Abstract
The deletion of the top3+ gene leads to defective nuclear division and lethality in Schizosaccharo myces pombe. This lethality is suppressed by concomitant loss of rqh1+, the RecQ helicase. Despite extensive investigation, topoisomerase III function and its relationship with RecQ helicase remain poorly understood. We generated top3 temperature-sensitive (top3-ts) mutants and found these to be defective in nuclear division and cytokinesis and to be sensitive to DNA-damaging agents. A temperature shift of top3-ts cells to 37°C, or treatment with hydroxyurea at the permissive temperature, caused an increase in ‘cut’ (cell untimely torn) cells and elevated rates of minichromosome loss. The viability of top3-ts cells was decreased by a temperature shift during S-phase when compared with a similar treatment in other cell cycle stages. Furthermore, the top3-ts mutant was not sensitive to M-phase specific drugs. These results indicate that topoisomerase III may play an important role in DNA metabolism during DNA replication to ensure proper chromosome segregation. Our data are consistent with Top3 acting downstream of Rqh1 to process the toxic DNA structure produced by Rqh1.
INTRODUCTION
Topoisomerase III is a type I topoisomerase engaged in breakage and reunion of single-stranded DNA. Eukaryotic topoisomerase III was first identified as a gene required for suppression of recombination between repeated genes in Saccharomyces cerevisiae (1). Partially purified topoisomerase III was found to be much less efficient than yeast DNA topoisomerases I or II in causing relaxation of supercoiled DNA substrates. In vivo studies demonstrated that yeast topoisomerase III has, at most, a weak activity in relaxing negatively supercoiled intracellular DNA (2). Null mutation of the top3+ gene in budding yeast results in various phenotypes including slow growth, hyper-recombination between repetitive DNA sequences, and a defect in sporulation (3).
The S.cerevisiae sgs1 mutant was isolated as a suppressor of the slow growth phenotype of the top3 deletion mutant (4). SGS1 is a member of the RecQ helicase family which includes fission yeast rqh1+ and human BLM, WRN and REQL4 helicases which are implicated in Bloom’s, Werner’s and Rothmund–Thomson syndromes, respectively (5–8). RecQ-family helicases possess 3′ to 5′ helicase activity and are required for the maintenance of genomic stability. In S.cerevisiae, sgs1 deletion mutants also show genomic instability including hyper-recombination of the rDNA locus and a reduced life span that correlates with the accumulation of extrachromosomal rDNA circles (9–12). Biochemically, Sgs1 physically interacts with Top3 and with Top2.
Several models have been postulated to explain the interactions between topoisomerase III and RecQ helicases: first, the Sgs1–Top3 complex is proposed to act as a reverse gyrase with a helicase-like domain and a type I topoisomerase (4). Secondly, topoisomerase III and RecQ helicase are thought play a role in recombination (4,13) and thirdly, it has been suggested that the Sgs1–Top3 complex acts at late stages of replication to facilitate the progression of the replication fork by combined unwinding activity of the helicase and decatenation activity of topoisomerase III (14,15).
In Schizosaccharomyces pombe, top3+ is essential for viability and plays a role in chromosome segregation (14). The top3-deleted spores germinate and divide several times, but fail to maintain long-term growth. The S.pombe RecQ family helicase, rqh1+, was initially isolated as a gene that complemented the rad12 and hus2 mutations (the latter found to be allelic), which showed sensitivity to both DNA-damaging agents and to the DNA synthesis inhibitor hydroxyurea (HU). The rqh1 null mutant is defective in the reversible recovery from the S-phase arrest, and in tolerating DNA damage during S-phase (16,17). Deletion of the rqh1+ gene suppresses the lethality of the top3 mutant in S.pombe, a result reminiscent of the situation in S.cerevisiae (14,18).
To further understand the functional roles of S.pombe topoisomerase III and its relationship to Rqh1 helicase, we generated top3 temperature-sensitive (top3-ts) mutants. The top3-ts mutants are sensitive to DNA-damaging agents, show increased levels of ‘cut’ (cell untimely torn) phenotypes and elevated rates of minichromosome loss following HU treatment. The viability of wild-type and rqh1-d mutant cells is sharply decreased by Rqh1 overproduction. The phenotypes of rqh1-overexpression strain are similar to those of top3-ts mutant. These results indicate that Top3 has an essential function in processing an intermediate DNA structure produced by Rqh1.
MATERIALS AND METHODS
Media and reagents
Complete and minimal growth media for fission yeast and chemical reagents were purchased from Difco and Sigma Aldrich. Plasmids were constructed by standard techniques (19) and general procedures for S.pombe genetics were carried out according to the methods of Alfa (20).
Yeast strains and plasmids
Yeast strains and plasmids used in this study are listed in Tables 1 and 2. The 6 kb EcoRV–NsiI DNA fragment of the wild-type top3+ gene derived from cosmid C16G5 (a gift from Rhian Gwilliam at Sanger Center) was cloned into the SmaI–PstI site of pUR19N to generate plasmid pMO19T3. The 3.1 kb XbaI–SalI DNA fragment of the top3+ gene was similarly cloned into pBluescripts II KS(+) and pBG2 to create pMO234 and pMO344, respectively. To disrupt the top3+ gene, the 1.7 kb PstI–EcoRI DNA fragment of pMO234 was replaced by the 2.2 kb LEU2+ gene of S.cerevisiae or the 1.8 kb ura4+ gene of S.pombe to generate pMO323 and pMO343, respectively. The 3.5 kb SalI–XbaI DNA fragment of pMO323 or the 2.6 kb MscI–ClaI DNA fragment of pMO343 was used to transform the diploid strain MO797. The resulting top3+/top3::LEU2+ and top3+/top3::ura4+ transformants were designated as MOL791 and MOU792, respectively.
