Abstract
In kinetoplastid flagellates such as Trypanosoma brucei, a small percentage of the thymine residues in the nuclear DNA is replaced by the modified base β-d-glucosyl-hydroxymethyluracil (J), mostly in repetitive sequences like the telomeric GGGTTA repeats. In addition, traces of 5-hydroxymethyluracil (HOMeUra) are present. Previous work has suggested that J is synthesised in two steps via HOMedU as an intermediate, but as J synthesising enzymes have not yet been identified, the biosynthetic pathway remains unclear. To test a model in which HOMeUra functions as a precursor of J, we introduced an inducible gene for the human DNA glycosylase hSMUG1 into bloodstream form T.brucei. In higher eukaryotes SMUG1 excises HOMeUra as part of the base excision repair system. We show that expression of the gene in T.brucei leads to massive DNA damage in J-modified sequences and results in cell cycle arrest and, eventually, death. hSMUG1 also reduces the J content of the trypanosome DNA. This work supports the idea that HOMeUra is a precursor of J, freely accessible to a DNA glycosylase.
INTRODUCTION
The nuclear DNA of kinetoplastid flagellates such as Trypanosoma and Leishmania contains the hypermodified base β-d-glucosyl-hydroxymethyluracil, called J (1,2). About 1% of the thymine (Thy) residues are replaced by J. The modification is predominantly found in repetitive DNA (3), being most abundant in the telomeric GGGTTA repeats where ∼50% of the total J is localised (4). J is absent from chromosome-internal and transcribed genes and the modification is a target for a specific binding protein (5). The function of J is not known.
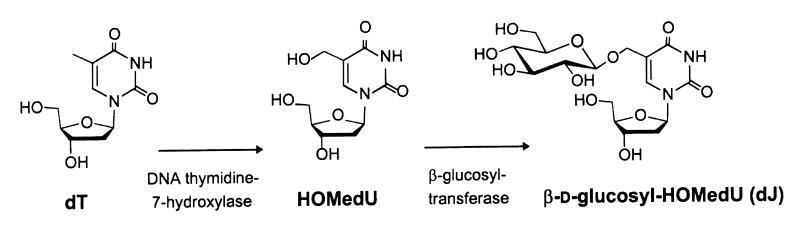
In addition to J, trypanosome DNA contains a small amount of 5-hydroxymethyluracil (HOMeUra) (<0.02% of total DNA) (6,7). In our current model for J biosynthesis (Fig. 1), 5-hydroxymethyldeoxyuridine (HOMedU) is the precursor of J (7). At the DNA level, thymidine (dT) is first converted to HOMedU which is then glucosylated. Support for this two-step pathway comes from the finding that exogenous HOMedU is randomly incorporated into the DNA and converted into J (7). Despite major efforts, the enzymes catalysing J biosynthesis have not been identified thus far. Direct support for the two-step pathway is therefore still missing. It is also not known whether the two steps are normally separated or coupled events. In the latter case HOMedU might only be an intermediate bound to the modifying machinery, not a ‘normal’ component of the DNA.
Figure 1.
Speculative two-step model for J biosynthesis. At the DNA level, thymidine is first converted to HOMedU by a putative thymidine 7-hydroxylase and then HOMedU is converted into J by a putative β-glucosyltransferase. From van Leeuwen et al. (7).
To test the two-step model for J biosynthesis, we introduced an enzyme into trypanosomes that excises the base HOMeUra out of the DNA. The enzyme used was the human single-strand-selective monofunctional uracil-DNA glycosylase, hSMUG1 (8). hSMUG1 is a component of the base excision repair (BER) system (reviewed in 9,10). It excises HOMeUra (11) and uracil (12) from single- and double-stranded DNA. The excision results in an abasic site, which is then further processed by other BER factors. hSMUG1 homologs are found in several higher eukaryotes (12) and its presence might be evolutionarily linked to DNA methylation (11,13). We were unable to detect homologous sequences in the trypanosome databases (S.Ulbert and P.Borst, unpublished results). In this paper we present the devastating effects of hSMUG1 on Trypanosoma brucei.
MATERIALS AND METHODS
Trypanosomes, culture conditions and nucleoside feeding experiments
Bloodstream form trypanosomes of strain 427 of T.brucei brucei (14) were cultured as described (15). For the nucleoside feeding experiments (7) thymidine was omitted from the medium. Tetracycline induction was performed with or without continuous nucleoside feeding for HOMedU and 5-bromodeoxyuridine (BrdU), respectively. Nucleosides were purchased from Sigma. Tetracycline (Roche) was used at 1 µg/ml or 5 ng/ml for high or low levels of induction, respectively. Tetracycline-free fetal calf serum was purchased from Clontech.
