Abstract
A 1536 channel oligonucleotide synthesizer, the MultiSyn, was developed with the capability to simultaneously synthesize 1536 oligonucleotides of 20mer length in 10 h. The instrument was designed to synthesize different sequences of various lengths in micro-wells and has synthesized oligonucleotides as long as 119 nt with reasonably good yields using CPG beads of 1000 Å pore size. The instrument consists of four 384 channel synthesis modules. Phosphoramidite chemistry was employed and step yields as high as 99.3% were achieved. The enhancement of oligonucleotide synthesis throughput is accomplished by increasing the spatial density of reaction wells. We have identified several parameters that are critical in achieving a good synthesis yield and negligible failure rate in small reaction wells. The coefficient of variation (CV) of product yields in 1536 reaction wells was 20%. The quality of the product was examined by capillary electrophoresis and mass spectrometry. The instrument has robustly synthesized oligonucleotides of various lengths for use as primers and probes for PCR amplifications, oligonucleotide microarrays and genotyping applications. This high throughput oligonucleotide synthesizer is a useful instrument for genomic applications, which require tens of thousands of probes or primers in a short time.
INTRODUCTION
Since the emergence of chemical synthesis methods for oligonucleotides (1–3), synthetic oligonucleotides have played a pivotal role in many research fields such as molecular biology, forensic science, medical diagnostics and many others. In recent years, with the advent of massively parallel analysis technologies such as the DNA microarrays, there has been an increasing demand for a large number of oligonucleotides. The demand for oligonucleotides has risen even more tremendously with the genome-wide single nucleotide polymorphisms validation and genotyping projects that require hundreds of thousands of oligonucleotides of different sequences (4,5). For genome-wide studies, cost of materials is a critical issue besides throughput. With the current oligonucleotide synthesis formats, the cost of oligonucleotide probes or primers can be prohibitively high for conducting genome-wide studies. Synthesis yield on a scale smaller than the current commercial preparation of 40 nmol is sufficient for many applications. For instance, PCR amplification usually requires a few to tens of picomoles of oligonucleotide primers. For oligonucleotide microarray fabrication each probe requires sub-picomole amounts of oligonucleotides per array. Several approaches have been developed to in situ synthesize oligonucleotide probes on glass slides for microarray applications (6–11). Other than the oligonucleotide microarray applications, many genomic studies often require hundreds of thousands of different oligonucleotides with quantities beyond the scale achievable by the aforementioned in situ synthesis method. A high throughput and low cost oligonucleotide synthesis method is still in need and will have a significant impact on genome studies.
In the quest to simultaneously synthesize a large variety of oligonucleotides (12), several types of multi-channel systems have been developed to synthesize around 100–200 oligonucleotides in a single run (13,14). All the currently available oligonucleotide synthesizers are based on the original or modified phosphoramidite chemistry developed by Beaucage and Caruthers (1) and Adams et al. (2).
Synthesis throughput can be improved by increasing the number of parallel syntheses. In standard 96-well microplates, the reaction wells have a diameter of ∼7 mm and the center-to-center distance is 9 mm. Several difficulties in synthesis may be encountered when a 384-well format is employed. For instance, cross-contamination between neighboring reaction wells, a large variation in yields in different wells, and splash of reaction reagents and synthesis supports out of the reaction wells under the influence of pressure/reagents. The aforementioned difficulties are more demanding in 1536-well plates where the well spacing is reduced to 2.25 mm, center-to-center. Increasing the number of parallel syntheses by the use of standard 384-well or 1536-well microplates is therefore technically challenging and requires new developments and breakthroughs in instrument design.
MATERIALS AND METHODS
Reagents
Standard phosphoramidite chemistry was used for oligonucleotide synthesis. The phosphoramidite reagents were purchased from Applied Biosystems (Foster City, CA) or ChemGenes (Ashland, MA). Anhydrous acetonitrile (5 p.p.m. water content) was purchased from Tedia (Fairfield, OH). Ammonium hydroxide (32%) was from Merck (Darmstadt, Germany). Methylamine was from Acros (Geel, Belgium). The solid-phase synthesis support, controlled pore glass (CPG), was purchased from Glen Research (Universal-Q Support; Sterling, VA).