Table 1. Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| CF199 | h– leu1-32 his3-D1 ura4-D18 ade6-M210 | From T. Cech |
| CF197 | h+ leu1-32 his3-D1 ura4-D18 ade6-M216 | From T. Cech |
| 1589 | h– leu1-32 his3-D1 ura4-D18 rqh1::ura4+ | From T. Enoch |
| MO134 | h– leu1-32 his3-D1 ura4-D18 ade6-M210 top3-134-ts | This study |
| MO252 | h– leu1-32 his3-D1 ura4-D18 ade6-M210 top3-252-ts | This study |
| MO316 | h– leu1-32 his3-D1 ura4-D18 top3::LEU2+ rqh1::ura4+ | This study |
| MORT132 | h– leu1-32 his3-D1 ura4-D18 ade6-M210 top3-134-ts rqh1::ura4+ | This study |
| MOL911 | h– leu1-32 his3-D1 ura4-D18 ade6-M210 containing plasmid pMO19T3 | This study |
| MO797 | h+/h– leu1-32/leu1-32 his3-D1/his3-D1 ura4-D18/ura4-D18 ade6-M210/ade6-M216 | This study |
| MOL791 | h+/h– leu1-32/leu1-32 his3-D1/his3-D1 ura4-D18/ura4-D18 ade6-M210/ade6-M216 top3+/top3::LEU2+ | This study |
| MOU792 | h+/h– leu1-32/leu1-32 his3-D1/his3-D1 ura4-D18/ura4-D18 ade6-M210/ade6-M216 top3+/top3::ura4 + | This study |
| TE788 | ade6-M210 containing Ch16 | From T. Enoch |
| TE786 | rqh1::ura4+ ura4-D18 ade6-M210 containing Ch16 | From T. Enoch |
| FY1497 | h+ leu1-32 ura4-DS/E ade6-M210 containing Ch16 | From R. Allshire |
| MO1841 | h– leu1-32 his3-D1 ura4-D18 ade6-M210 top3-134-ts containing Ch16 | MO134 × FY1497 |
| MO1825 | Same with MO1841 | MO134 × FY1497 |
Table 2. Plasmids used in the study.
| Plasmid |
Constructions |
Reference |
| pMO19T3 | pUR19N containing a 6 kb EcoRV–NsiI fragment of the top3+ gene | This work |
| pMO234 | pBluescripts II ks(+) containing a 3.1 kb XbaI–SalI fragment of the top3+ gene | This work |
| pMO344 | pBG2 containing a 3.1 kb XbaI–SalI fragment of the top3+ gene | This work |
| pMO323 | top3::LEU2+ construct | This work |
| pMO343 | top3::ura4+ construct | This work |
| pTE436 | rqh1::ura4+ construct | From T. Enoch |
| pMO322 | pBG2 containing 5′-420 bp and 3′-230 bp flanking fragments only | This work |
To make a top3rqh1 double mutant, top3+/top3::LEU2+ diploid MOL791 was transformed with SacI–SphI-linearized pTE436 plasmids (a gift from Tarmar Enoch at Harvard Medical Center). The correct transformants were confirmed by Southern blot, sporulated on ME plates and the resulting tetrads were dissected on YES plates. The top3::LEU2+ rqh1::ura4+ haploid colonies were selected by replica plating onto minimal medium lacking uracil and leucine. The resulting double mutant was designated as MO316.
Construction of top3-ts alleles
To create top3-ts mutants by plasmid shuffling, hydroxylamine (HA) mutagenesis (21) and error-prone PCR-based mutagenesis (22) methods were used as described but with some minor modifications. Briefly, top3+/top3::LEU2+ diploid MOL791 was transformed with pMO19T3 containing the wild-type top3+ gene. The resulting positive colonies were sporulated on ME plates at 25°C and then top3::LEU2+ haploid cells containing the pMO19T3 were selected (named as MOL911). To specifically mutagenize top3+ gene, 10 µg of pMO344 containing wild-type top3+ was treated with 1 M HA for 5 or 10 min and then was used to transform MOL911. The generated transformants were replica plated onto two 5-fluoroorotic acid (5-FOA) plates to remove pMO19T3. Replica plates were incubated at 25 and 37°C.
For error-prone PCR-based mutagenesis, the 5′- and 3′-flanking DNA fragments of 420 and 230 bp were amplified by PCR using primers (TOP3-Sph1-6175, 5′-GGAACTTACACTAATTTAGCATGCTCTA; TOP3-Pst1-5780, 5′-AAAAATGACTGCAGTGTA) and (TOP3-Pst1-3540, 5′-TAACGC AGACGTGCAGAGTCTACC; TOP3-Sac1-3330, 5′-GTG TATTGTAACTGAGCTCGTCTTTTCTGA), respectively. The resulting DNA fragments were digested with either SphI–PstI or PstI–SacI restriction enzymes and inserted together into the SphI–SacI site of pBG2 to generate plasmid pMO322. Random mutations in the top3+ gene were generated by Mn2+-buffered error-prone PCR. The PCR mixtures contained 100 pmol of each oligonucleotide (TOP3-CT-R, 5′-GACGATATTAAGTCGACTTG; TOP3-Sph1-6175), 50 ng of pMO19T3 DNA as a template, 0.1 mM MnCl2, 2.5 mM MgCl2, 0.25 mM dNTP mixtures, 10 mM Tris–HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, and 5 U Taq polymerase (Promega). The PCR products and PstI-linearized pMO322 were cotransformed into MOL911. Total transformants produced by homologous recombination between the gapped plasmids and mutagenized PCR products were replica-plated onto two 5-FOA plates to exclude the ura4+ marked top3+ plasmid and replica plates were incubated at 25 or 37°C.
Colonies showing temperature-sensitive growth were identified and the plasmids containing the corresponding mutagenized top3 genes were extracted from and transferred into the integration vector Yiplac128. The resulting plasmids were linearized with MscI restriction enzyme and then transformed into the top3+/top3::ura4+ diploid strain MOU792. The cells which the mutated top3 gene was inserted into original top3+ locus were selected by Southern blot, sporulated, and ura4+/LEU2+ colonies were selected. The resulting colonies were subjected to 5-FOA plates to remove the ura4+ marker by homologous recombination (23).