Insertion of the hSMUG1 gene into T.brucei
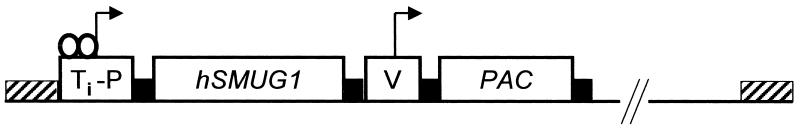
The coding sequence for hSMUG1 was amplified using PCR on plasmid pGEX6P1, which contains the hSMUG1 cDNA as a GST fusion (11). The primers used contained a HA epitope tag at either the 5′ or 3′ end. The gene was cloned into the vector pHD615 PAC that is based on the plasmid pHD615 (16), but contains a puromycin acetyltransferase resistance gene instead of a hygromycin resistance gene. The inducible promoter of this construct contains a tetracycline operator (Fig. 2). The cell line used for transfection of the final construct (pHDhSMUG1) was HN TET. These cells have the tetracycline repressor gene from the construct pHD 449 (16) in the α/β-tubulin gene array and a hygromycin phosphotransferase resistance gene in the 221 VSG gene expression site as well as a neomycin resistance gene in the VO2 VSG gene expression site (17). HN TET cells were continuously grown in phleomycin at 2 µg/ml to select for the tetracycline repressor gene. Bloodstream form transfection was carried out as described (18). Drug selection of the hSMUG1 transfectants was done using puromycin (Sigma) at 0.1 µg/ml and phleomycin at 2 µg/ml to select for the hSMUG1 construct and for the tetracycline repressor gene, respectively. The HA tag was situated at either the N- or the C-terminus of the hSMUG1 gene. Transfectants for both constructs were cloned out and further investigated. As the position of the HA tag did not seem to have any influence on the behaviour of the cells upon tetracycline induction, all the experiments reported here were done using trypanosomes expressing the N-terminally HA tagged hSMUG1.
Figure 2.
Schematic drawing of the construct pHDhSMUG1 used to yield inducible expression of hSMUG1. The construct is based on pHD615 (16). Ti-P, tetracycline-inducible promoter; V, constitutive VSG promoter; hSMUG1, human single-strand-selective monofunctional uracil-DNA glycosylase; PAC, puromycin acetyltransferase; black boxes, trypanosome-specific RNA processing signals (for details see 16); hatched boxes, flanks for integration into the non-transcribed rRNA spacer in opposite orientation to the direction of endogenous transcription. The solid black line represents pGEM vector sequence. The two open circles are tetracycline repressor molecules derived from transcription of the repressor gene in the α/β-tubulin arrays (data not shown). Binding of the repressor is inhibited by tetracycline.
In vitro BER assays
To prepare crude cell lysates, 3 × 108 trypanosomes were harvested from in vitro cultures and washed in phosphate-buffered saline. The pellet was resuspended in 500 µl of lysis buffer (10 mM HEPES, pH 7.6, 1 mM EDTA, 50% glycerol, supplemented with the complete protease inhibitor mixture, EDTA-free, from Roche) and left on ice for 30 min. Subsequently the cells were disrupted by douncing for 10 strokes and three sonications using a Sonifier™ B-30 cell disrupter (output control 5, intensity 50%, five pulses). The crude cell lysate was frozen in small aliquots. Aliquots of 0.1–1 µl of the extracts were used for the in vitro assays. The oligonucleotides used in the BER assays were a 24mer containing four HOMeUra residues at position 5, 11,17 and 23 (5) and a 26mer containing one uracil residue at position 14 (Sigma). The oligonucleotides were 32P-labelled at the 5′ end and annealed to a 5-fold molar excess of unlabelled complementary strand containing a guanine residue opposite the modified base. One unit of the PBS1 uracil glycosylase inhibitor (New England Biolabs) was used to inhibit the endogenous uracil-DNA glycosylase.
The cell lysate was incubated with 100 fmol of the labelled oligonucleotides in 50 mM HEPES, pH 7.5, 20 mM NaCl, 1 mg/ml BSA, 1 mM EDTA and 1 mM DTT. The reaction (final volume 15 µl) was carried out at 37°C for 1 h. To break abasic sites, NaOH was added to a final concentration of 0.1 M and the samples were incubated at 90°C for 10 min. Then an equal volume of gel loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue) was added. An 18% polyacrylamide gel containing urea at 7 M was used to separate the reaction products and an image was generated using a Fuji BAS reader.
Cell cycle analysis
Trypanosomes were fixed in formaldehyde (19) and transferred to a microscope slide. The slides were dehydrated using 70, 90 and 100% ethanol and the DNA was stained with DAPI. The nuclei and kinetoplasts of the trypanosomes were counted under a fluorescence microscope. Between three and five counts were performed per cell type and at least 100 cells were analysed per count. The DNA measurement by flow cytometry was performed as described (20).
