Abstract
Molecular beacons (MBs) are hairpin-like fluorescent DNA probes that have single-mismatch detection capability. Although they are extremely useful for many solution-based nucleic acid detections, MBs are expensive probes for applications that require the use of a large number of different DNA probes due to the high cost and tedious procedures associated with probe synthesis and purification. In addition, since both ends of MB probes are covalently modified with chromophores, they do not offer the flexibility for fluorophore change and the capability for surface immobilization through free DNA ends. In this report, we describe an alternative form of MB, denoted tripartite molecular beacon (TMB), that may help overcome these problems. A TMB uses an unmodified oligodeoxyribonucleotide that forms a MB-like structure with two universal single-stranded arms to bring on a universal pair of oligodeoxyribonucleotides modified separately with a fluorophore and a quencher. We found that TMBs are as effective as standard MBs in signaling the presence of matching nucleic acid targets and in precisely discriminating targets that differ by a single nucleotide. TMBs have the necessary flexibility that may make MBs more affordable for various nucleic acid detection applications.
INTRODUCTION
Molecular beacons (MBs) are single-stranded DNA probes useful for solution-based nucleic acid detection (1). A typical MB, with a fluorophore attached at the 5′ end and a quencher at the 3′ end, forms a hairpin-like, stem–loop structure by itself (Fig. 1A). In this closed structure form, the fluorophore is located within a short distance of the quencher and the energy absorbed by the fluorophore is not emitted as fluorescence but transferred to the quencher and released as heat. As a result, the system is unable to fluoresce strongly on its own. When a target nucleic acid is introduced, the formation of the rigid helical structure between the loop of the MB and the target causes the dissociation of the hairpin stem and the separation of the fluorophore from the quencher. Since the distantly located quencher is no longer able to absorb the energy from the excited fluorophore, the open state MB emits strong fluorescence.
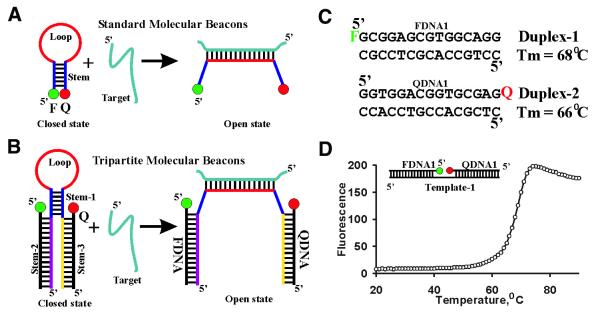
Figure 1.
Standard MBs and TMBs. (A) A standard MB consists of a probe domain (in red) flanked by two short self-complementary DNA sequences (in blue). A fluorophore (F) is covalently linked to the end of one arm and a quencher (Q) is covalently attached to the other. The short distance between F and Q in the closed state permits high-efficiency fluorescence quenching. When a nucleic acid target is introduced, the formation of a loop–target duplex structure causes the separation of F from Q in the open state. This leads to intensive fluorescence signaling. (B) In a TMB, F and Q are covalently linked to two separate short oligodeoxyribonucleotides (denoted FDNA and QDNA) which hybridize strongly with corresponding single-stranded arms (one in purple and one in yellow) extended beyond the short hairpin stem (stem-1, shown in blue). F and Q are still located in close proximity in the closed state, permitting high-efficiency quenching of the fluorescence. In the presence of the nucleic acid target (in sea green), closed state TMB is transformed into the open state, which emits a very strong fluorescence signal. (C) Design of FDNA and QDNA. High GC contents make duplex-1 and duplex-2 very stable (melting points of 68 and 66°C were observed, respectively). The melting points were determined by following the absorbance change of relevant DNAs (100 nM) in solutions containing 500 mM NaCl, 3.5 mM MgCl2 and 10 mM Tris–HCl (pH 8.3). The top strand of duplex-1 was used as FDNA1 and the top strand of duplex-2 was used as QDNA1. F, fluorescein; Q, DABCYL [4-(4-dimethylaminophenylazo)benzoic acid]. (D) A thermal denaturation profile of the linear duplex assembled using FDNA1, QDNA1 and a DNA template [template 1, with the sequence of d(CCTGCCACGCTCCGCTCTCGCACCGTCCACC)]. The following concentrations were used for the DNA oligodeoxyribonucleotide: 100 nM for FDNA1, 200 nM for template-1, 300 nM for QDNA1. Other conditions are described in the Materials and Methods.
