relative to duplex substrates (8). G4 DNA unwinding activity almost certainly contributes to the function of SGS1 in the maintenance of two G-rich genomic domains, the rDNA and the telomeres. SGS1 is required for recombination-dependent telomere maintenance in cells lacking telomerase (13–15). In addition, Sgs1p localizes to the nucleolus, where rDNA is transcribed and rRNA biogenesis occurs; sgs1-deficient strains are characterized by nucleolar fragmentation and production of rDNA circles.
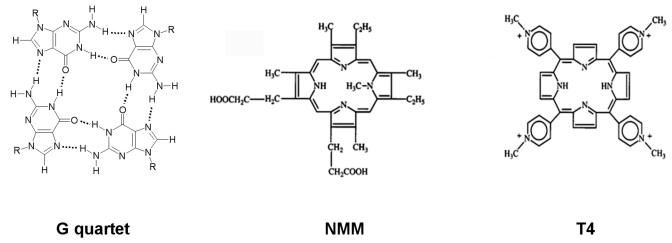
Figure 1.
G4 DNA and porphyrins that bind it. A G quartet and porphyrin derivatives NMM and T4 (also referred to as TMPyP4 or H2TMPyP) are shown drawn to scale.
Both BLM and another human RecQ family member, WRN, are able to unwind HJ, which represent an intermediate in homologous recombination (10,16). This has led to suggestions that BLM and WRN reduce the level of recombinational repair by disrupting potentially recombinogenic molecules that arise after replication fork arrest (10,16). Some cooperation between BLM and WRN is supported by evidence of their physical interaction in vivo (17). However, complete redundancy of function would not account for the contrasting patterns of genetic instability that characterize BLM- and WRN-deficient cells.
In order to understand RecQ family helicase function in maintaining genomic stability, we have sought to learn more about the substrate specificity of these enzymes, and to identify compounds which inhibit helicase activity on specific classes of substrates and could be used as probes of function in vivo. Here we report that both BLM and S.cerevisiae Sgs1p preferentially unwind G4 DNA relative to HJ. This preferential unwinding can be explained by a 15-fold difference in binding affinities for G4 DNA relative to HJ. The high affinities of BLM and Sgs1p for G4 DNA (kD = 5 nM) predict that a specialized and conserved domain mediates this interaction. We further show that the anionic porphyrin N-methyl mesoporphyrin IX (NMM; Fig. 1) is a highly specific inhibitor of G4 DNA unwinding, and provide a model for how inhibition occurs. These data suggest that the principle function of BLM and Sgs1p may be to provide the replication apparatus with an unstructured template, rather than to promote recombinational repair of a collapsed fork.
MATERIALS AND METHODS
Preparation of enzymes and substrates
BLM and Sgs1p were purified as described (18,19), with several modifications. The BLM expression construct pJK1 was transformed into strain BCY123 (20) for isogenic expression of Sgs1p and BLM. Starter cultures of BCY123 harboring C-terminal His6-tagged expression plasmids for Sgs1p (pRS222, expressing a polypeptide 869 amino acids in length, including residues 400–1268 which span the conserved helicase domains) or BLM (pJK1, expressing full-length human BLM, 1417 amino acids) were grown in SC-Ura media at 30°C to saturation, then inoculated into 6 l of complete media (1% yeast extract, 2% peptone) supplemented with 2% (v/v) lactic acid and 3% (v/v) glycerol, at final pH 6.5. Expression was induced at OD600 = 0.25 by the addition of galactose to a final concentration of 2% (w/v). Cells were incubated 12 h longer, opened, and recombinant proteins purified by nickel chelate chromatography.
Oligonucleotides TP (TGGACCAGACCTAGCAGCTATGGGGGAGCTGGGGAAGGTGGGAATGTGA); J1 (GACGCTGCCGAATTCTGGCGTTAGGAGATACCGATAAGCTTCGGCTTAA); J2 (CTTAAGCCGAAGCTTATCGG TATCTTGCTTACGACGCTAGCAAGTGATC); J3 (TGATCACTTGCTAGCGTCGTAAGCAGCTCGTGCTGTCTAGAGACATCGA); J4 (ATCGATGTCTCTAGACAGCACG AGCCCTAACGCCAGAATTCGGCAGCGT); H1 [GCATCGGCTTCCCAACTAGCT10]; and K1 [T30GCTAGTTGGGAAGCCGATGC] were synthesized at the Keck facility (Yale University). G4 DNA was formed from TP and its structure confirmed as described previously (7). Synthetic HJ were formed by annealing equimolar amounts of J1, J2, J3 and J4, and the partial-duplex substrate H1/K1 by annealing equimolar amounts of H1 and K1. All DNA substrates were gel-purified prior to use. DNA recovery was quantified by measuring the A260 of denatured DNA. Molar amounts of G4 DNA refer to moles of four-stranded structures (1 mol of G4 = 4 mol of single strands). HJ and H1/K1 were stored in TE (10 mM Tris, 1 mM EDTA, pH 7.4), and G4 DNA was stored in TE containing 10 mM KCl. Substrates were end labeled with [γ-32P]ATP and T4 polynucleotide kinase (Promega) for binding and unwinding assays.