DNA analysis
Trypanosomes were incorporated into low melting point agarose blocks (2.5 × 107 cells/block) and digested with proteinase K (Merck) for 48 h. The blocks were loaded on a standard 1% agarose gel that was run overnight at 10 V. Alkaline gel electrophoresis, blotting and hybridisation were done according to Sambrook et al. (21). Alkaline gels were run under the same conditions as neutral gels. The probe for the telomeric repeats was an oligonucleotide with the sequence (TAGGGT)4 and the α/β-tubulin probe has been described (22). The anti-J-DNA immunoblot was carried out as described (23). After stripping, the blot was rehybridised with a probe for the α/β-tubulin genes and the J content of the cells was calculated based on DNA loading.
RESULTS
Construction of trypanosomes containing an inducible hSMUG1 gene
To test whether HOMedU is an accessible intermediate in the biosynthesis of J we introduced a DNA glycosylase that is able to remove HOMeUra from DNA (11) into bloodstream form T.brucei. As the expression of a DNA repair enzyme against HOMeUra could be harmful to a trypanosome, we used the tetracycline system (24) to control the transcription of hSMUG1. The trypanosome line HN TET contains the gene for the tetracyline repressor (see Materials and Methods). This cell line was used to insert a copy of the human hSMUG1 under the control of a tetracycline-inducible promoter (Fig. 2). The site of integration was the rDNA array (16). HN TET trypanosomes are referred to as wild-type cells.
Functional hSMUG1 is made in the transfectant upon tetracycline induction
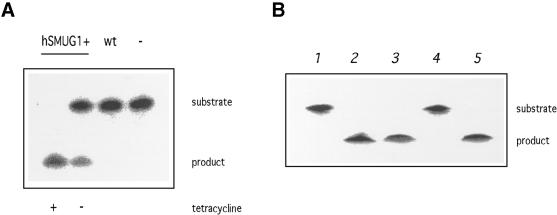
To investigate the functionality of our constructs, we determined the hSMUG1 activity in lysates prepared from trypanosomes cultured with or without tetracycline. In vitro assays were performed using radiolabelled oligonucleotides with HOMedU at defined positions. Incubation with functional hSMUG1 leads to the excision of the modified base and an abasic site. Treatment with NaOH at 0.1 M and heat breaks the oligonucleotide at the abasic site and makes the glycosylase activity detectable on a sequencing gel. As shown in Figure 3A, no activity against HOMedU was detectable in wild-type trypanosomes. In contrast, hSMUG1 transfectants showed a high activity upon tetracycline induction (Fig. 3A). Even without induction the intensity of the signal corresponding to the cut oligonucleotide was ∼2% of the samples from trypanosomes cultured with tetracycline (calculated after testing a range of extract concentrations; data not shown). The uninduced activity was still present when serum was used that contained no traces of tetracycline (data not shown), suggesting that this activity resulted from leaky background transcription independent of tetracycline.
Figure 3.
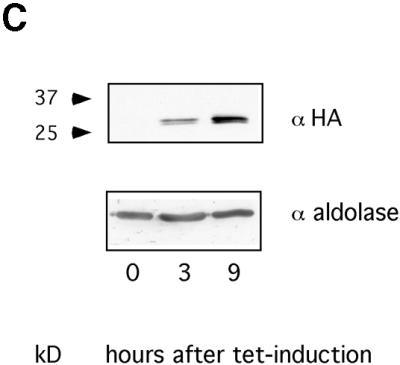
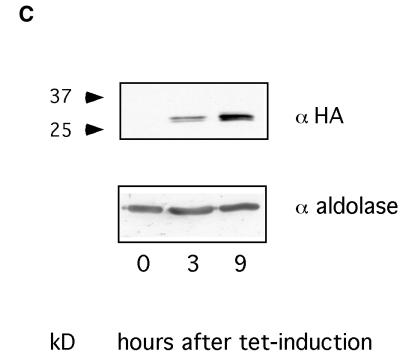
(A) In vitro BER assay using crude cell lysates on a duplex oligonucleotide containing HOMedU paired with G (the HOMedU:A base pair yielded similar results; not shown). The substrate band represents the full-length oligonucleotide, the product band is the cut oligonucleotide after excision of the HOMeUra residue. The lane on the right (–) is the substrate without lysate. (B) In vitro BER assay using crude cell lysates on a duplex oligonucleotide containing uracil paired with G. Lane 1, substrate without lysate; lane 2, wild-type cells; lane 3, hSMUG1 transfectants with tetracycline; lane 4, wild-type cells with UGI; lane 5, hSMUG1 transfectants with tetracycline and UGI. (C) Western blot analysis of the hSMUG1 transfectants. The blot was incubated with an anti-HA tag antibody, stripped, and incubated with an anti-aldolase antibody as a loading control. 3 × 106 cell equivalents were loaded per lane.