MBs have significant advantages over linear probes (2,3). They serve as simple fluorescent reporters for specific nucleic acid targets in hybridization assays without the need to separate the probe–target complex from excess probes. The signaling specificity is very high and similar nucleic acid targets that differ only by a single mismatch or deletion can be distinguished with precision. The fluorescence reporting is very sensitive and fluorescence enhancement up to two orders of magnitude can be observed when a matching target is introduced. Because of all these, MBs have been used in a variety of nucleic acid-based detections. For example, MBs were used to monitor the synthesis of specific nucleic acids (4–7), to set up one-step assays to identify single-nucleotide variations in DNA (8–11) and to detect specific RNA molecules within living cells (12,13).
Despite the aforementioned advantages, standard MBs have drawbacks. First, MBs are expensive to make. Each MB has to be individually synthesized in order to covalently link the fluorophore and quencher moieties onto a specific DNA probe. Each synthesized MB needs to be rigorously purified to remove failed sequences. It is particularly important to eliminate probes that have a fluorophore attached but lack the quencher because these molecules will cause high background fluorescence. Secondly, covalent integration of fluorophore and quencher with DNA offers no flexibility in fluorophore change. For situations where two or more DNA probes with identical DNA sequences but with different fluorophores need to be used, multiple syntheses and purifications have to be carried out. Thirdly, for applications that involve surface immobilization (such as DNA microarrays and optical biosensors), MBs have to be immobilized on the surface. Since most fluorophores can be photo-bleached relatively easily, extreme care is needed during the immobilization process to prevent the photo bleaching of the fluorophore. Moreover, since both the 5′ and 3′ ends of a MB are occupied, traditional ways of arraying DNA oligonucleotide onto solid supports through terminal modifications cannot be applied to MBs and other attachment methods have to be used (14–18). Considering all these problems, the application of MBs in situations where it is desirable to detect hundreds or even thousands of different nucleic acid targets simultaneously or separately can become difficult and expensive. It would be too costly to construct a DNA chip that consists of a large number of different MB probes. Thus, there remains a real need for a novel MB format that can help solve the problems associated with standard MBs and make MBs easy to use and more affordable for applications that demand a large number of probes. In this report, we describe an alternative form of MB that may help overcome some of the aforementioned problems. More specifically, we have reformulated a MB into a tripartite DNA assembly, which we termed a ‘tripartite molecular beacon’ (TMB). A TMB uses an unmodified DNA oligonucleotide that forms a MB-like structure with two universal single-stranded arms to bring on a universal pair of DNA oligonucleotides modified separately with a fluorophore and a quencher. In the first stage of our assessment described herein, we found that TMBs were as effective as standard MBs in signaling the presence of matching nucleic acid targets and in precisely discriminating targets that differ by a single nucleotide. With the new formulation, MBs have now the necessary flexibility that should help make them more affordable for various high throughput applications.
MATERIALS AND METHODS
Oligodeoxyribonucleotides
Normal and modified oligodeoxyribonucleotides were all prepared by automated DNA synthesis using standard cyanoethylphosphoramidite chemistry (Keck Biotechnology Resource Laboratory, Yale University; Central Facility, McMaster University). MBs used for our studies contained fluorescein as the fluorophore and/or 4-(4-dimethylaminophenylazo)benzoic acid (DABCYL) as the quencher. Fluorescein and DABCYL were placed on the 5′ and 3′ ends of the relevant oligodeoxyribonucleotides, respectively. 5′-Fluorescein and 3′-DABCYL DNAs were synthesized using 5′-fluorescein phosphoramidite and 3′-DABCYL-derivatized controlled pore glass (Glen Research, Sterling, VA).