Unwinding and binding reactions
Unwinding assays were carried out in 20 µl reactions containing 50 mM Tris–HCl, pH 7.4, 2 mM MgCl2, 2 mM ATP, 50 mM NaCl, and 100 µg/ml BSA, 1 mM DTT, 1 ng enzyme, and 10 fmol 32P-labeled DNA at 37°C for 30 min unless otherwise indicated. BLM and Sgs1p were treated with 0.5 mM DTT for 5 min on ice prior to addition. Reactions were stopped by adding 5 mM EDTA, pH 8.0 and 0.5 mg/ml proteinase K (final concentrations), and deproteinized by incubation for 10 min at 37°C. Products were analyzed by electrophoresis on 8% polyacrylamide gels in 0.5× TBE containing 10 mM KCl. Binding reactions were carried out in 300 mM NaCl, 50 mM Tris pH 7.4, 2 mM MgCl2, 2 mM ATP-γ-S, 1 mM DTT, 10 fmol of 32P-labeled DNA and indicated quantities of enzyme, and incubated at room temperature for 20 min. Following the addition of 0.1 vol of 50% glycerol, binding reactions were loaded onto a 5% native gel in 0.5× TBE, 10 mM KCl, and free nucleic acids and nucleoprotein complexes were resolved by electrophoresis at 6.5 V/cm for 4 h at room temperature. Binding was quantified by phosphoimaging, and kD values calculated as averages from at least three separate experiments. The inhibitors NMM and meso-tetra(N-methyl-4-pyridyl)porphine tetra tosylate (T4; Frontier Scientific, Logan, UT; Fig. 1) were made up as aqueous solutions. The ki was determined as the concentration of inhibitor required to reduce unwinding by 50%.
ATPase assays
For ATPase assays, 50 µl reactions were incubated under standard unwinding conditions and then aliquoted. One portion (10 µl) was used for quantification of unwinding as above, and the remainder assayed for release of free inorganic phosphate (21). In brief, 5, 10 or 20 µl of the reaction was combined with 800 µl of color reagent [0.03% (w/v) malachite green (Sigma) and 1.05% ammonium molybdate in 1 N HCl and 0.1% (v/v) Triton X-100], incubated for 1 min at room temperature, and then the reaction was stopped by the addition of 100 µl of 34% citric acid. A650 readings were compared with a standard curve prepared from 1 mM KH2PO4 in 0.01 N H2SO4.
RESULTS
BLM preferentially unwinds G4 DNA, not HJ
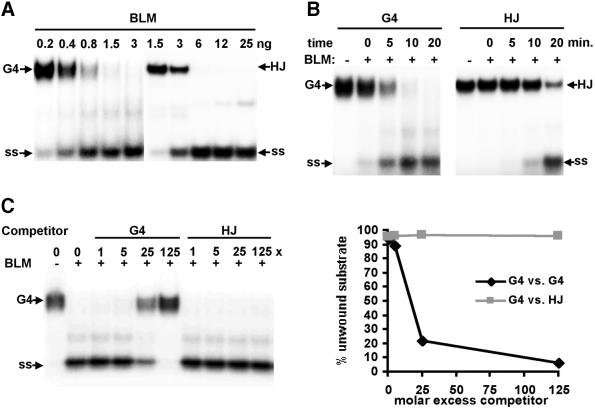
Human BLM helicase and S.cerevisiae Sgs1p both display at least a 15-fold preference for unwinding G4 DNA relative to standard duplex DNA substrates (7,8). BLM can also unwind HJ (10,11), raising the question of which substrate—if either—is unwound preferentially. We assayed substrate preference by enzyme titration, kinetics and direct competition. As shown in Figure 2A, 50% of the G4 DNA substrate (10 fmol) was unwound by 0.8 ng of BLM, while at least 7-fold more enzyme was required to unwind a comparable amount of HJ substrate. The rate of unwinding of G4 DNA was 4-fold more rapid than the unwinding of HJ (Fig. 2B). As further evidence of substrate preference, unlabeled HJ was not able to compete with G4 DNA unwinding by BLM, even at a 125-fold molar excess of competitor (Fig. 2C).
Figure 2.