Besides HOMeUra, hSMUG1 also excises uracil, especially when mispaired with G. This activity is insensitive to the uracil glycosylase inhibitor (UGI) of Bacillus subtilis bacteriophage PBS1, which inhibits uracil-DNA glycosylase of both bacterial and eukaryotic origins (25). Figure 3B shows that T.brucei contains an active uracil-DNA glycosylase (lane 2), which is completely inhibited by UGI (lane 4). Extracts from trypanosomes expressing hSMUG1 in contrast showed a high activity against uracil that was not inhibited by UGI (Fig. 3B, lane 5). We conclude that the HA-tagged hSMUG1 that is produced in the transfected trypanosomes has both enzymatic activities characteristic of hSMUG1, excision of uracil and of HOMeUra.
We measured the time course of hSMUG1 expression after tetracycline induction on a protein blot using antibodies directed against the HA tag. As shown in Figure 3C, a band of the correct size (11) could be detected after 3 h and a maximum signal was reached after 9 h.
hSMUG1 causes cell cycle arrest and cell death in T.brucei, dependent on the level of HOMedU in the DNA
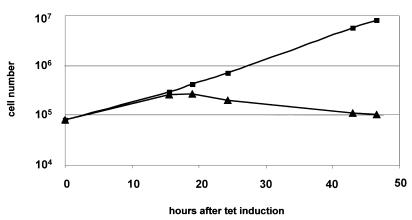
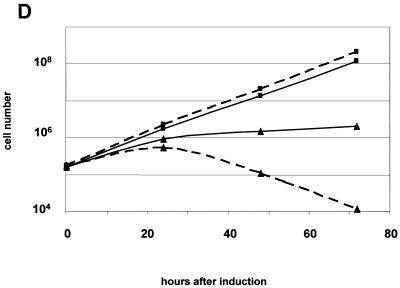
The hSMUG1 transfectants showed no significant difference in growth compared to wild-type trypanosomes (data not shown). However, addition of tetracycline to the medium killed the cells. After maximally two cell divisions the culture stopped growing and the cells started to die (Fig. 4). The cells did not survive in tetracycline concentrations >5 ng/ml (data not shown). This indicates that even slight induction above the leaky expression of hSMUG1 is lethal to the trypanosome.
Figure 4.
Growth curve of hSMUG1 transfectants with (triangles) and without (squares) tetracycline (1 µg/ml).
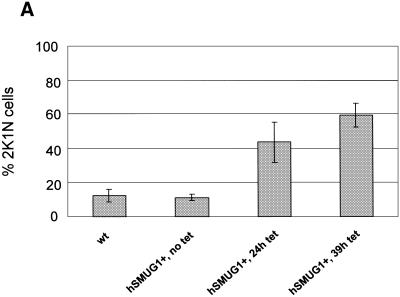
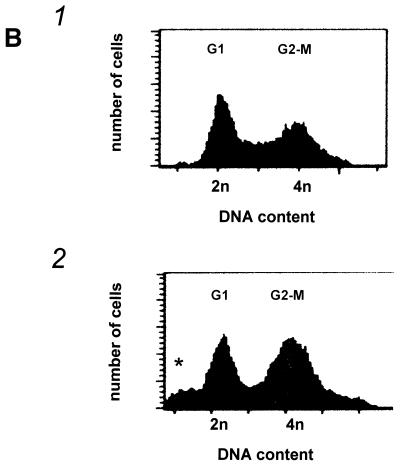
To assess how the trypanosomes died, we investigated the influence of hSMUG1 on the cell cycle pattern. In T.brucei, progression through the cell cycle can be determined by counting the number of kinetoplasts (a structure containing mitochondrial DNA networks) and nuclei within one cell. As the kinetoplast initiates the S and G2 phases earlier than the nucleus (26), a cell with two kinetoplasts and one nucleus (2K/1N) is in the G2 phase of the cell cycle. In wild-type cells and hSMUG1 transfectants without tetracycline the number of 2K/1N cells was ∼10% (Fig. 5A), whereas cultures expressing high levels of hSMUG1 showed an increase in 2K/1N cells to ∼50%. Thus, a large part of the culture was stalled in the G2 phase of the cell cycle. To verify these results, we measured DNA content by flow cytometry. Figure 5B shows that the number of cells in the G2/M fraction increased relative to the G1 fraction. In addition, cells with less than the diploid amount of DNA (dying or dead) were present.
Figure 5.