UV, methyl methane-sulfonate (MMS) and HU sensitivity tests
Two top3-ts mutants and the parental (control) strain CF199 were grown in YES medium to mid-log phase and the cell number counted using a hemocytometer. To analyze the sensitivity to HU and MMS, appropriate numbers of cells in the exponential growth phase were spread onto YES plates with or without DNA-damaging agents of various concentrations in duplicate. To test UV sensitivity, cells were spread on YES agar plates and then irradiated with different doses of UV light in duplicate. Plates were incubated either at 25°C or at 25°C following 12 h incubation at 37°C. The number of viable colonies in each plate treated with the DNA-damaging agents was normalized with respect to that of an untreated control for each temperature regime.
Minichromosome loss assay
The minichromosome loss rate of the rqh1-d mutant was determined with TE788 and TE786 and for the top3-ts mutants with FY1497 and MO1841. The loss rate of minichromosome Ch16 from wild-type and mutant cells was determined by two independent methods (16,17) with slight modifications. In both methods, the number of cells changed from ade+ to ade– by the loss of Ch16 during one generation was ascertained. Each cell culture was either untreated or treated with 10 mM HU at 30°C and appropriate numbers of cells were spread onto YES plates not supplemented with adenine. The plates of MO1841 and FY1497 were prepared in duplicate and one of the plates incubated at 37°C for 12 h before being transferred to 25°C. The other plate was incubated directly at 25°C. The plates of TE786 and TE788 were incubated at 30°C. After 5–7 days incubation, the proportion of red colonies was calculated. The loss rates estimated by the two methods were averaged.
Cell viability test after G1 cell cycle arrest
Exponentially growing cells were washed three times with EMM media lacking a nitrogen source, resuspended in the same media at 2 × 106 cells/ml and then incubated at 25°C for 16 h. After changing to fresh YES media, cell aliquots were then sampled at 30 min intervals, transferred to 37°C for 2 h and the appropriate number of cells were then spread onto YES plates. An aliquot of the same cells was fixed for FACS analysis or septation index counts. The relative colony-forming units were calculated by comparison with the number of colonies evident when plates were incubated at 25°C for 2 h after cell cycle restart.
Sensitivity test to thiabendazole (TBZ) and N-tosyl-l-phenylalanine chloromethyl ketone (TPCK)
Cells were grown to mid-log phase, washed with fresh YES medium, and then resuspended at a concentration of 1 × 107 cells/ml. In duplicate, serial 10-fold dilutions were spotted onto YES plates containing the relevant agent and onto a control plate. TBZ plates contained 10 µg/ml TBZ. TPCK plates were composed of 1% DMSO, 5 µg/ml phloxine B and 120 µM TPCK. As before, each plate was incubated at 25°C or at 25°C following 12 h incubation at 37°C.
The preparation of Rqh1 overexpression strains
The pREP41-HAN vector was used for Rqh1 overexpression. Cells bearing rqh1-overproducing plasmid were first grown in EMM medium containing 4 µM thiamine (transcription repressed), washed three times with EMM medium, and further grown in EMM lacking thiamine for 20 h (transcription induced at ∼14–16 h). The overexpression of HA-Rqh1 at 20 h was confirmed by western blot using anti-HA antibody (Boehringer Mannheim).
RESULTS
Isolation of top3-ts mutants
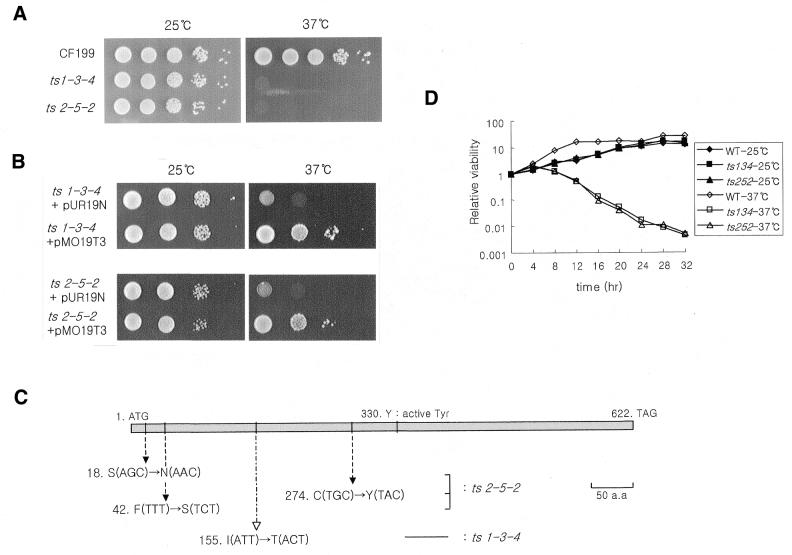
Using the methodology described (see Materials and Methods) we generated two independent top3-ts alleles on multi-copy plasmids designated as top3-ts134 and top3-ts252 (Fig. 1A). The wild-type top3+ gene compensated for the temperature-sensitive phenotype, whereas the empty vector did not (Fig. 1B), indicating that the temperature-sensitive phenotype of the mutants resulted from mutation of the top3+ gene. To confirm this, the DNA sequences of the mutated top3 genes were analyzed (Fig. 1C). Only one amino acid change, Ile155 to Thr, was evident in top3-ts134 (silent mutations occurred at several sites). Ile155 is conserved between S.pombe and S.cerevisiae Top3, human Top3α, and Escherichia coli Top1. In top3-ts252, three amino acid changes were identified Ser18 to Asn, Phe42 to Ser and Cys274 to Tyr. Both mutated top3 genes were cloned into an integration vector (see Materials and Methods) and stable temperature-sensitive mutants were constructed. The strains whose original top3+ gene was replaced by the mutated top3 gene were renamed MO134 (top3-ts134) and MO252 (top3-ts252). The viability of two temperature-sensitive mutants gradually decreased following a temperature shift (Fig. 1D).
Figure 1.