Preferential unwinding and binding of G4 DNA by BLM. (A) Native gel analysis of products of unwinding of 32P-labeled G4 DNA and HJ substrates by BLM. Amounts of enzyme per reaction are indicated above the autoradiograph. Arrows indicate G4 and HJ substrates and single-stranded unwinding products (ss). (B) Kinetics of unwinding of 32P-labeled G4 DNA and HJ, measured by native gel analysis. Products of reactions containing 5 ng of BLM were incubated at 37°C for the times indicated. (C) Unwinding of 32P-labeled G4 DNA in the presence of unlabeled G4 DNA or HJ competitor. Reactions containing 1 ng of BLM were incubated in the presence of the indicated molar excess of competitor and resolved on a native gel (left). Data are graphed on the right.
Sgs1p preferentially unwinds G4 DNA, not HJ
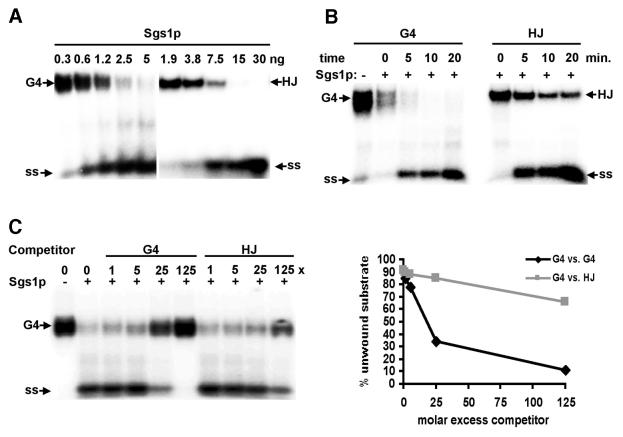
Both BLM (10) and WRN (16) unwind HJ, but the activity of Sgs1p had not been tested on this substrate. Using standard assay conditions, we showed that Sgs1p does unwind HJ, but is several-fold more active on G4 DNA (Fig. 3A). The rate of G4 DNA unwinding was 3-fold more rapid than unwinding of HJ (Fig. 3B), and unlabeled HJ competed poorly in direct competition assays (Fig. 3C). Thus, G4 DNA is the preferred substrate for Sgs1p.
Figure 3.
Preferential unwinding of G4 DNA by Sgs1p. (A) Native gel analysis of products of unwinding of 32P-labeled G4 DNA and HJ substrates by Sgs1p. Amounts of enzyme per reaction are indicated above the autoradiograph. Arrows indicate G4 and HJ substrates and single-stranded unwinding products (ss). (B) Kinetics of unwinding of 32P-labeled G4 DNA and HJ, measured by native gel analysis. Products of reactions containing 5 ng of Sgs1p were incubated at 37°C for the times indicated. (C) Unwinding of 32P-labeled G4 DNA in the presence of unlabeled G4 DNA or HJ. Reactions containing 5 ng of Sgs1p were incubated in the presence of the indicated molar excess of competitor and resolved on a native gel (left). Data are graphed on the right.
The recombinant Sgs1p used in these experiments lacked several hundred residues from both the N- and C-termini. Nonetheless, it is comparably active on G4 DNA to full- length BLM (compare Figs 2A and 3A). Thus, the domains responsible for G4 DNA unwinding must map to the central conserved region of these helicases.
Binding affinity determines substrate preference
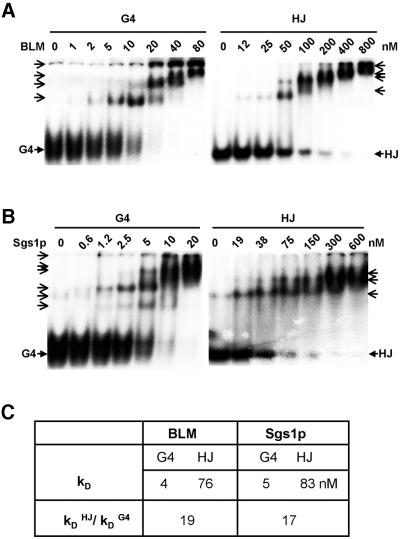
To ask if preferential unwinding of G4 DNA reflects preferential binding to this structure, we measured binding affinities by gel mobility shift assays. As shown in Figure 4A, BLM bound G4 DNA tightly (kD = 4 nM), while binding to HJ was much weaker (kD = 76 nM). Similarly, Sgs1p bound G4 DNA tightly (kD = 5 nM), and bound poorly to HJ (kD = 83 nM; Fig. 4B). In all cases, slowly migrating protein–DNA complexes appear at high protein concentrations, which may reflect helicase aggregation on the DNA substrates. As summarized in Figure 4C, the affinity of BLM for G4 DNA is 19-fold greater than its affinity for HJ, and, similarly, the affinity of Sgs1p for G4 DNA is 17-fold greater than its affinity for HJ. Differences in binding affinity may largely account for the substrate preference documented in unwinding assays.