Unmodified DNA oligonucleotides were purified by 10% preparative denaturing (8 M urea) polyacrylamide gel electrophoresis, followed by elution and ethanol precipitation. 5′-Fluorescein- and/or 3′-DABCYL-modified oligodeoxyribonucleotides were purified by reverse-phase high-pressure liquid chromatography using a Beckman-Coulter HPLC System Gold with a 168 Diode Array detector. The HPLC column was an Agilent Zorbax ODS C18 column, 4.5 × 250 mm (5 µm). A two-buffer system was used with buffer A being 0.1 M triethylammonium acetate (pH 6.5) and buffer B being pure acetonitrile. The best separation was achieved by using a non-linear elution gradient (10% B for 10 min, 10–40% B in 65 min) at a flow rate of 0.5 ml/min. The main peak was found to have very strong absorption at both 260 and 491 nm. The DNA within central two-thirds of peak-width was collected and dried under vacuum. Purified oligodeoxyribonucleotides were dissolved in water and their concentrations were determined spectroscopically. All chemical reagents were purchased from Sigma.
Fluorescence measurements
The following concentrations were used for various oligodeoxyribonucleotides (if not otherwise specified): 100 nM for fluorophores, 200 nM for hairpin DNA, 300 nM for quenchers and 600 nM for complementary DNA target. All measurements were made in 500 µl solutions containing 500 mM NaCl, 3.5 mM MgCl2 and 10 mM Tris–HCl (pH 8.3). The fluorescence of all MB mixtures was measured on a Cary Eclipse Fluorescence Spectrophotometer (Varian) with excitation at 490 nm and emission at 520 nm.
For obtaining the thermal denaturation profile of a particular reaction mixture, the DNA solution was heated to 90°C for 5 min, and the temperature was then decreased from 90 to 20°C at a rate of 1°C/min. A reading was made automatically for every 1°C decrease.
RESULTS
Design of TMBs
We hypothesized that a MB can be reformatted into a three-component system, as shown in Figure 1B, which we termed a tripartite molecular beacon (TMB). The fluorophore is now attached onto the 5′ end of a small oligodeoxyribonucleotide (FDNA) and the quencher is linked to the 3′ end of another small oligodeoxyribonucleotide (QDNA). The third oligonucleotide (LDNA) is a standard, unmodified oligodeoxyribonucleotide that consists of five sequence segments. The 5′ domain (shown in purple) is complementary to the FDNA and together they form an intermolecular stem, designated as stem-2. The 3′ segment (shown in yellow) is complementary to the QDNA and together they form another intermolecular stem, designated as stem-3. The two short sequence motifs (shown in blue) next to the FDNA and QDNA binding domains are self-complementary and are able to form an intramolecular stem, designated as stem-1. The probe sequence segment (shown in red) is complementary to an external nucleic acid target (shown in sea green). In the absence of a target sequence, the LDNA adopts the closed state structure and the tripartite DNA assembly does not fluoresce strongly. In the presence of the target sequence, the TMB is converted to the open state, the fluorophore moves away from the quencher and, consequently, fluorescence is emitted.
It is important that stem-2 and stem-3 are very stable so that the FDNA and QDNA anneal strongly with LDNA. This is particularly true for the temperature range from 20 to 50°C within which nucleic acid hybridizations are usually carried out. The strong interaction between FDNA and LDNA as well as between QDNA and LDNA can be achieved by having a high GC content in both stems. Figure 1C lists two GC-rich, 15-bp duplexes, duplex-1 and duplex-2, that have an observed Tm of 68 and 66°C, respectively (calculated from the melting curves determined using absorption spectroscopy with a solution containing 10 mM Tris–HCl, pH 8.3 at 23°C, 0.5 M NaCl, 3.5 mM MgCl2 and 0.1 µM of DNA). The use of such stable duplexes as stem-2 and stem-3 was expected to strongly link FDNA and QDNA with LDNA.
The ability of QDNA to quench the fluorescence of FDNA was tested with a linear duplex assembly in which a template DNA engages both FDNA1 and QDNA1 in helical structure formation to bring the fluorophore and the quencher into close proximity. As seen in Figure 1D, the fluorescence of the DNA mixture was very low when FDNA1 and QDNA1 were fully assembled onto the DNA template at a low temperature range. At high temperatures, FDNA1 and QDNA1 were dissociated from the template and strong fluorescence was observed. This indicates that closely located QDNA1 can efficiently quench the fluorescence of FDNA1. An apparent Tm of 68°C was derived with the data shown in Figure 1D. It is noteworthy that the maximal fluorescence of the mixture was found to be at 74°C, beyond which the fluorescence intensity decreased linearly. Examination of the solution containing only FDNA1 (or containing both FDNA1 and QDNA1) revealed the same pattern (data not shown), suggesting that FDNA1 was completely dissociated from template-1 at 74°C and the fluorescence decrease beyond 74°C simply reflected the temperature dependence of the intrinsic fluorescence of FDNA1.