Synthesizer design and construction
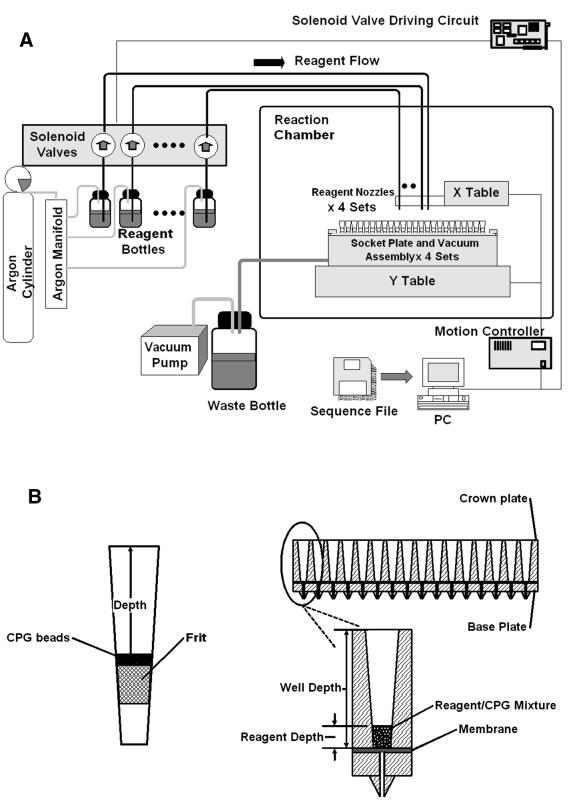
The 1536 channel synthesizer consists of four 384 channel synthesis modules. As depicted in Figure 1A, each module consists of three major components.
Figure 1.
(A) Schematic diagram of the 1536 channel oligonucleotide synthesizer. The synthesizer consists of four sets of 384 channel synthesis modules. All the synthesis modules are controlled by a motion and reagent delivery control system. (B) Schematic diagrams illustrating one design of a reaction well consisting of a filter tip with a frit to retain the CPG beads and another design of a high density reaction assembly consisting of a crown plate, a CPG bead- retaining medium (membrane) and a base plate.
A reaction plate assembly and vacuum assembly: a tailor-made 384-well microplate assembly compliant with the standard microplate format serves as a reaction cartridge in which each well is an independent reaction vessel. After each synthesis step, the wells are drained by a vacuum pump.
A reagent reservoir and tubing: a set of 12 or more reagent bottles hold the reagents, which are transported in Teflon tubing from the bottles to the reagent valve assembly.
A reagent valve assembly: a set of solenoid valves (LFVX050840A; The Lee Co., Westbrook, CT), ejection nozzles and a fast-response driving circuit are integrated to allow the ejection volume to be adjusted to as little as 2 µl/ejection. All reagent nozzles are arranged in order and assembled on a nozzle mount. The mount is designed to hold 20 different nozzles and is fixed to one of the translation stages. The current design uses 12 valves in each module so that, in addition to the deoxyribonucleoside phosphoramidites (A, G, C and T), two additional labeling reagents, Aminolink (ABI 402872) and C18-spacer (ChemGenes CLP-9765), can be used.
All four 384-well synthesis modules are mounted on a translation stage and the nozzle mounts are seated on another translation stage. The two translation stages together make up a two-dimensional (2-D) motion device that allows the nozzles to cover the whole reaction plate assemblies. A menu-driven software program, written for the Microsoft Windows operating system, controls the 2-D reagent delivery system. The menu-driven program allows users to import sequence information in batch format from a plain text file. The computer program reads the sequence information, generates a synthesis map and carries out the syntheses in a fully automatic way. The computer program performs synthesis by the map and does not issue ejection commands to unused reaction wells when less than 1536 reaction wells are needed. The four synthesis modules, nozzles and translation stages are placed in an airtight container under a positive argon pressure of 0.5 p.s.i. to provide an inert atmosphere for synthesis.