Figure 4.
Preferential binding of BLM and Sgs1p to G4 DNA. (A) Gel mobility shift analysis of BLM binding to 32P-labeled G4 DNA and HJ at indicated protein concentrations. Positions of free G4 DNA and HJ are indicated by solid arrows. Open arrows indicate protein–DNA complexes. (B) Gel mobility shift analysis of Sgs1p binding to 32P-labeled G4 DNA and HJ at indicated protein concentrations. Symbols as in (A). (C) Summary of binding affinity, comparing kD for each helicase binding to G4 DNA and HJ, and showing relative affinities of binding to G4 DNA and HJ.
NMM specifically inhibits G4 DNA unwinding
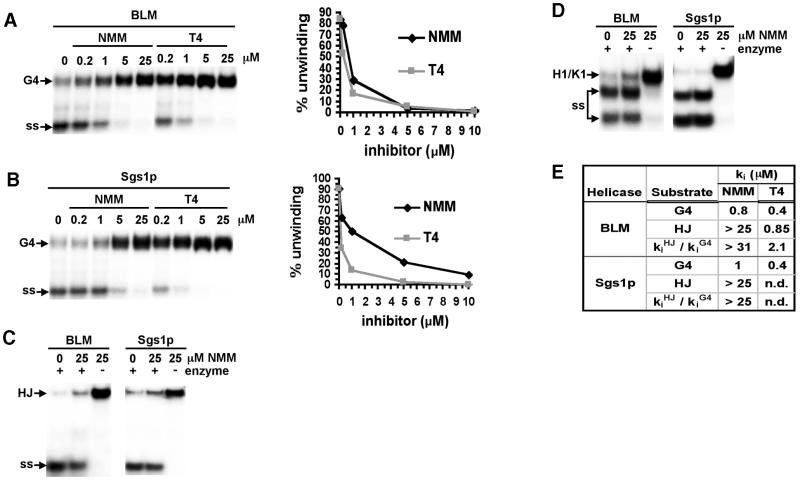
Two porphyrin derivatives, NMM and the related compound T4 (Fig. 1), have been shown to bind to G4 DNA (22–25), and NMM has proven to be a specific inhibitor of G4 DNA unwinding by E.coli RecQ helicase (26). Therefore, we assayed the effects of these compounds on DNA unwinding by the eukaryotic RecQ helicases, BLM and Sgs1p. Low concentrations of either NMM or T4 completely inhibited G4 DNA unwinding by both BLM helicase (NMM, ki = 0.8 µM; T4, ki = 0.4 µM; Fig. 5A) and Sgs1p (NMM, ki = 1 µM; T4, ki = 0.4 µM; Fig. 5B).
Figure 5.
Inhibition of G4 DNA unwinding by NMM and T4. (A) Unwinding of 32P-labeled G4 DNA by BLM was assayed in the absence and presence of NMM and T4 at indicated concentrations, and products resolved on native gels (left). Data are graphed on the right. Symbols as in Figure 2. (B) Unwinding of 32P-labeled G4 DNA by Sgs1p, assayed and graphed as in (A). (C) Unwinding of 32P-labeled HJ by BLM and Sgs1p in the presence of NMM. (D) Unwinding of 32P-labeled partial duplex substrate H1/K1 by BLM and Sgs1p in the presence of NMM. (E) Summary of analysis of inhibition, comparing ki (concentration of inhibitor required to diminish unwinding activity 50%) for each enzyme and substrate.
Strikingly, inhibition by NMM is specific for G4 DNA. As shown in Figure 5C, unwinding of HJ by BLM and Sgs1p was not inhibited by relatively high concentrations of NMM (25 µM), while that same concentration of T4 completely abrogated HJ unwinding by both enzymes. Further analysis (data not shown) demonstrated that T4 is comparably potent as an inhibitor of BLM unwinding of HJ (ki = 0.8 µM) and G4 DNA (ki = 0.4 µM). Additional evidence of the substrate specificity of NMM was obtained in experiments which analyzed inhibition of partial-duplex DNA unwinding (Fig. 5D): 25 µM NMM had no effect on unwinding of double-stranded DNA (dsDNA) by either BLM or Sgs1p, whereas the same concentration of T4 completely inhibited unwinding. The inhibition data, summarized in Figure 5E, show that NMM is a potent and highly specific inhibitor of G4 DNA unwinding by RecQ helicases, and that low concentrations of NMM do not affect unwinding of HJ or partial-duplex substrates.