It is apparent from Figure 1D that the properly annealed FDNA1 and QDNA1 have low fluorescence within the temperature range of 20–60°C. For example, the fluorescence intensity at 37°C was only ∼10% higher than that at 20°C. Even at 50°C, the solution had a fluorescence that was only ∼40% higher than that at 20°C. Therefore, the TMBs constructed with these two oligodeoxyribonucleotides were expected to behave similarly to the standard MBs within the normal temperature range used in most nucleic acid hybridization experiments.
It is well known that DNA sequences with high GC/AT ratios are prone to the formation of a variety of different secondary structures. Therefore, the use of binding arms with high GC content can be a cause for secondary structure concerns. However, the usual appearance of the melting curve of the linear duplex assembly suggests that the two high GC-containing binding arms do not behave abnormally in the presence of fully complementary FDNA and QDNA, indicating that the two sequences that we have designed are adequate for TMB constructions.
Comparison of standard MBs and TMBs
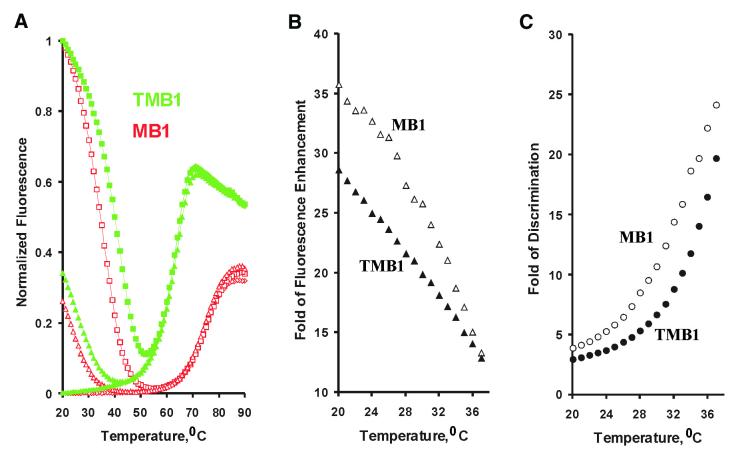
To test whether a TMB has a detection performance similar to a standard MB, we constructed TMB1 and compared its properties with a previously studied MB (4), named here as MB1. TMB1 and MB1 share the same hairpin loop and stem sequences while TMB1 has extra binding arm sequences shown in Figure 1C. Thermal denaturation profiles (Fig. 2A) were obtained for both TMB1 (filled data points in green) and MB1 (open data points in red) under three sets of conditions: in the absence of target (in diamonds), with a perfectly matched target (in squares) and in the presence of a mismatch target (in triangles). The melting curves of MB1 and TMB1 had similar appearances under each condition. In the absence of a nucleic acid target, the fluorescence in both systems experienced a rapid drop when the fully denatured solution was cooled to pass the point at which the intramolecular stem started to form, bringing the fluorophore and the quencher into close proximity. The intensity reduction significantly slowed down at ∼60°C for MB1 and at ∼50°C for TMB1 when most of the beacon molecules existed in the closed structure state. When the match target was used, the fluorescence intensity reached a minimal value at 52°C for TMB1 and at 56°C for MB1. A further decrease in temperature resulted in a rapid fluorescence increase indicative of the formation of duplex structures between the DNA target and the probe sequence and the dissociation of the internal stem (1–3). The fluorescence increase was highly specific since another target that contained a single mismatch caused a much smaller fluorescence increase in both systems.
Figure 2.