(A) The growth comparisons of two top3-ts candidates. top3::ura4+ haploids containing a mutated top3 gene on a plasmid were compared at 25 and 37°C. (B) The growth defects of both top3-ts candidates are rescued by introducing a wild-type top3+ gene on plasmid pMO19T3 at the restrictive temperature. (C) Amino acids changes of top3-ts mutants. The top3-ts134 was derived from the error-prone PCR method. top3-ts252 was made by HA mutagenesis. (D) The wild-type and mutant cell cultures incubated at the permissive and restrictive temperatures were plated onto YES plates. The resulting colonies were normalized to the number of cells on the control plates at zero point.
The top3-ts mutants show abnormal nuclear structure and cell morphology
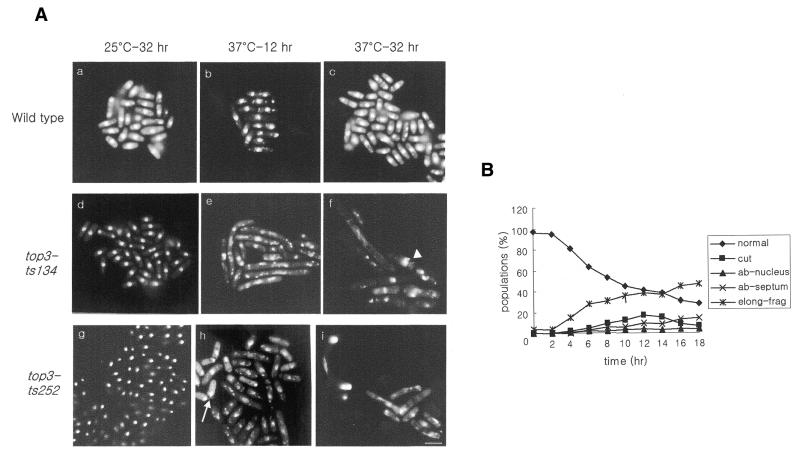
To explore the cellular functions of top3+, the cell morphology and nuclear structure of top3-ts mutants were examined at the restrictive temperature (Fig. 2A). The top3-ts134 and top3-ts252 cells showed a normal growth pattern at the permissive temperature (Fig. 2A panels d and g), but, upon temperature shift to the restrictive temperature, cells gradually elongated and curved like a bow (Fig. 2A panels e, f, h and i). DAPI-staining material was fragmented over time, spreading out into the cytoplasm (Fig. 2A panels f and i). Consistent with the loss of nuclear integrity, cells accumulated with abnormal and multiple septa, and many anucleated cells or cells with misplaced nuclei were observed at later time points. These phenotypes are consistent with the previously reported phenotype of top3 deletion spores following germination (14). The kinetics of accumulation of various phenotypes were followed following the shift to the restrictive temperature (Fig. 2B). It is apparent that the nuclear material showed abnormal morphology soon after the temperature shift, followed by cell elongation and aberrant cytokinesis. These defects suggested that top3+ is involved in the maintenance of chromosome stability and that loss of Top3 function results in delayed and aberrant cell division.
Figure 2.
(A) Cellular and nuclear morphologies of top3-ts mutants. Parental strain CF199 (panels a–c) and top3-ts mutant strains MO134 (panels d–f) and MO252 (panels g–i) were grown at the permissive temperature (25°C) and then shifted to the restrictive temperature (37°C). The arrowhead in panel f indicates an abnormal septum. The arrow in panel h indicates a ‘cut’ cell. Bar, 10 µm. (B) Cells with each abnormality were counted at 2 h intervals. cut, ‘cut’ phenotype; ab-nucleus, anucleated or off-centered nuclei; ab-septum, multiple septa or a septum formed at one side of cell; elong-frag, elongated cells with fragmented nuclei.
The top3-ts mutants are sensitive to DNA-damaging agents
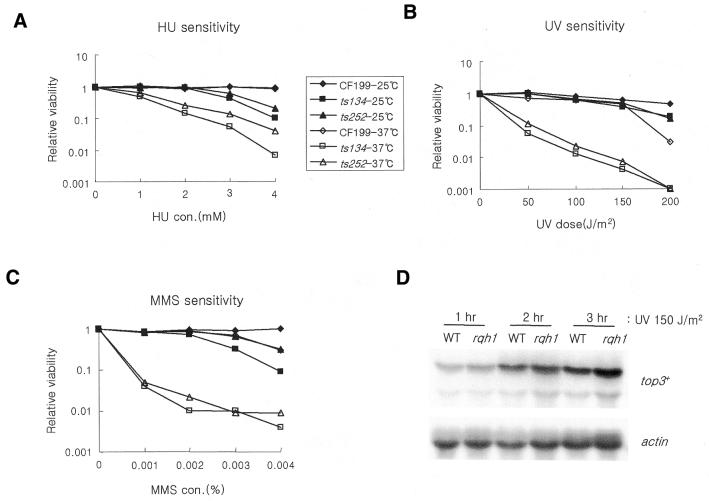
Rqh1 has been implicated in DNA damage responses (16) and top3+ interacts genetically with rqh1+. We have used the top3-ts mutants to study the role of top3+ in DNA damage responses independently of rqh1-d mutants. This has not been achieved in S.pombe previously because the top3 null mutant is not viable. Compared with the wild-type cell, both top3-ts strains were sensitive to a range of DNA-damaging agents at the restrictive temperature (Fig. 3A–C). In addition to colonogenic sensitivity to HU (Fig. 3A), top3-ts mutants also formed very small colonies on plates containing HU even at the permissive temperature (data not shown). In S.cerevisiae, the top3 deletion mutant was also sensitive to DNA-damaging agents, especially to HU (24).
Figure 3.