Two types of reaction plates have been designed and used in the synthesis. Figure 1B shows the two types of 384-well reaction plate assembly. The first design of the 384-well reaction plate assembly consists of a socket plate to hold 384 independent pipette tips fitted with filter frits (FA113-N-20; Porex Bio Products Group, CA). A gasket between the socket plate and the vacuum assembly ensures a tight seal. The vacuum assembly is connected to a vacuum ballast, which also serves as the waste container.
The second type of reaction plate assembly, also shown in Figure 1B, consists of three parts, a crown plate, a CPG bead-retaining medium and a base plate. The crown plate consists of conical holes of ∼2 mm diameter at the bottom of the hole and a depth of 15 mm. The CPG bead-retaining medium must meet the requirements that the gas flow rate is lower than the fluid flow rate at the applied suction head for draining. Hydrophilic membranes usually have bubble point pressures higher than the vacuum head applied for draining and are ideal for the purpose. To facilitate product collection without cross-contamination between wells, the base plate is shaped so that the exit of each well tapers to a cone shape.
Synthesis procedure
We have synthesized oligonucleotides of various sequences and lengths using the synthesis modules described above. The CPG synthesis support is suspended in a 1:1 (v/v) mixture of chloroform/dibromomethane and then pipetted into the reaction wells. For 5 nmol scale syntheses, 0.2 mg of CPG is suspended in 10 µl of the solvent mixture in each well. The solvent is drained by applying a vacuum to the reaction wells and followed by a 30 min argon purge to the synthesizer chamber just before the synthesis starts.
The phosphoramidite synthesis chemistry consists of four stages, namely detritylation, coupling, capping and oxidation. The synthesis parameters are as follows. For detritylation, 27 µl of 3% trichloroacetic acid in dichloromethane is injected into each well. The detritylation reaction goes on for 10 s and is followed by a wash with 120 µl of acetonitrile (CH3CN). To achieve the highest coupling efficiency, the ejection of 4.5 µl of phosphoramidite and 11.5 µl of tetrazole is performed twice at each coupling stage. Each reaction is allowed to proceed for 45 s before the reagents are drained away. Capping reagent (6.5 µl) is then injected into each well, left for 10 s and drained away. Then 9 µl of the oxidation reagent is delivered to each reaction well, left for 25 s and drained away. Because of the small diameter of the reaction wells (∼2.2 mm), 4 µl of reagent is enough to cover the CPG beads. After the oxidation stage, the reaction wells are washed with CH3CN and the synthesis reactions cycle again from the detritylation stage. Reagent consumption is about the same for synthesis scales between 5 and 20 nmol and is listed in Table 1. With the reagent amounts listed in Table 1, the yield increases linearly with the amount of CPG beads. Therefore, the most cost effective synthesis scale is 20 nmol for the 1536 channel synthesizer. Each synthesis cycle takes 30 min and the synthesis of 1536 oligonucleotides of 20 nt length is completed in ∼10 h.
Table 1. Reagent consumption for 5–20 nmol synthesis scale per reaction well.
| Reagent | Quantity |
|---|---|
| Phosphoramidites | 9 µl ≈ 0. 36 mg |
| Activators (tetrazole) | 23 µl ≈ 0.72 mg |
| Capping solution | 6.5 µl |
| Oxidizer | 9 µl |
| Detritylation solution | 27 µl |
| Acetonitrile | 320 µl |
| Total waste per cycle | 395 µl |
The post-synthesis procedures are as follows. After synthesis is completed, the reaction wells are washed extensively with CH3CN to remove residual reagents in the reaction wells. The oligonucleotides are cleaved from the solid support with the reagents and procedures recommended by the CPG bead manufacturer. The reaction is allowed to proceed at room temperature for 5–60 min before the reaction plate assembly is mounted on a collection plate. The two plates are placed in a centrifuge (RT-7; Sorvall, Newton, CT) to spin down the crude product. The collection plate is sealed and incubated at 55°C for 8 h or overnight to remove the protecting groups from the oligonucleotides. The products are cooled to room temperature before the seal is removed.