Comparison of TMB1 with MB1. (A) Fluorescence intensity was measured as a function of temperature in the absence of a nucleic acid target for MB1 (unfilled red diamonds) and for TMB1 (filled green diamonds), in the presence of the match target [d(TACTCTTATATCATATTTGGTGTTTGCTTT)] for MB1 (unfilled red squares) and for TMB1 (filled green squares), as well as in the presence of a single-mismatch target [d(TACTCTTATATCATc TTTGGTGTTTGCTTT), the small letter indicates the single base mutation relative to the match target] for MB1 (unfilled red triangles) and for TMB1 (filled green triangles). TMB1 is made of FDNA1, QDNA1 (sequences shown in Fig. 1C) and LDNA1 with a sequence of d(CCTGCCACGCTCCGCGCGAGC CACCAAATATGATATGCTCGCCTCGCACCGTCCACC) (FDNA1-binding sequence shown in bold, QDNA1-binding domain indicated in italic and self-complementary motifs underlined). MB1 has the sequence of F-d(GCGAGCCACCAAATATGATATGCTCGC)-Q (F, Fluorescein; Q, DABCYL). The fluorescence intensity was normalized for each system as [(FT°C) – (F20°C, no-target)] / [(F20°C, match) – (F20°C, no-target)] where (FT°C) is the fluorescence reading of a solution at any designated temperature, (F20°C, no-target) and (F20°C, match) are the readings at 20°C for the samples containing no target and the match target, respectively. Experimental details are given in the Materials and Methods. (B) Signal-to-background fluorescence ratio, calculated as (FT°C, match) / (FT°C, no-target), is plotted for MB1 (open triangles) and for TMB1 (filled triangles). (C) Single nucleotide discrimination capability, calculated as (FT°C, match – FT°C, no-target) / (FT°C, mismatch – FT°C, no-target), is plotted for MB1 (open circles) and TMB1 (filled circles). (FT°C, match), (FT°C, no-target) and (FT°C, mismatch) are the fluorescence readings for the samples containing match target, no target and mismatch target all at same temperature.
There were two visible differences in the temperature profiles of the two MBs. First, MB1 had an apparent Tm of 74.5°C that was 12°C higher than that of TMB1 (62.5°C). The observed melting point of MB1 is in excellent agreement with the calculated Tm of 74.2°C (in 1 M NaCl) by the M-fold program (http://bioinfo.math.rpi.edu/∼mfold/dna/form1.cgi). The smaller Tm observed for TMB1 is likely due to two reasons: (i) the stem-2 and stem-3 had an observed Tm of 68°C in the linear duplex described above, therefore it is not possible for the TMB1 system to have an observed Tm above 68°C; (ii) the assumed base pair at the outside edge of the stem-1 in TMB1 was very likely not formed due to the overcrowding at the location where stem-1, stem-2 and stem-3 meet. With the assumption that this base pair is not formed, the M-fold program predicts a Tm of 63.0°C for TMB1 (in 1 M NaCl), which matches quite well with the observed melting point of 62.5°C. Several other TMBs have also been examined for melting points and the observed Tm values were consistent with the assumption that the periphery base pairs were not formed (data not shown).
The second difference between the two MB systems is that TMB1 had the unique appearance at a high temperature range (from 74 to 90°C) that was similar to that observed in the linear duplex (Fig. 1D) and was indicative of complete dissociation of FDNA1 from its complementary partner.
The effectiveness of MBs can be judged by their signal-to-noise (S/N) ratio and their single-mismatch discrimination ability. The S/N can be simply calculated as the ratio of fluorescence intensity in the presence of the match target over the intensity in the absence of any target (i.e. fold of fluorescence enhancement), and single-mismatch discrimination ability can be taken as the ratio of the net fluorescence intensity (Ftarget – Fno-target) obtained with the match target and with the single-mismatch target (i.e. fold of discrimination). Figure 2B and C plot these two ratios for both MB1 (open data points) and TMB1 (filled data points) as a function of temperature between 20 and 37°C. For MB1, a maximum of ∼35-fold in S/N was observed at 20°C and the S/N ratio decreased at a linear rate of 1.4-fold/°C when the temperature increased. For TMB1, the maximal S/N was slightly smaller at ∼28-fold and the S/N ratio decreased at a slower pace with a near linear rate of ∼0.95-fold/°C between 20 and 37°C.