The sensitivity of top3-ts mutants to HU (A) and DNA-damaging agents, UV (B) and MMS (C). The sensitivity of wild-type and top3-ts mutant strains MO134 and MO252 was measured by colony formation on YES plates containing the indicated concentrations of HU and MMS either at the permissive temperature or at the restrictive temperature for 12 h followed by incubation at 25°C. The relative viability of each strain (25 and 37°C) was calculated by dividing the number of viable cells on plates containing the damaging agents by the number of cells on untreated control plates. (D) The relative levels of top3+ transcripts in wild-type (WT) and rqh1-d mutant 1589 (R1) after UV irradiation were examined by northern blot. After irradiation, total RNA was extracted at 1 h intervals and probed with top3 cDNA and an actin gene as a loading control.
We also analyzed the level of top3+ transcript in response to UV irradiation. Top3 mRNA levels increased within 1 h of 150 J/m2 UV irradiation. This response was also evident, and possibly slightly elevated, in an rqh1-d mutant background (Fig. 3D). The DNA damage sensitivity of top3-ts mutants, combined with the DNA damage inducibility of the top3+ transcript implies an involvement of top3+ in the DNA damage response, possibly during repair. It is likely that Top3 and Rqh1 function in the same DNA damage response pathway(s) because the damage sensitivities of the top3rqh1 double mutant is equivalent to that of the rqh1-d single mutant (14).
The top3-ts mutant is defective in recovery from S-phase arrest by HU
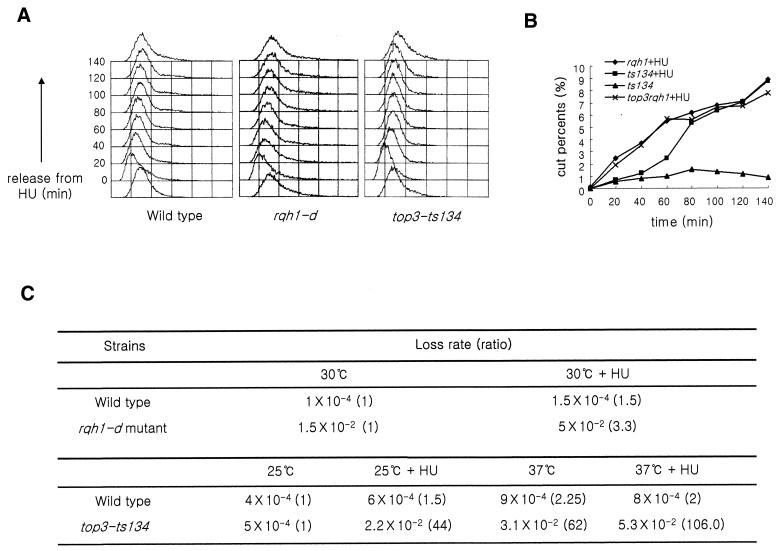
A previous report demonstrated that the rqh1 null mutant was HU sensitive and defective in recovery from S-phase arrest (17). Therefore, we examined if the HU sensitivity of top3-ts mutants was similarly related to a recovery defect. Flow cytometry analysis (Fig. 4A) showed that the wild-type, rqh1-d mutant, and top3-ts mutant could resume and complete bulk DNA replication with similar kinetics following release from HU arrest at 37°C. However, subsequent chromosome segregation in both rqh1 null and top3-ts mutants was defective, indicating that, while bulk DNA replication appeared complete, the chromosomes were either incompletely replicated or remained entangled (Fig. 4B). At 140 min after HU release, both rqh1-d and top3-ts mutants showed an elongated cell morphology and an increased frequency of ‘cut’ phenotypes when compared with either controls without HU treatment (Fig. 4B) or to top3+rqh1+-treated controls (data not shown) (17).
Figure 4.
Analysis of DNA contents and the percentage of cells showing the ‘cut’ phenotype following release from HU arrest. (A) After S-phase arrest (10 mM HU at 37°C for 2 h) cells were fixed for FACS analysis at 20 min intervals. (B) rqh1-d, top3rqh1 double and top3-ts mutant cells were treated with 10 mM HU as above and then incubated at 30°C for 4 h in fresh YES media. Cells were sampled every 20 min for DAPI staining. The proportions of cells showing the ‘cut’ phenotype were plotted against the incubation times after HU removal. (C) The minichromosome loss rates per generation for rqh1 and top3-ts mutants.
We also examined the top3rqh1 double mutant in the same assay (Fig. 4B). When released from HU arrest, the number of cells with the ‘cut’ phenotype also increased with kinetics similar to the rqh1-d. These observations indicate that Rqh1 and Top3 are required for the same late step of DNA replication.
Both Top3 and Rqh1 are required for the fidelity of chromosome segregation
Using a minichromosome loss assay (see Materials and Methods) we measured the loss rate of a non-essential minichromosome in wild-type, top3-ts, and rqh1-d mutant cells (Fig. 4C). The rqh1-d mutant exhibited a 75-fold increased frequency of Ch16 loss compared with wild-type controls. This was further increased 3.3-fold following 4 h of treatment with 10 mM HU. These observations imply that rqh1-d mutant cells are defective in chromosome segregation and that this is exacerbated after a temporary block in S-phase.
The loss rates of top3-ts mutant cells were elevated at the permissive temperature by HU treatment (44-fold, compared with wild-type, 1.5-fold) and increased with either a 12 h incubation at the restrictive temperature (62-fold, compared with wild-type cells, 2.2-fold) or transient HU treatment (106-fold, compared with wild-type, 2-fold). Since rqh1-d and top3-ts mutants showed very low viability after the HU treatment and top3-ts cells lose viability after a temperature shift, the minichromosome loss rates probably underestimate the mis-segregation problem.