Product analysis and quantification
The purity of the synthesized oligonucleotides was measured by capillary electrophoresis (CE) and mass spectrometer. The CE data were acquired using a Beckman (Fullerton, CA) P/ACE System 2100 with UV detection at 254 nm. Capillary column, separation gel and running buffer were obtained from Beckman (ssDNA 100-R kit). The capillary had an inner diameter of 100 µm and a total length of 27 cm (with a 20 cm inlet-to-detector length). The peak area in the electrophoretogram was measured to calculate the step yield of oligonucleotide synthesis.
The mass spectra of oligonucleotides were measured using MALDI-TOF mass spectrometers (Voyager DE-STR; PerSpective Biosystems, Framingham, MA). The sample preparations and spectrum measurements were performed according to the instructions provided by the mass spectrometer manufacturer. To measure synthesis yield, oligonucleotides synthesized in 384-well modules were transferred to 384-well UV-transmissible plates (catalog no. 781801; Greiner, Frickenhausen, Germany) to measure the absorbance over the range 200–400 nm with a multi-well spectrophotometer (SpectraMax Plus 384; Molecular Devices, Sunnyvale, CA). The optical absorbance at 260, 280 and 320 nm were taken to measure the amount of products.
RESULTS
Product yield variation
A measure for the performance of a multi-channel synthesizer is the variation of product yield in different wells. Maps illustrating the yield variation of 1536 oligonucleotides in four sets of 384-well synthesis module are shown in Figure 2A. The average product yield is 0.93 OD for a nominal 5 nmol oligonucleotide synthesis scale. The absorbance measurements showed that oligonucleotides were present in every well in the plates. Figure 2A was used as a guide for instrument alignment. As it is evident in Figure 2A that there is no spatial bias of product yield in the 384-well synthesis modules, it indicates that the variation is random and the instrument was in alignment. No CPG bead loss occurred during synthesis and no synthesis failure was observed. A histogram of product yield of 1536 oligonucleotides in four sets of 384-well synthesis modules is shown in Figure 2B. The standard deviation of yield was calculated to be 0.19 OD or 20% coefficient of variation (CV). When the same synthesizer was loaded with 96-well reaction plates in place of 384-well plates, a smaller CV of 15% was observed (data not shown). These results suggest that it is more difficult to maintain a small yield variation in highly parallel and small volume synthesis.
Figure 2.
(A) Optical density map of 1536 individual oligonucleotides in 384 × 4 synthesis modules. The length of the oligonucleotides ranges from 19 to 24 nt. The average length is ∼21 nt. The average optical density and the standard deviation are indicated for each synthesis module. (B) A histogram of optical density of all the 1536 oligonucleotides shown in (A). The average optical density is 0.93 and the standard deviation is 0.19 for the 1536 oligonucleotides. The calculation was based on the measured values and no correction for oligonucleotide length and sequences was made.
The observed yield variation was a compound error of various sources such as the error of pipetting volume, error of spectrophotometry measurement, extinction coefficient difference of the four nucleotides that make up the oligonucleotide sequences, variation of CPG quantity and others. These variations together with the variations introduced by the synthesizer, which are governed by many instrumental parameters, produced the ensemble yield variation.
Several undesirable effects can occur in a parallel synthesis apparatus when synthesis support dislodging occurs in wells. These effects can downgrade the performance of the synthesizer or even result in synthesis failure for the entire plate. The effects are more significant in highly parallel synthesizers such as the ones with the 384-well synthesis format than with the 96-well format. The current design eliminates dislodging of the synthesis support (CPG) during the reactions (14).