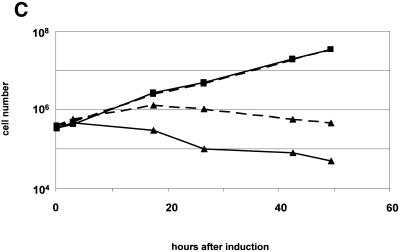
(A) Percentage of cells with two kinetoplasts and one nucleus in cultures of trypanosomes exposed to various levels of endogenous hSMUG1. (B) Measurement of the DNA content of trypanosomes using flow cytometry. (1) Wild-type cells; (2) hSMUG1 transfectants after 40 h in tetracycline (1 µg/ml). Sub-G1 cells are marked with an asterisk. (C and D) Effect of nucleoside feeding on hSMUG1 induction. (C) Growth curve of hSMUG1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with HOMedU (200 µM for 48 h). Dotted lines are non-fed control cells. (D) Growth curve of hSMUG1 transfectants with (triangles) or without (squares) tetracycline (1 µg/ml). The full lines represent trypanosomes that were fed with BrdU (100 µM for 72 h). Dotted lines are non-fed control cells.
To test whether the lethal effect of hSMUG1 could be increased by raising the level of HOMedU in the DNA, we grew the trypanosomes in medium containing the nucleoside HOMedU for about six generations. This HOMedU is incorporated into the DNA and converted into J (7). Without tetracycline, the hSMUG1 transfectants showed no significant difference in growth in medium containing HOMedU at 200 µM compared to wild-type cells (Fig. 5C). This indicates that the endogenous HOMedU is already in excess to the low amounts of hSMUG1 produced by leaky transcription. However, when tetracycline was added, hSMUG1 cells that were HOMedU fed showed a stronger growth defect (Fig. 5C). They hardly doubled in number and started to die earlier than the control (non-fed) hSMUG1 trypanosomes.
We also analysed the effect of BrdU feeding on trypanosome death by hSMUG1 induction. Growing trypanosomes for eight to nine generations in BrdU at 100 µM leads to a replacement of 15–20% of the Thy residues. BrdU cannot be converted into HOMedU and growth in BrdU decreases the J level about 3-fold (7). Furthermore, BrdU is not removed from DNA by repair (G.W.Teebor, unpublished observation). Figure 5D shows that induction of hSMUG1 in these cells had much less effect on cell growth than induction in trypanosomes that had not been pre-grown in BrdU. The BrdU-fed trypanosomes also reacted to hSMUG1, but the damage caused by the enzyme was less, allowing the cells to survive. The culture even continued growing at a low rate. Taken together these results indicate that the growth inhibitory effect of hSMUG1 on trypanosomes is strictly dependent on the level of HOMedU in the trypanosome DNA.
Expression of hSMUG1 causes DNA damage in T.brucei
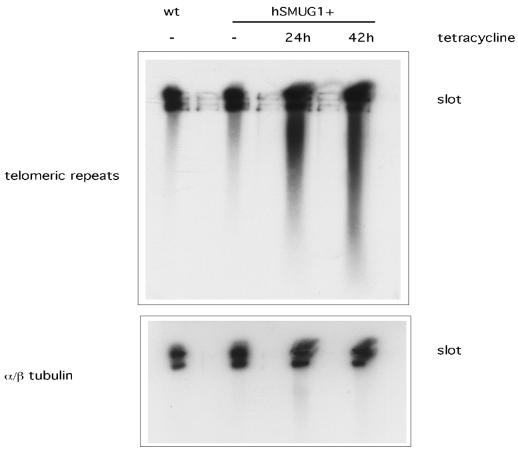
The rapid decrease in trypanosome multiplication after hSMUG1 induction suggested that the cells were overwhelmed by DNA damage. To verify this, cells were incorporated into agarose blocks, digested with proteinase K and loaded on a standard agarose gel run at low voltage. The gel was transferred to a nitrocellulose membrane and probed for the telomeric GGGTTA repeats, which contain high amounts of J. As can be seen in Figure 6, the DNA of wild-type cells and uninduced hSMUG1 transfectants remained mostly in the slot. After tetracycline induction the DNA of the hSMUG1 transfectants was fragmented and migrated to a substantial extent into the gel. The fraction of DNA migrating into the gel was further increased when an alkaline gel was used (data not shown). As abasic sites in the DNA are alkali-labile, DNA fragmentation increases under alkaline conditions. We also investigated the chromosome-internal α/β-tubulin gene arrays, which do not contain J (23), and found that the tubulin genes migrated only marginally into the gel compared to the telomeric repeats (Fig. 6), even when an alkaline gel was used (data not shown).
Figure 6.
Agarose gel electrophoresis on wild-type cells and hSMUG1 transfectants cultured with and without tetracycline. Whole cells were cast in agarose blocks, digested with proteinase K and subjected to electrophoresis. The gel was run at a low voltage, blotted and hybridised with a telomeric repeat probe. Subsequently, the membrane was stripped and hybridised with a probe for the α/β-tubulin gene array.