The viability of cells in S-phase slightly decreased with a rise in temperature
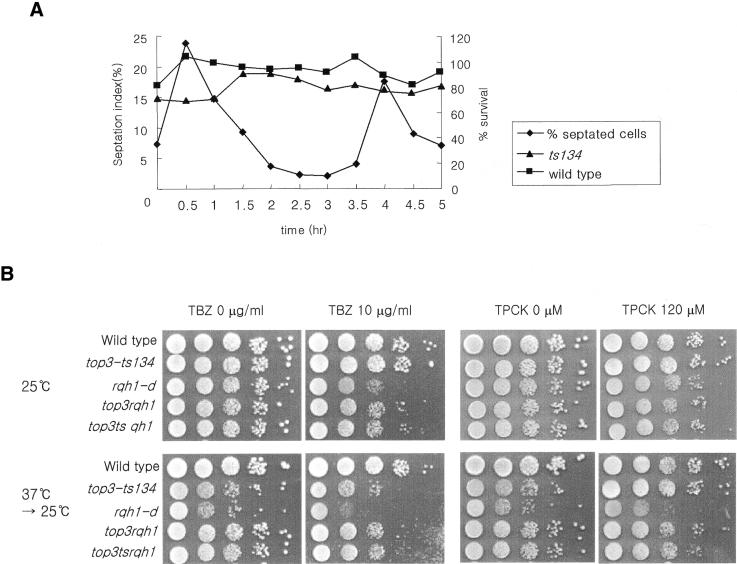
Because the top3-ts134 mutant is sensitive to HU, defective in recovery after S-phase arrest and loses Ch16 dramatically after HU treatment, we examined at which point in the cell cycle the top3-ts mutant cells lost viability. Cells were synchronized in G1-phase by nitrogen starvation and aliquots were transferred to 37°C for 2 h at 30 min intervals. The viability was plotted with the septation index of the unshifted culture (Fig. 5A). In S.pombe there is a temporal correlation between septation and S-phase and so the septation index is taken as a measure of the number of replicating cells. The viability of the top3-ts mutant was modestly reduced by a transient temperature shift during G1–S-phase, but not by a shift during G2. Although these differences are modest [which is not unexpected given the fact that top3-ts cells lose viability slowly following a shift of an asynchronous cell culture to 37°C (data not shown)], these results are consistent with the lethality of the top3-ts mutant being due to defects during DNA replication.
Figure 5.
(A) Wild-type and top3-ts134 mutant cells were arrested in G1 by nitrogen starvation and released into nitrogen-containing media to restart the cell cycle. Upon release, at 30 min intervals, aliquots of cells were transferred to 37°C for 2 h. Cell viability was then checked by plating cells at 25°C. An aliquot was used for counting the septation index at the permissive temperature. (B) The sensitivity of the indicated mutant cells to TBZ and TPCK. Wild-type, the top3-ts mutant strain MO134, rqh1, and top3ts rqh1, and top3rqh1 double mutant cells were serially diluted and spotted onto duplicate plates contained either 10 µg/ml TBZ or 120 µM TPCK, and onto control plates. One copy of each plate was incubated at 25°C and another was transferred to 25°C after incubation at 37°C for 12 h.
Chromosome mis-segregation is not due to defects in the anaphase promoting complex (APC) or microtubules
Explanations for chromosome mis-segregation defects in top3-ts mutant cells could be that Top3 functions to maintain the architecture of the centromere, or that it affects microtubule interactions with chromosomes, and has an uncharacterized function in APC-catalyzed anaphase initiation. To examine these possibilities, we tested top3-ts134 mutants for sensitivity to the microtubule-destabilizing agent TBZ and the protease inhibitor TPCK.
At 25°C, TBZ and TPCK had little or no effect on growth of top3-ts, rqh1-d and rqh1top3 double mutant cells (Fig. 5B). A 12 h transient shift to the restrictive temperature similarly had little or no effect on top3-ts or rqh1top3 double mutants in the presence of TBZ or TPCK. A modest decrease in viability was observed for rqh1 null cells after this treatment in the presence of TPCK. These results provide strong evidence that the chromosome mis-segregation defect of top3-ts mutant is not due to defects in M-phase.
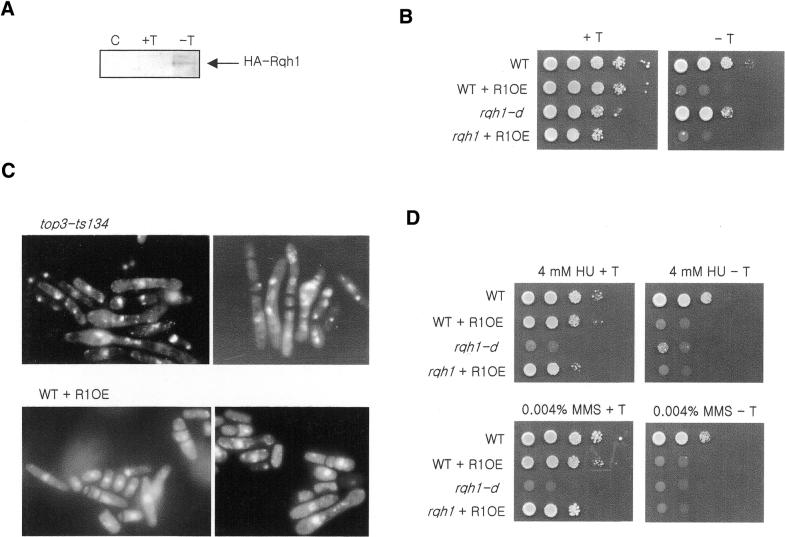
Rqh1 overexpression causes cell inviability
Rqh1 and Top3 are thought to function together, and to biochemically interact. Our data demonstrate similarities in some phenotypes when Rqh1 is lost or Top3 function is impaired. However, loss of the rqh1 gene is not lethal, whereas loss of the top3 gene is. Because the top3rqh1 double mutant is viable (14,18) and sgs1 mutants suppress the slow growth of top3-d mutant in S.cerevisiae, it has thus been suggested (4) that Top3 resolves potentially lethal DNA structures that are produced by Rqh1. If this is the case, we hypothesized that Rqh1 overexpression may itself be a lethal event, as it may perturb the balance of Rqh1–Top3 functions, causing a phenotype similar to that seen with the loss of top3 function. Indeed, when we expressed rqh1 from an inducible exogenous promoter (Fig. 6A), both wild-type and rqh1-d mutants were unable to grow (Fig. 6B). The morphology of rqh1-overexpressing cells was reminiscent of top3-ts mutant cells at the restrictive temperature (Fig. 6C). They were elongated, had double or triple septa, abnormal nuclei and showed ‘cut’ phenotypes. The percentage of ‘cut’ cells was, however, slightly lower than that seen for top3-ts mutant cells at 37°C. The rqh1-overexpressing cells also showed an increase in ‘cut’ cells when they were transiently treated with HU, reminiscent of top3-ts cells (data not shown). From these results, we suggest that Top3 has a role in the maintenance of an optimum level of Rqh1 activity or in resolving structures created by Rqh1 helicase, and may function downstream of Rqh1.