Product quality
The product quality can be determined by its step yield and purity. Although the step yield of an oligonucleotide synthesis is influenced by its nucleotide sequence, the average step yield was measured by using a unit segment of 17mer. The unit segment sequence, tgcctcggacattaagt (12), was used to carry out the syntheses of oligonucleotides of various lengths. The segment sequence contains all 16 possible permutations of consecutive base tandems. The average step yield thus obtained can be representative of the results of various oligonucleotide sequences. The quantity of the full-length product was determined by capillary electrophoresis (CE) (15). Figure 3 shows CE separation results of crude oligonucleotide products with lengths from 17 to 119 nt. The major peak in each electrophoretogram is the full-length product whereas the minor peaks preceding it are truncated products. The average step yield of the 17mer oligonucleotide was 99.3%. The exact step yield of long oligonucleotides was difficult to calculate owing to the large number of truncated product peaks but was estimated to be ∼98.8% for the 51mer, 98.3% for the 85mer and 98.1% for the 119mer oligonucleotides. For synthesis of oligonucleotides longer than 119 nt, CPG beads of 2000 Å pore were used and synthesis was done with 96-well format rather than 384-well format to have enough quantity for practical use.
Figure 3.
Capillary electrophoresis of synthesis products of various lengths: (A) 17mer; (B) 51mer; (C) 85mer; (D) 119mer. The sequences are tgcctcggacattaagt, (tgcctcggacattaagt)3, (tgcctcggacattaagt)5 and (tgcctcggacattaagt)7, respectively.
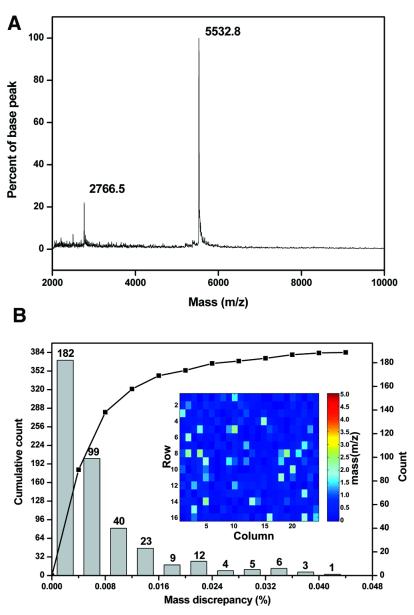
In oligonucleotide synthesis, several factors contribute to the purity of the final product. Side reactions such as depurination may occur during chemical synthesis of oligonucleotides (16,17). Incomplete post-synthesis deprotection may leave protection groups such as benzyl, isobutyl and acetyl groups on the nucleotides (18). Deleted or n – 1 products exist in greater proportion in long oligonucleotides due to an incomplete capping reaction and other reactions (19,20). Modified phosphoramidite chemistry to alleviate the above synthesis defects have been described or proposed in the literature reports (21–23). Different applications require oligonucleotides of different purity levels. For instance, applications such as gene synthesis and site-directed mutagenesis require very high quality oligonucleotides without the deleted (n – 1) products. Genotyping by mass spectrometry requires oligonucleotides with complete removal of the protection groups on the nucleotides. On the other hand, the synthesis defects described above are less critical to primers for PCR amplification. Oligonucleotides for DNA microarrays can be free of truncated products by removing them after the oligonucleotide probes are immobilized to the solid substrate through the 5′ terminal linker moiety of the oligonucleotide (24,25). In order to synthesize oligonucleotides for various applications including those demanding the highest purity products in a high throughput format, we are currently investigating methods such as those described in literature reports to reduce or eliminate the aforementioned synthesis defects. The quality of oligonucleotides can best be examined by mass spectrometry for synthesis defects. Figure 4A shows the mass spectrum of an 18mer oligonucleotide. The observed molecular weight of the base peak (5532.8 Da) matches the expected value (5533.7 Da) within the accuracy range of the mass spectrometry measurement with external calibration method for oligonucleotides (26). The additional peak at 2766.5 Da (m/z) is the [M+2H]2+ molecule that had gained two protons during the desorption process. Automated high throughput mass spectrometry analysis of the products in a 384-well synthesis module was performed with internal calibration standards to assess the quality and the accuracy of the synthesis. Figure 4B shows a map of molecular weight discrepancy between the observed and the expected for the oligonucleotides in a 384-well synthesis module. All the 384 oligonucleotides in a synthesis module have a molecular weight discrepancy <0.044% of the calculated molecular weight as shown in Figure 4B. The largest molecular weight discrepancy was 2.5 Da but when the same sample was picked for measurement again, the discrepancy was reduced to <1 Da. This observation indicates that the small discrepancies between the observed and the calculated molecular weights can be attributed to the automated process, which inevitably compromises the effects of matrix homogeneity (27) as well as other MALDI-TOF instrument related errors. The results show that the side reactions during chemical synthesis have been minimized and the synthesizer produced high quality products with correct sequences.