NMM traps BLM on the G4 DNA substrate, where the enzyme consumes ATP without unwinding DNA
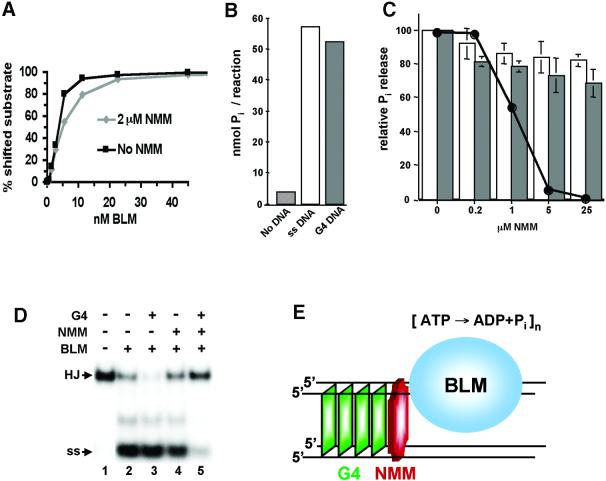
NMM could, in principle, inhibit G4 DNA unwinding by preventing binding to DNA. To address this possibility, we measured BLM binding to G4 DNA in the absence and presence of NMM. Comparison of binding curves showed that 2 µM NMM (2 × ki) did not interfere with enzyme–substrate complex formation (Fig. 6A). Alternatively, NMM could inhibit the hydrolysis of ATP that is necessary to drive translocation. To test this possibility, we assayed ATP hydrolysis in the presence and absence of inhibitor and G4 DNA or single-stranded DNA (ssDNA). BLM did not hydrolyze ATP in the absence of DNA, but both G4 DNA (50 nM) and ssDNA (200 nM) caused comparable stimulation of the enzyme’s ATPase activity, consistent with the equivalent concentrations of 3′ ends in these reactions (Fig. 6B). NMM did cause a small reduction in Pi release in reactions containing either G4 DNA or ssDNA substrates, but Pi release was only modestly reduced even at NMM concentrations which completely inhibited G4 DNA unwinding (Fig. 6C). Therefore, inhibition of ATPase activity does not account for inhibition of unwinding.
Figure 6.
Effect of NMM on G4 DNA binding and ATPase activity of BLM. (A) NMM does not alter BLM binding affinity for G4 DNA. Graph of results of gel mobility shift assays measuring binding of BLM to 32P-labeled G4 DNA, in the absence (squares) or presence (diamonds) of 2 µM NMM (2 × ki). (B) G4 DNA and ssDNA stimulate BLM ATPase activity. Pi release was measured in reactions containing no DNA (striped bar), 200 nM ssDNA (open bar) or 50 nM G4 DNA (shaded bar). (C) NMM does not alter BLM ATPase activity. Pi release was measured in reactions containing 200 nM ssDNA (open bars), or 50 nM G4 DNA (shaded bars). An unwinding inhibition curve (black line) derived from electrophoretic analysis of the same reactions is superimposed on the bar graphs. (D) The NMM–G4 DNA complex inhibits HJ unwinding. Unwinding of 32P-labeled HJ (1 pmol) by BLM (0.5 pmol) was assayed in the presence of unlabeled G4 DNA (0.2 pmol, lane 3); NMM (200 pmol, lane 4) or NMM–G4 DNA (lane 5). (E) Mechanism of inhibition of G4 unwinding by NMM. A helicase approaches the G4 structure from the 3′ end and encounters the masked face of the G4 stack. BLM continues to hydrolyze ATP but is unable to unwind or to dissociate from the substrate.
These observations suggested that the helicase could become trapped on the NMM–G4 DNA complex, unable to unwind and unable to dissociate. If so, then the presence of the NMM–G4 DNA complex should prevent BLM unwinding other substrates. This was tested by assaying the unwinding of 32P-labeled HJ in reactions containing G4 DNA alone, NMM alone, or the NMM–G4 DNA complex (Fig. 6D). BLM unwound the labeled HJ (1 pmol) almost to completion (Fig. 6D, lane 2), and unwinding was unaffected by the presence of low concentrations of unlabeled G4 DNA (0.2 pmol, or a 1:5 molar ratio of G4 DNA:HJ; Fig. 6D, lane 3) or 10 µM NMM (Fig. 6D, lane 4). However, when both G4 DNA (0.2 pmol) and NMM (10 µM) were added prior to the addition of enzyme, unwinding was severely impeded (Fig. 6D, lane 5). Thus, the helicase becomes effectively immobilized on the NMM–G4 DNA complex, unable to move along the G4 DNA or moving off to another substrate, but consuming ATP in a futile attempt to translocate. This model for inhibition is shown in Figure 6E.