Figure 6.
Preparation and examination of rqh1-overexpression strains. (A) Rqh1 was fused to three HA epitopes in the pREP41-HAN vector. After induction for 20 h by the removal of thiamine, HA-Rqh1 was detected by western blot analysis using anti-HA antibody. (C) Extract of cells harboring pREP41-HAN control vector induced for 20 h without thiamine; (+T) uninduced pREP41-HR1; (–T) pREP41-HR1 induced for 20 h in the absence of thiamine. The 150 kDa HA-Rqh1 band was detected only in induced pREP41-HR1 extracts. (B) Spotting assay to examine the cell growth and damage sensitivity of Rqh1-overexpressing strains. When Rqh1 was overexpressed (–T) in the wild-type and rqh1-d mutant, cell growth was blocked. (C) Cell morphologies of top3-ts and rqh1-overexpression strains. The top3-ts mutant was grown for 32 h at the restrictive temperature. The rqh1-overexpression strain was induced for 20 h in the absence of thiamine. Both cultures showed ‘cut’ phenotypes, abnormal cytokinesis and aberrant nuclear structures. (D) The sensitivity to HU and MMS of the rqh1 null mutant was complemented by the repressed pREP41-HR1 (+T).
We also attempted to examine the sensitivity of rqh1-overproducing cells to DNA-damaging agents (Fig. 6D). However, when rqh1 was overexpressed from the pREP41 promoter in wild-type or rqh1-d mutant cells, viability was lost. When the nmt promoter was repressed by the addition of thiamine (Fig. 6D, left panels), cells were viable and the DNA damage sensitivity of the rqh1 null mutant disappeared. These results indicate that the low level expression of Rqh1 from the repressed nmt promoter could compensate for the damage sensitivity of the rqh1-d mutant.
DISCUSSION
Goodwin et al. (14) examined the phenotypic consequences of germinating top3 deleted spores and observed abnormal nuclei, multiple septa and elongated cells. However, they were not able to directly examine the role of Top3 in DNA damage repair or chromosome stability because the only viable top3 defective cells examined were, by necessity, also deleted for the rqh1 gene. In our study, we have examined the phenotype of top3 deficient cells by creating top3-ts mutants. Following a temperature shift to the restrictive temperature, we see morphological phenotypes that are very similar to those reported by Goodwin et al. (14). The percentage of normal looking cells decreases within a few generations and an increase is seen in cells with an abnormal septum, mispositioned nuclei and ‘cut’ nuclei. Cells also elongate and ultimately many cells appear to contain fragmented DAPI-staining material.
Top3 is required for chromosome stability
By studying minichromosome stability in top3-ts mutants we have been able to demonstrate that S.pombe top3+ is required for the fidelity of chromosome segregation. After a brief shift to the restrictive temperature, we found that minichromosome loss rates were significantly increased. Interestingly, both at the permissive temperature and a brief temperature shift to 37°C, the loss rate of the minichromosome was also increased following a transient incubation with HU, which arrests cells in S-phase. This result is reminiscent of the data from Stewart et al. (17), who demonstrated increased chromosome loss in rqh1 null cells after HU treatment. Because Stewart et al. (17) concluded that rqh1-d mutant cells were unable to effectively recover viability following a transient arrest in S-phase, we examined this recovery response in more detail. Indeed, we observed that, like rqh1 null cells, top3-ts cells at the restrictive temperature were ultimately able to complete bulk DNA synthesis, but that this did not represent intact, accurately or completely replicated DNA because the subsequent mitosis and cytokinesis were delayed and ultimately resulted in numerous mitotic defects.
Evidence for an essential function of Top3 during S-phase
On the basis of the sensitivity of top3-ts mutants to the S-phase inhibitor HU, we also examined the point in the cell cycle at which top3-ts cells lost viability at the restrictive temperature. Our data indicate that, if cells proceed through S-phase with impaired Top3 function, viability is modestly decreased. However, if Top3 function is impaired in G2 for several hours (by a transient temperature shift), viability is not lost. While these effects are quite subtle—which is consistent with the fact that top3-ts cells continue cycling for several generations and lose viability quite slowly at the restrictive temperature—we feel that, in concert with the HU sensitivity of top3-ts mutants and the fact that transient S-phase delay greatly increases minichromosome loss rates, these data point to an essential function for S.pombe Top3 in S-phase. A recent report revealed that top3 deletion mutants were highly sensitive to HU and the level of TOP3 mRNA peaked at late G1-phase in S.cerevisiae, suggesting a specific role of Top3 in S-phase (24).
Minichromosome loss mutants fall into a number of classes, including DNA replication and repair genes, genes affecting centromere function and genes affecting the progress though mitosis such as those involved with the APC. In order to provide evidence to support our hypothesis that Top3 function is important in S-phase and not directly within mitosis, we examined the sensitivity of top3-ts mutants to microtubule polymerization inhibitor TBZ and to the proteosome inhibitor TPCK. Since top3-ts has increased minichromosome loss at the restrictive temperature, we reasoned that, should top3+ be required for centromere architecture or have an uncharacterized role within mitotic progression, it should (like many mutants affecting mitotic processes) be significantly sensitive to either TBZ or TPCK. In fact, we found no evidence of such sensitivity at either the permissive temperature or combined with a transient temperature shift. We thus conclude that it is most likely that the essential function for top3+ is during S-phase.