Figure 4.
(A) Mass spectrum of an 18mer oligonucleotide. The sequence is tgtaaaacgacggccagt. The calculated molecular weight is 5533.7 Da. An additional [M+2H]2+ peak is present in the spectrum and its measured molecular weight also matches the expected molecular weight. (B) A histogram together with a cumulative curve showing the mass accuracy of oligonucleotide synthesis in a 384-well synthesis module. All of the oligonucleotides in a 384-well synthesis module have a measured mass (m/z) discrepancy <0.044% of the calculated mass. The mass measurements were done with two internal calibration standards. The inset is a quality assessment map of 384 individual oligonucleotides in a synthesis module. The pseudo-color encoded cells indicate the discrepancy between the measured and the expected mass (m/z).
DISCUSSION
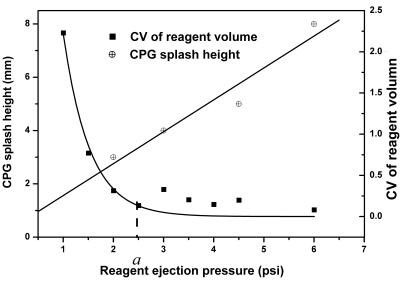
The results show that we have achieved high throughput, high product quality and lower variation in product yield. Several key parameters have been identified and implemented to achieve the results. These parameters include the design of reaction wells, the layout of the reagent delivery pipeline and the selection of the CPG-retaining medium. As described in the Materials and Methods, the system pressure and the valve opening time determine the reagent injection volume. We tuned the system pressure, the ejection speed and the quantity of the reagents to minimize variation in reagent injection volume and splash of CPG. As shown in Figure 5, the CPG splash height and the CV of reagent volume are dependent on the reagent ejection pressure. The suitable ejection pressure is determined by choosing the one that yields the minimum CV of reagent volume while keeping the CPG splash below a reasonable height. The CPG splash height can be inspected visually and was measured with a ruler. In Figure 5, point a indicates the pressure that yields an injection volume CV of 0.1 with a CPG splash height of 3 mm. The reaction wells were designed accordingly to have a suitable height to retain the CPG beads inside the wells during the synthesis reactions. The parameter is important to avoid splash loss of CPG beads in the reaction wells.
Figure 5.
Experimental results of CPG bead splash height as well as the coefficient of variation (CV) of reagent ejection volume as a function of reagent ejection pressure. The optimal ejection pressure is selected based on having an acceptable ejection CV with reasonable bead splash height. In this figure, point a indicates the ejection pressure that gives rise to a CV of 10%.
While loss of CPG beads causes zero yields in individual reaction wells, the CPG-retaining medium has a crucial effect on the step yield of each well and hence the yield variation of the final products. For a plate with multiple reaction wells, if some wells are not drained completely before the addition of the next synthesis reagent, the residual reagent interferes with the synthesis chemistry, resulting in low product yield, zero yield or unwanted side-product in those wells. To minimize these effects, it is critical to select a bead-retaining medium with a high bubble point pressure. Hydrophilic membranes have bubble point pressures higher than the vacuum head applied for draining. Using such membranes as the bead-retaining medium, wells that drain faster than the others do not allow gas flow-through and the suction head is maintained over those wells that still contain reagent.