It was shown previously that excessive BER in mammalian cells leads to double-strand breaks (27). As the number of abasic sites arising from the action of hSMUG1 exceeds the capacity of the endogenous repair machinery, a fragmentation of sequences with a high HOMedU level is the consequence. The massive abundance of double-strand breaks and alkali-labile sites in hSMUG1 cells after tetracycline induction correlates with the effect of hSMUG1 on growth. Our interpretation of these results is that high expression of hSMUG1 leads to excessive DNA repair. The result is a DNA damage response of the cells that leads to cell cycle arrest and eventually to death. Our results also confirm that HOMedU is absent from transcribed, chromosome-internal genes as there is no DNA damage detectable in the α/β-tubulin array.
Interestingly, there was no significant difference detectable between wild-type cells and hSMUG1 transfectants without tetracycline (Fig. 6), although these trypanosomes show a low level of functional hSMUG1 (Fig. 3A). This means that the endogenous BER system of the trypanosomes can cope with the ‘extra repair’ due to hSMUG1 (confirming the growth curve in Fig. 4), until it exceeds a certain limit. This limit is presumably set by the endogenous BER factors of the trypanosome (such as AP endonucleases, DNA polymerase β, etc.) that become limiting after tetracycline induction.
hSMUG1 interferes with J biosynthesis
As HOMedU has been shown to be involved in J biosynthesis (7), we analysed the influence of hSMUG1 on the J level. The J content of the cells was determined by a dot-blot analysis using a polyclonal anti-J antibody. Twenty-four hours after tetracycline addition the hSMUG1 trypanosomes had about four times less J than non-induced or wild-type cells (Fig. 7). As the trypanosomes can maximally complete two cell divisions in that period (Fig. 4) we conclude that hSMUG1 is removing most of the available HOMedU made after DNA replication, thereby inhibiting de novo J synthesis.
Figure 7.
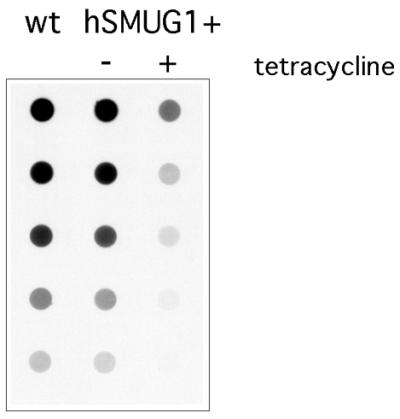
J measurement on genomic DNA of wild-type trypanosomes and hSMUG1 cells cultured with and without tetracycline. Aliquots of 50 ng of DNA were diluted in 1:1 steps and spotted onto a membrane which was then incubated with an anti-J antibody. The membrane was stripped and hybridised with a probe for the α/β-tubulin gene array to calculate the difference in J level corrected for DNA loading (data not shown).
It is unlikely that hSMUG1 excises J directly, as we have shown by in vitro assays with purified, recombinant hSMUG1 that the enzyme does not excise J from duplex or single-stranded oligonucleotides (S.Ulbert, L.Eide, E.Seeberg and P.Borst, manuscript in preparation). We also investigated the effect of a low tetracycline induction over a longer period of time on J levels. Incubation of the hSMUG1 transfectants in tetracycline at 5 ng/ml for 5 days resulted in a 90% decrease in the J level and a slower growth of the cells (data not shown). We have not succeeded in decreasing the J level further for technical reasons. Tetracycline is unstable in culture systems and we were unable to maintain lower tetracycline levels than 5 ng/ml over a long timespan. Even with a daily refreshment of tetracycline the cells started to pick up normal growth after ∼5 days and the J content was approximating wild-type levels again (data not shown).
DISCUSSION
Trypanosoma brucei and related kinetoplastid flagellates contain the modified base J in their nuclear DNA. Whereas the amount and distribution of J has been determined in detail, little is known as yet about its biosynthesis. We have previously shown by nucleoside-feeding experiments that exogenous HOMedU is incorporated in trypanosome DNA and converted into J (7). This suggests but does not prove that free HOMedU is an obligatory intermediate in J biosynthesis, and led to the model shown in Figure 1. However, all attempts to find the glucosyltransferase mediating the conversion of HOMedU into J have failed thus far and direct support for the scheme in Figure 1 is therefore still lacking. To test whether HOMedU is an intermediate that is freely accessible to a DNA glycosylase able to take out the base we have introduced such a glycosylase, hSMUG1, into T.brucei.
Expression of hSMUG1 in trypanosomes led to a dramatic loss of viability. This effect was dependent on the level of HOMedU in the DNA. Raising the HOMedU content led to a stronger effect, i.e. the cells died much earlier; lowering it by feeding BrdU reduced the sensitivity of the transfectants. We conclude from these results that hSMUG1 killed the cells by excising HOMeUra.