Top3 is involved in DNA repair
Both the top3-ts mutants were sensitive to the DNA-damaging agents UV and MMS at the restrictive temperature. This sensitivity is similar to that seen for the rqh1-d single mutant and for top3rqh1 double mutants. Previously, in S.pombe it has not been possible to interpret the observation that rqh1-d single and rqh1top3 double mutants have the same sensitivity to DNA-damaging agents because it was not known if a top3 mutant alone was DNA damage sensitive. Our data demonstrate that this is the case, and thus allow the conclusion that Top3 is involved in DNA damage survival, that the top3-ts mutant is epistatic with the rqh1-d mutant and thus Top3 acts in the same DNA repair pathway(s) as Rqh1, as characterized by Murray et al. (16). In support of the contention that Top3 has a significant role in the DNA damage responses, we also observed that top3+ transcript levels were significantly induced by DNA-damaging agents. In addition, Chakraverty et al. (24) demonstrated that top3 deletion mutants were defective in Rad53 phosphorylation following DNA damage during S-phase in S.cerevisiae, suggesting the role of Top3 in DNA damage response during S-phase.
Conclusions
Several models have been proposed to explain the function of the Top3–Sgs1 complex in S.cerevisiae. One possibility is that the complex acts at the termination step of replication (15,25). Another possibility is that the Top3–Sgs1 complex is involved in the processing of structures arising at stalled replication forks (26). Our results are consistent with both these possibilities, which are not themselves mutually exclusive. The high frequency of ‘cut’ phenotypes and the elevated minichromosome loss rate we see in the top3-ts mutant after a transient temperature shift or following HU treatment support a role in the termination step of replication by implying that Top3 is involved in chromosome decatenation and separation. However, top3-ts and rqh1 mutants are both sensitive to DNA-damaging agents, which is consistent with a role in the repair of stalled replication forks. It is, of course, possible that Top3–Rqh1 play roles in both processes or that incorrect repair at sites of stalled replication forks can cause an irreversible catenation event in the absence of Top3. Significant further work to explore these possibilities will be necessary to understand the role of Top3 and Rqh1.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs T. Enoch, T. Cech, R. Allshire and R. Gwilliam for providing yeast strains and plasmids. We also thank Professors A. M. Carr, O. Hwang, H. Kooh and J. B. Kim for their invaluable comments on this manuscript. This research was supported in part by a grant from the Korea Science and Engineering Foundation, grant 1999-2-209-015-3 (to S.D.P.) and by a BK21 Research Fellowship from the Ministry of Education, Republic of Korea (M.O. and S.D.P.).
REFERENCES
- 1.Wallis J.W., Chrebet,G., Brodsky,G., Rolfe,M. and Rothstein,R. (1989) A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell, 58, 409–419. [DOI] [PubMed] [Google Scholar]
- 2.Kim R.A. and Wang,J.C. (1992) Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J. Biol. Chem., 267, 17178–17185. [PubMed] [Google Scholar]
- 3.Gangloff S., Zou,H. and Rothstein,R. (1996) Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J., 15, 1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 4.Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- 6.Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S., Martin,G.M., Mulligan,J. and Schellenberg,G.D. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 7.Yamagata K., Kato,J., Shimamoto,A., Goto,M., Furuichi,Y. and Ikeda,H. (1998) Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA, 95, 8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nature Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- 9.Myung K., Datta,A., Chen,C. and Kolodner,R.D. (2001) SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nature Genet., 27, 113–116. [DOI] [PubMed] [Google Scholar]
- 10.Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- 11.Sinclair D.A., Mills,K. and Guarente,L. (1997) Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science, 277, 1313–1316. [DOI] [PubMed] [Google Scholar]
- 12.Watt P.M., Hickson,I.D., Borts,R.H. and Louis,E.J. (1996) SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics, 144, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duguet M. (1997) When helicase and topoisomerase meet! J. Cell Sci., 110, 1345–1350. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin A., Wang,S.W., Toda,T., Norbury,C. and Hickson,I.D. (1999) Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res., 27, 4050–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watt P.M., Louis,E.J., Borts,R.H. and Hickson,I.D. (1995) Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell, 81, 253–260. [DOI] [PubMed] [Google Scholar]
- 16.Murray J.M., Lindsay,H.D., Munday,C.A. and Carr,A.M. (1997) Role of Schizosaccharomyces pombe RecQ homolog, recombination and checkpoint genes in UV damage tolerance. Mol. Cell. Biol., 17, 6868–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart E., Chapman,C.R., Al-Khodairy,F., Carr,A.M. and Enoch,T. (1997) rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J., 16, 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maftahi M., Han,C.S., Langston,L.D., Hope,J.C., Zigouras,N. and Freyer,G.A. (1999) The top3(+) gene is essential in Schizosaccharomyces pombe and the lethality associated with its loss is caused by Rad12 helicase activity. Nucleic Acids Res., 27, 4715–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Alfa C. (1993) Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 21.Sikorski R.S. and Boeke,J.D. (1991) In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol., 194, 302–318. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y.C., Min,S., Gim,B.S. and Kim,Y.J. (1997) A transcriptional mediator protein that is required for activation of many RNA polymerase II promoters and is conserved from yeast to humans. Mol. Cell. Biol., 17, 4622–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothstein R. (1991) Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–301. [DOI] [PubMed] [Google Scholar]
- 24.Chakraverty R.K., Kearsey,J.M., Oakley,T.J., Grenon,M., de La Torre Ruiz,M.A., Lowndes,N.F. and Hickson,I.D. (2001) Topoisomerase III acts upstream of Rad53p in the S-phase DNA damage checkpoint. Mol. Cell. Biol., 21, 7150–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothstein R. and Gangloff,S. (1995) Hyper-recombination and Bloom’s syndrome: microbes again provide clues about cancer. Genome Res., 5, 421–426. [DOI] [PubMed] [Google Scholar]
- 26.Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays, 21, 286–294. [DOI] [PubMed] [Google Scholar]