High throughput oligonucleotide synthesis can be achieved using the instrument described in this article. Each 384-well synthesis module provides a throughput of approximately 800 oligonucleotides of 20mer length per 24 h if operated two shifts per day. A 1536 channel synthesizer consisting of four 384-well synthesis modules can produce more than 3000 oligonucleotides in a day. Some immediate applications of the 1536 channel oligonucleotide synthesizer include synthesizing PCR primers and probes for genome-wide studies as well as oligonucleotide microarray production. The synthesizer has been applied to synthesize more than 100 000 oligonucleotides for DNA microarray and SNP genotyping applications. Using the current design of 384-well synthesis modules, the addition of an extra synthesis assembly for a 12 reagent synthesis process requires only 12 additional reagent delivery solenoid valves and three additional reagent distribution valves. Since it allows addition of extra synthesis modules, the design is flexible for further enhancement of throughput. Finally, the design of the instrument and its operation software allow using synthesis formats such as 48-well, 96-well and 384-well plates to accommodate different synthesis scales.
Acknowledgments
ACKNOWLEDGEMENTS
The assistance of Dr Chiun Gung Juo for mass spectrometry measurements is greatly appreciated. We gratefully acknowledge the grant support provided by the National Science Council (NSC-89-2318-B-022-M51 and NSC-90-2318-B-001-005-M51) and the Medical Biotechnology Program of Academia Sinica, Taipei, Taiwan, ROC.
REFERENCES
- 1.Beaucage S.L. and Caruthers,M.H. (1981) Deoxynucleotide phosphoramidite. Tetrahedron Lett., 22, 1859–1862. [Google Scholar]
- 2.Adams S.P., Galluppi,G.R., Holder,S.B., Kavka,K.S. and Wykes,E.J. (1983) Hindered diakylamino nucleoside phosphite reagents in the synthesis of 2 DNA 51-mers. J. Am. Chem. Soc., 105, 661–663. [Google Scholar]
- 3.Roget A., Bazin,H. and Teoule,R. (1989) Synthesis and use of labelled nucleoside phosphoramidite building blocks bearing a reporter group: biotinyl, dinitrophenyl, pyrenyl and dansyl. Nucleic Acids Res., 17, 7643–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai E. (2001) Application of SNP technologies in medicine: lessons learned and future challenges. Genome Res., 11, 927–929. [DOI] [PubMed] [Google Scholar]
- 5.Roberts L. (2000) Human genome research. SNP mappers confront reality and find it daunting. Science, 287, 1898–1899. [DOI] [PubMed] [Google Scholar]
- 6.Pease A.C., Solas,D., Sullivan,E.J., Cronin,M.T., Holmes,C.P. and Fodor,S.P. (1994) Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl Acad. Sci. USA, 91, 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Southern E.M., Case-Green,S.C., Elder,J.K., Johnson,M., Mir,K.U., Wang,L. and Williams,J.C. (1994) Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucleic Acids Res., 22, 1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard A.P., Kaiser,R.J. and Hood,L.E. (1996) High-density oligonucleotide arrays. Biosens. Bioelectron., 11, 687–690. [Google Scholar]
- 9.Hughes T.R., Mao,M., Jones,A.R., Burchard,J., Marton,M.J., Shannon,K.W., Lefkowitz,S.M., Ziman,M., Schelter,J.M., Meyer,M.R. et al. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nature Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 10.Singh-Gasson S., Green,R.D., Yue,Y., Nelson,C., Blattner,F., Sussman,M.R. and Cerrina,F. (1999) Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nature Biotechnol., 17, 974–978. [DOI] [PubMed] [Google Scholar]
- 11.Beier M. and Hoheisel,J.D. (2002) Analysis of DNA-microarrays produced by inverse in situ oligonucleotide synthesis. J. Biotechnol., 94, 15–22. [DOI] [PubMed] [Google Scholar]