Analysis of the DNA in the trypanosomes expressing hSMUG1 revealed severe DNA damage due to an accumulation of double-strand breaks and abasic sites. It has been shown previously that overexpression of a DNA glycosylase can lead to DNA damage rather than increased, complete BER (28). An increase in the initial step of BER, the excision of a base, needs to be accompanied by an increase in the levels of the other BER factors. Otherwise the BER system becomes imbalanced (29), leading to genome instability and decreased viability. In our case we introduce a DNA glycosylase which attacks a genome in which one of its substrates is present in excess, as has been done by Kavli et al. (30) for Escherichia coli and Mi et al. (27) for mammalian cells. This should result in the accumulation of abasic sites that lead to single strand breaks. Double strand breaks are then the consequence of two adjacent single-strand breaks on both strands, as we detected in the telomeres of T.brucei.
It is unlikely that the trypanosomes died because of the decrease in J level caused by hSMUG1, as we know that bloodstream form T.brucei can tolerate much lower levels of J, as low as 5% of the wild-type (31). We rather think that hSMUG1 kills the trypanosomes by overwhelming DNA damage resulting in cell cycle arrest and death. The cells can deal with the damage as long as hSMUG1 is limiting and not induced. This shows that the endogenous BER system can process the abasic sites produced by low level excision of HOMedU.
It is remarkable that a 3-fold reduction in J in trypanosomes grown in BrdU prevents cell death by hSMUG1 induction. Under these conditions, <20% of all dT in the DNA is replaced by BrdU, suggesting that the putative thymidine hydroxylase, which converts dT into HOMedU, could be inhibited by BrdU in DNA, resulting in a substantially decreased rate of HOMedU synthesis and, hence, much less HOMeUra excision by hSMUG1.
The excision of HOMeUra by hSMUG1 resulted in a substantial decrease in the level of J in trypanosome DNA. In a parallel study we investigated the role of J as a potential target for different DNA glycosylases (S.Ulbert, L.Eide, E.Seeberg and P.Borst, manuscript in preparation). We found that J is not recognised by hSMUG1 and, therefore, conclude that the J level of the hSMUG1 transfectants decreased as a consequence of the excision of HOMeUra, supporting the role of HOMeUra as a precursor of J.
Because all our attempts to detect a glucosyltransferase in trypanosome extracts have failed thus far, alternative routes for the biosynthesis of J have been considered. Glucosylation might be coupled to hydroxylation in a concerted reaction and the possibility has even been raised that hydroxylation is energetically driven by the glucosylation reaction (S.Beverley, personal communication). These schemes have become less attractive, as we have shown that the HOMedU used for J synthesis appears to be freely accessible to hSMUG1. The two-step synthesis of J presented in Figure 1 remains therefore a plausible working hypothesis and we are continuing our efforts to identify the enzymes involved.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Paul-André Genest, Rudo Kieft, Henri van Luenen, Cristiane Toaldo, Zhong Yu and Hein te Riele for critical reading of this manuscript and Adrian Begg, Marcel van Vugt and Stephen Beverley (Washington University) for discussing experiments. We thank Christine Clayton (ZMBH Heidelberg) for sending us the plasmids pHD449 and pHD615 and Nico Meeuwenoord and Jaques H. van Boom (University of Leiden) for DNA oligonucleotides containing HOMedU. This work was supported by a grant from the Boehringer Ingelheim Fonds to S.U. and by the Netherlands Foundation for Chemical Research (CW), with financial aid from the Netherlands Organisation for Scientific Research (NWO) to P.B.