DISCUSSION
We have shown that Sgs1p and BLM preferentially unwind G4 DNA relative to HJ substrates. Preferential unwinding reflects binding affinity: both BLM and Sgs1p bind G4 DNA tightly (kD = 5 nM), and display more than 15-fold lower affinity for HJ. Binding and unwinding activities were comparable in assays of full-length recombinant BLM (1417 residues) and truncated recombinant Sgs1p (869 residues), which contains the conserved central helicase domain but lacks both N- and C-terminal sequences (18). This maps G4 DNA recognition to the central helicase domain. While a number of structural domains have been identified that confer sequence-specific binding of polypeptides to duplex DNA, very little is yet known about protein domains that can determine the interaction with G4 DNA. The conserved central helicase domain of BLM and Sgs1p is a promising candidate for further analysis.
The potential of telomeric DNA to form G4 structures has fueled the idea that low molecular weight compounds that bind G4 DNA might prove useful in the treatment of disease (27). The porphyrin NMM is a substrate-specific inhibitor for RecQ family helicases, and may be a prototype for the design of compounds with such therapeutic applications. NMM specifically inhibits unwinding of G4 DNA by both BLM and Sgs1p (ki = 0.8 µM), but does not affect unwinding of HJ and dsDNA (ki >> 25 µM). NMM is similarly active in inhibiting G4 DNA unwinding by E.coli RecQ (26). The specificity of NMM for G4 DNA is not shared by all porphyrin derivatives. T4 (see Fig. 1) binds to G4 DNA and has been used for in vivo studies directed at understanding G4 DNA function (28,29). While low concentrations of T4 do inhibit G4 DNA unwinding (ki = 0.4 µM), comparable concentrations also inhibit unwinding of HJ and partial-duplex DNA substrates. This lack of specificity limits the potential usefulness of T4 in vivo. In contrast to most known compounds that interact with G4 DNA, NMM contains carboxyl groups which have the potential to be negatively charged at neutral pH. This may explain the unusual specificity of NMM: while positive charges will in principle stabilize drug–DNA interactions, they might diminish the ability of a compound to discriminate between nucleic acids in general and G4 DNA.
NMM inhibits G4 DNA unwinding without significantly affecting binding or ATP hydrolysis. This provides a framework for understanding how BLM and Sgs1p bind to and unwind G4 DNA. Both BLM and Sgs1p require a 3′ single-stranded tail >4 nt in length in order to unwind a G4 DNA substrate (7,8), but neither binds well to ssDNA (kD > 200 nM; data not shown). Therefore, these helicases may contain one domain that recognizes G4 DNA structure, and an independent domain that moves along the ssDNA tail until it reaches the 3′-most G quartet, where it begins unwinding. Porphyrins appear to interact with G4 DNA by stacking upon the terminal G quartet (30). If a porphyrin inhibitor obscured the 3′ face of the G4 barrel, the helicase would be unable to unwind the substrate.
It was surprising that, in the presence of inhibitor, BLM remains bound to its substrate and continues to hydrolyze ATP—analogous to a car spinning its wheels in mud or snow. Nonetheless, uncoupling of ATP hydrolysis from helicase function has also been reported for other helicases. When confronted with an inappropriate substrate, such as an RNA: DNA hybrid or psoralen-crosslinked DNA, RecBC function is thwarted but the enzyme does not dissociate from its substrate and continues futilely to hydrolyze ATP (31,32). In addition, certain mutations in PcrA produce enzyme which has lost the capacity to unwind DNA, but can still bind DNA and hydrolyze ATP (33–35). NMM may therefore prove useful in analyzing the translocation and unwinding mechanisms of RecQ family helicases.
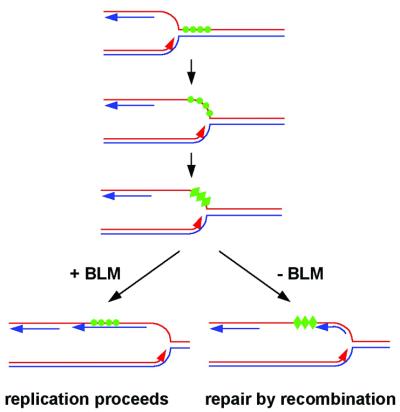
How does G4 DNA unwinding contribute to the functions of BLM and Sgs1p in the maintenance of genomic stability in vivo? Just as triplet repeats appear to become structured during transient DNA denaturation that occurs during lagging strand replication (36), a G-rich template strand could form structures stabilized by G quartets. As shown in Figure 7, these structures would normally be removed by BLM or Sgs1p, a function consistent with the physical and genetic associations documented for these helicases with topoisomerases (37–44) and RPA (45). Moreover, BLM is an intrinsic part of the BASC complex which is thought to recognize abnormal DNA structures and damaged DNA (46), and it localizes to PML bodies, along with DNA repair factors (47,48). Key to the model in Figure 7 is the proposal that BLM and Sgs1p function proactively to prevent fork arrest by unwinding topological obstacles to replication. In contrast, WRN may function primarily to promote recombinational rescue of DNA synthesis once a fork has stalled, consistent with its localization to stalled forks (16). Future experiments will test this possibility.