- 12.Sindelar L.E. and Jaklevic,J.M. (1995) High-throughput DNA synthesis in a multichannel format. Nucleic Acids Res., 23, 982–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lashkari D.A., Hunicke-Smith,S.P., Norgren,R.M., Davis,R.W. and Brennan,T. (1995) An automated multiplex oligonucleotide synthesizer: development of high-throughput, low-cost DNA synthesis. Proc. Natl Acad. Sci. USA, 92, 7912–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rayner S., Brignac,S., Bumeister,R., Belosludtsev,Y., Ward,T., Grant,O., O’Brien,K., Evans,G.A. and Garner,H.R. (1998) MerMade: an oligodeoxyribonucleotide synthesizer for high throughput oligonucleotide production in dual 96-well plates. Genome Res., 8, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren W.J. and Vella,G. (1993) Analysis of synthetic oligodeoxyribonucleotides by capillary gel electrophoresis and anion-exchange HPLC. Biotechniques, 14, 598–606. [PubMed] [Google Scholar]
- 16.Septak M. (1996) Kinetic studies on depurination and detritylation of CPG-bound intermediates during oligonucleotide synthesis. Nucleic Acids Res., 24, 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eadie J.S. and Davidson,D.S. (1987) Guanine modification during chemical DNA synthesis. Nucleic Acids Res., 15, 8333–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boal J.H., Wilk,A., Harindranath,N., Max,E.E., Kempe,T. and Beaucage,S.L. (1996) Cleavage of oligodeoxyribonucleotides from controlled-pore glass supports and their rapid deprotection by gaseous amines. Nucleic Acids Res., 24, 3115–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temsamani J., Kubert,M. and Agrawal,S. (1995) Sequence identity of the n–1 product of a synthetic oligonucleotide. Nucleic Acids Res., 23, 1841–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen D., Yan,Z., Cole,D.L. and Srivatsa,G.S. (1999) Analysis of internal (n–1)mer deletion sequences in synthetic oligodeoxyribonucleotides by hybridization to an immobilized probe array. Nucleic Acids Res., 27, 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwiatkowski M., Nilsson,M. and Landegren,U. (1996) Synthesis of full-length oligonucleotides: cleavage of apurinic molecules on a novel support. Nucleic Acids Res., 24, 4632–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krotz A.H., Cole,D.L. and Ravikumar,V.T. (1999) Synthesis of an antisense oligonucleotide targeted against C-raf kinase: efficient oligonucleotide synthesis without chlorinated solvents. Bioorg. Med. Chem., 7, 435–439. [DOI] [PubMed] [Google Scholar]
- 23.Yu D., Tang,J., Iyer,R.P. and Agrawal,S. (1994) Diethoxy n,n-diisopropyl phosphoramidite as an improved capping reagent in the synthesis of oligonucleotides using phosphoramidite chemistry. Tetrahedron Lett., 35, 8565–8568. [Google Scholar]
- 24.Guo Z., Guilfoyle,R.A., Thiel,A.J., Wang,R. and Smith,L.M. (1994) Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res., 22, 5456–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaucage S.L. (2001) Strategies in the preparation of DNA oligonucleotide arrays for diagnostic applications. Curr. Med. Chem., 8, 1213–1244. [DOI] [PubMed] [Google Scholar]
- 26.Tang K., Shahgholi,M., Garcia,B.A., Heaney,P.J., Cantor,C.R., Scott,L.G. and Williamson,J.R. (2002) Improvement in the apparent mass resolution of oligonucleotides by using 12C/14N-enriched samples. Anal. Chem., 74, 226–231. [DOI] [PubMed] [Google Scholar]
- 27.Garcia B.A., Heaney,P.J. and Tang,K. (2002) Improvement of the MALDI-TOF analysis of DNA with thin-layer matrix preparation. Anal. Chem., 74, 2083–2091. [DOI] [PubMed] [Google Scholar]