REFERENCES
- 1.Gommers-Ampt J., van Leeuwen,F., de Beer,A.L.J., Vliegenthart,J.F.G., Didzdaroglu,M., Kowalak,J.A. and Borst,P. (1993) Beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan Trypanosoma brucei. Cell, 75, 1129–1136. [DOI] [PubMed] [Google Scholar]
- 2.van Leeuwen F., Taylor,M.C., Mondragon,A., Moreau,H., Gibson,W., Kieft,R. and Borst,P. (1998) Beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl Acad. Sci. USA, 95, 2366–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Leeuwen F., Kieft,R., Cross,M. and Borst,P. (2000) Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Biochem. Parasitol., 109, 133–145. [DOI] [PubMed] [Google Scholar]
- 4.van Leeuwen F., Wijsman,E.R., Kuyl-Yeheskiely,E., van der Marel,G.A., van Boom,J.H. and Borst,P. (1996) The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res., 24, 2476–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross M., Kieft,R., Sabatini,R., Wilm,M., de Kort,M., van der Marel,G.A., van Boom,J., van Leuuwen,F. and Borst,P. (1999) The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J., 18, 6573–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gommers-Ampt J., Teixeira,A.J.R., van de Werken,G., van Djk,W. and Borst,P. (1993) The identification of hydroxymethyluracil in DNA of Trypanosoma brucei. Nucleic Acids Res., 21, 2039–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Leeuwen F., Kieft,R., Cross,M. and Borst,P. (1998) Biosynthesis and function of the modified DNA base β-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Cell. Biol., 18, 5643–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haushalter K.A., Stukenberg,P.T., Kirschner,M.W.and Verdine,G.L. (1999) Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol., 9, 174–185. [DOI] [PubMed] [Google Scholar]
- 9.Schärer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- 10.Krokan H.E., Nilsen,H., Skorpen,F., Otterlei,M. and Slupphaug,G. (2000) Base excision repair of DNA in mammalian cells. FEBS Lett., 476, 73–77. [DOI] [PubMed] [Google Scholar]
- 11.Boorstein R.J., Cummings,A., Marenstein,D.R., Chan,M.K., Ma,Y., Neubert,T.A., Brown,S.M. and Teebor,G.W. (2001) Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem., 276, 41991–41997. [DOI] [PubMed] [Google Scholar]
- 12.Nilsen H., Haushalter,K.A., Robins,P., Barnes,D.E., Verdine,G.L. and Lindahl,T. (2001) Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J., 20, 4278–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boorstein R.J., Chiu,L.N. and Teebor,G.W. (1989) Phylogenetic evidence of a role for 5-hydroxymethyluracil-DNA glycosylase in the maintenance of 5-methylcytosine in DNA. Nucleic Acids Res., 17, 7653–7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross G.A.M. (1975) Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology, 71, 393–417. [DOI] [PubMed] [Google Scholar]
- 15.Hirumi H. and Hirumi,K. (1989) Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder layers. J. Parasitol., 75, 985–989. [PubMed] [Google Scholar]
- 16.Biebinger S., Wirtz,E., Lorenz,P. and Clayton,C. (1997) Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol., 85, 88–112. [DOI] [PubMed] [Google Scholar]
- 17.Rudenko G., Chaves,I., Dirks-Mulder,A. and Borst,P. (1998) Selection for activation of a new variant surface glycoprotein gene expression site in Trypanosoma brucei can result in deletion of the old one. Mol. Biochem. Parasitol., 95, 97–109. [DOI] [PubMed] [Google Scholar]
- 18.Carruthers V.B., van der Ploeg,L.H. and Cross,G.A. (1993) DNA-mediated transformation of bloodstream-form Trypanosoma brucei. Nucleic Acids Res., 21, 2537–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaves I., Zomerdijk,J., Dirks-Mulder,A., Dirks,R.W., Raap,A.K. and Borst,P. (1998) Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 12328–12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muñoz-Jordan J.L. and Cross,G.A. (2001) Telomere shortening and cell cycle arrest in Trypanosoma brucei expressing human telomeric repeat factor TRF1. Mol. Biochem. Parasitol., 114, 169–181. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Ulbert S., Chaves,I. and Borst,P. (2001) Expression site activation in Trypanosoma brucei with three marked variant surface glycoprotein gene expression sites. Mol. Biochem. Parasitol., 120, 225–235. [DOI] [PubMed] [Google Scholar]
- 23.van Leeuwen F., Wijsman,E.R., Kieft,R., van der Marcel,G.A., van Boom,J.H. and Borst,P. (1997) Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev., 11, 3232–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wirtz E. and Clayton,C. (1995) Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science, 268, 1179–1183. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z. and Mosbaugh,D.W. (1989) Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J. Biol. Chem., 264, 1163–1171. [PubMed] [Google Scholar]
- 26.Matthews K.R. and Gull,K. (1994) Cycles within cycles: the interplay between differentiation and cell division in Trypanosoma brucei. Parasitol. Today, 10, 473–476. [DOI] [PubMed] [Google Scholar]
- 27.Mi L.J., Chaung,W., Horowitz,R., Teebor,G.W. and Boorstein,R.J. (2001) Excessive base excision repair of 5-hydroxymethyluracil from DNA induces apoptosis in Chinese hamster V79 cells containing mutant p53. Carcinogenesis, 22, 179–186. [DOI] [PubMed] [Google Scholar]
- 28.Glassner B.J., Rasmussen,L.J., Najarian,M.T., Posnick,L.M. and Samson,L.D. (1998) Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl Acad. Sci. USA, 95, 9997–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frosina G. (2000) Overexpression of enzymes that repair endogenous damage to DNA. Eur. J. Biochem. 267, 2135–2149. [DOI] [PubMed] [Google Scholar]
- 30.Kavli B., Slupphaug.G., Mol,C.D., Arvai,A.S., Petersen,S.B., Tainer,J.A. and Krokan,H.E. (1996) Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J., 15, 3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 31.Cross M., Kieft,R., Sabatini,R., Dirks-Mulder,A., Chaves,I. and Borst,P. (2002) J binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol. Microbiol., in press. [DOI] [PubMed] [Google Scholar]