Figure 7.
Model of the function of BLM or Sgs1p in replication. A replication fork approaches a G-rich region (green circles), and opening of the DNA duplex allows the G-rich strand to form G4 DNA (green triangles, third line). Either BLM or Sgs1p can recognize and unwind the G4 DNA, to produce a usable template strand (bottom line, left). In the absence of BLM or Sgs1p, G4 DNA persists, stalling replication. Replication restart by recombination results in the hyper-recombination characteristic of BLM- or SGS1-deficient cells.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the Maizels Laboratory and Stu Linn and Gerry Smith for useful discussions. This research was supported by the US National Institutes of Health grants R01 GM39799 and P01 CA16038.
REFERENCES
- 1.Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- 2.Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 3.Kamath-Loeb A.S., Shen,J.C., Loeb,L.A. and Fry,M. (1998) Werner syndrome protein. II. Characterization of the integral 3′→5′ DNA exonuclease. J. Biol. Chem., 273, 34145–34150. [DOI] [PubMed] [Google Scholar]
- 4.Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nature Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- 5.Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- 6.Sinclair D.A., Mills,K. and Guarente,L. (1997) Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science, 277, 1313–1316. [DOI] [PubMed] [Google Scholar]
- 7.Sun H., Karow,J.K., Hickson,I.D. and Maizels,N. (1998) The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem., 273, 27587–27592. [DOI] [PubMed] [Google Scholar]
- 8.Sun H., Bennett,R.J. and Maizels,N. (1999) The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res., 27, 1978–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Brabant A.J., Ye,T., Sanz,M., German,I.J., Ellis,N.A. and Holloman,W.K. (2000) Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry, 39, 14617–14625. [DOI] [PubMed] [Google Scholar]
- 10.Karow J.K., Constantinou,A., Li,J.L., West,S.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohaghegh P., Karow,J.K., Brosh,R.M.,Jr, Bohr,V.A. and Hickson,I.D. (2001) The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res., 29, 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry M. and Loeb,L.A. (1999) Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- 13.Huang P., Pryde,F.E., Lester,D., Maddison,R.L., Borts,R.H., Hickson,I.D. and Louis,E.J. (2001) SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol., 11, 125–129. [DOI] [PubMed] [Google Scholar]
- 14.Cohen H. and Sinclair,D.A. (2001) Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl Acad. Sci. USA, 98, 3174–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson F.B., Marciniak,R.A., McVey,M., Stewart,S.A., Hahn,W.C. and Guarente,L. (2001) The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J., 20, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Constantinou A., Tarsounas,M., Karow,J.K., Brosh,R.M., Bohr,V.A., Hickson,I.D. and West,S.C. (2000) Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep., 1, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Kobbe C., Karmakar,P., Dawut,L., Opresko,P., Zeng,X., Brosh,R.M.,Jr, Hickson,I.D. and Bohr,V.A. (2002) Colocalization, physical and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem., 277, 22035–22044. [DOI] [PubMed] [Google Scholar]
- 18.Bennett R.J., Sharp,J.A. and Wang,J.C. (1998) Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem., 273, 9644–9650. [DOI] [PubMed] [Google Scholar]
- 19.Karow J.K., Chakraverty,R.K. and Hickson,I.D. (1997) The Bloom’s syndrome gene product is a 3′–5′ DNA helicase. J. Biol. Chem., 272, 30611–30614. [DOI] [PubMed] [Google Scholar]
- 20.Caron P.R., Watt,P. and Wang,J.C. (1994) The C-terminal domain of Saccharomyces cerevisiae DNA topoisomerase II. Mol. Cell. Biol., 14, 3197–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanzetta P.A., Alvarez,L.J., Reinach,P.S. and Candia,O.A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem., 100, 95–97. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Geyer,C.R. and Sen,D. (1996) Recognition of anionic porphyrins by DNA aptamers. Biochemistry, 35, 6911–6922. [DOI] [PubMed] [Google Scholar]
- 23.Ren J. and Chaires,J.B. (1999) Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry, 38, 16067–16075. [DOI] [PubMed] [Google Scholar]
- 24.Anantha N.V., Azam,M. and Sheardy,R.D. (1998) Porphyrin binding to quadrupled T4G4. Biochemistry, 37, 2709–2714. [DOI] [PubMed] [Google Scholar]
- 25.Arthanari H., Basu,S., Kawano,T.L. and Bolton,P.H. (1998) Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res., 26, 3724–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X. and Maizels,N. (2001) Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res., 29, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mergny J.L. and Helene,C. (1998) G-quadruplex DNA: a target for drug design. Nature Med., 4, 1366–1367. [DOI] [PubMed] [Google Scholar]
- 28.Izbicka E., Nishioka,D., Marcell,V., Raymond,E., Davidson,K.K., Lawrence,R.A., Wheelhouse,R.T., Hurley,L.H., Wu,R.S. and Von Hoff,D.D. (1999) Telomere-interactive agents affect proliferation rates and induce chromosomal destabilization in sea urchin embryos. Anticancer Drug Des., 14, 355–365. [PubMed] [Google Scholar]
- 29.Izbicka E., Barnes,L.D., Robinson,A.K., Davidson,K.K., Lawrence,R.A. and Hannibal,G.T. (2001) Alterations in DNA repair and telomere maintenance mechanism affect response to porphyrins in yeast. Anticancer Res., 21, 1899–1903. [PubMed] [Google Scholar]
- 30.Han H., Langley,D.R., Rangan,A. and Hurley,L.H. (2001) Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc., 123, 8902–8913. [DOI] [PubMed] [Google Scholar]
- 31.Karu A.E., MacKay,V., Goldmark,P.J. and Linn,S. (1973) The recBC deoxyribonuclease of Escherichia coli K-12. Substrate specificity and reaction intermediates. J. Biol. Chem., 248, 4874–4884. [PubMed] [Google Scholar]
- 32.Karu A.E. and Linn,S. (1972) Uncoupling of the recBC ATPase from DNase by DNA crosslinked with psoralen. Proc. Natl Acad. Sci. USA, 69, 2855–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soultanas P., Dillingham,M.S., Wiley,P., Webb,M.R. and Wigley,D.B. (2000) Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J., 19, 3799–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dillingham M.S., Wigley,D.B. and Webb,M.R. (2000) Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry, 39, 205–212. [DOI] [PubMed] [Google Scholar]
- 35.Dillingham M.S., Soultanas,P., Wiley,P., Webb,M.R. and Wigley,D.B. (2001) Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl Acad. Sci. USA, 98, 8381–8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Usdin K. and Woodford,K.J. (1995) CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res., 23, 4202–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watt P.M., Louis,E.J., Borts,R.H. and Hickson,I.D. (1995) Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell, 81, 253–260. [DOI] [PubMed] [Google Scholar]
- 39.Johnson F.B., Lombard,D.B., Neff,N.F., Mastrangelo,M.A., Dewolf,W., Ellis,N.A., Marciniak,R.A., Yin,Y., Jaenisch,R. and Guarente,L. (2000) Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells. Cancer Res., 60, 1162–1167. [PubMed] [Google Scholar]
- 40.Wu L., Davies,S.L., North,P.S., Goulaouic,H., Riou,J.F., Turley,H., Gatter,K.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product interacts with topoisomerase III. J. Biol. Chem., 275, 9636–9644. [DOI] [PubMed] [Google Scholar]
- 41.Hu P., Beresten,S.F., van Brabant,A.J., Ye,T.Z., Pandolfi,P.P., Johnson,F.B., Guarente,L. and Ellis,N.A. (2001) Evidence for BLM and Topoisomerase IIIα interaction in genomic stability. Hum. Mol. Genet., 10, 1287–1298. [DOI] [PubMed] [Google Scholar]
- 42.Fricke W.M., Kaliraman,V. and Brill,S.J. (2001) Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem., 276, 8848–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett R.J. and Wang,J.C. (2001) Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: studies of fusion proteins. Proc. Natl Acad. Sci. USA, 98, 11108–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett R.J., Noirot-Gros,M.F. and Wang,J.C. (2000) Interaction between yeast sgs1 helicase and DNA topoisomerase III. J. Biol. Chem., 275, 26898–26905. [DOI] [PubMed] [Google Scholar]
- 45.Brosh R.M. Jr, Li,J.L., Kenny,M.K., Karow,J.K., Cooper,M.P., Kureekattil,R.P., Hickson,I.D. and Bohr,V.A. (2000) Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem., 275, 23500–23508. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Cortez,D., Yazdi,P., Neff,N., Elledge,S.J. and Qin,J. (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev., 14, 927–939. [PMC free article] [PubMed] [Google Scholar]
- 47.Sanz M.M., Proytcheva,M., Ellis,N.A., Holloman,W.K. and German,J. (2000) BLM, the Bloom’s syndrome protein, varies during the cell cycle in its amount, distribution and co-localization with other nuclear proteins. Cytogenet. Cell Genet., 91, 217–223. [DOI] [PubMed] [Google Scholar]
- 48.Bischof O., Kim,S.H., Irving,J., Beresten,S., Ellis,N.A. and Campisi,J. (2001) Regulation and localization of the bloom syndrome protein in response to DNA damage. J. Cell Biol., 153, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]