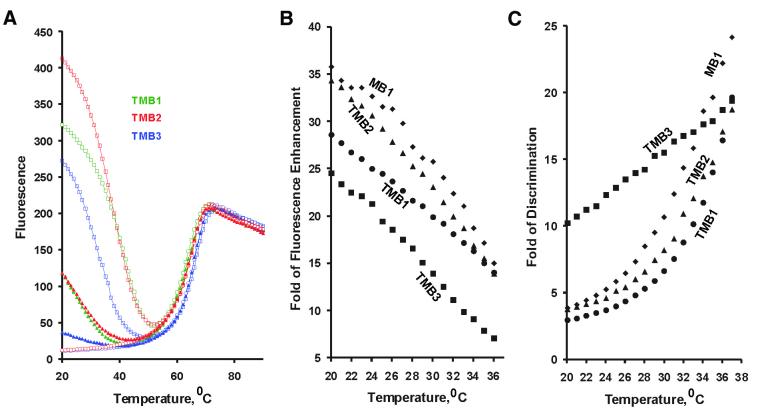
MB1 and TMB1 had a comparable capability for single nucleotide discrimination as well (Fig. 3C) although MB1 held a slight edge again over TMB1 at all temperatures. Both MB1 and TMB1 contain two 6-nt self-complementary motifs that are designed to form the desired 6-bp intramolecular hairpin stem. However, as discussed above, the periphery base pair in the hairpin stem of TMBs was likely not able to form due to the steric hindrance. We speculated that this might be the reason for the slightly reduced performance of TMB1. To test this hypothesis, we made two new TMBs, TMB2 and TMB3, that had the same sequence of TMB1 except an extra AT or GC pair as the periphery base pair in stem-1. Figure 3A shows the relevant thermal denaturation profiles of the two new TMBs, Figure 3B plots the S/N ratio of TMB1, TMB2, TMB3 and MB1, and Figure 3C compares their discrimination capability. TMB2, with a periphery AT pair, had a much improved S/N ratio that approached that seen with MB1. It also had a slightly improved discrimination capability over TMB1. TMB3, with an extra GC pair, had a smaller S/N ratio but had a robust single-nucleotide discrimination capability in the entire temperature range of 20–37°C. These data suggest that properly designed TMBs can have comparable capabilities as standard MBs in specifically recognizing nucleic acid targets.
Figure 3.
TMBs with an additional periphery base pair. (A) Both TMB2 [d(CCTGCCACGCTCCGCaGCGAGCCACCAAATATGATATGCTCGCtCTC GCACCGTCCACC)] and TMB3 [d(CCTGCCACGCTCCGCgGCGAGCCACCAAATATGATATGCTCGCcCTCGCACCGTCCACC)] have the same sequence as TMB1 except for the base insertions (shown in small letters; FDNA1-binding sequence shown in bold, QDNA1-binding domain indicated in italic and self-complementary motifs underlined). Fluorescence intensity was measured as a function of temperature in the absence of nucleic acid target (diamonds), in the presence of the match target (squares) and as well as in the presence of a mismatch target (triangles). Match and mismatch target nucleic acid sequences are given in Figure 2. (B) Signal-to-background fluorescence ratio, and (C) single nucleotide discrimination capability for TMB2 (filled triangles), TMB3 (filled squares), as well as for MB1 (filled diamonds) and TMB1 (filled circles).
General utility of the common FDNA and QDNA pair
Since FDNA and QDNA are not directly involved in target binding, they can be used as a universal fluorophore/quencher pair to construct any MB with a standard DNA oligonucleotide (LDNA) as long as FDNA and QDNA do not affect the formation of the intended hairpin structure of LDNA. This may make TMBs an alternative and cost-effective form of MB for applications that require a large number of probes since there is no need to covalently modify every probe with a fluorophore and a quencher.
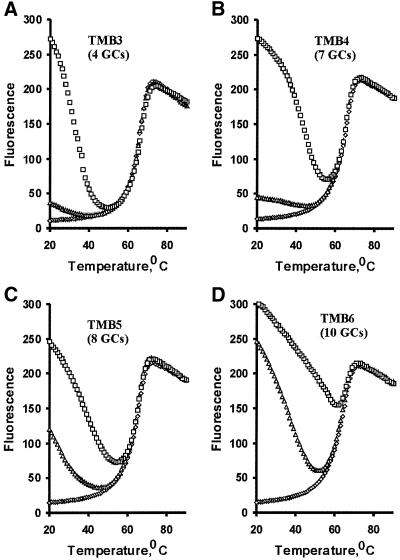
To demonstrate the general utility of a single set of FDNA and QDNA for multiple MB assembling, we have constructed three more TMBs, TMB4, TMB5 and TMB6, using three different LDNA molecules and the common FDNA1 and QDNA1 pair. Each LDNA was designed to form a hairpin structure with the universal set of stem-1, stem-2 and stem-3 but with a unique 15-nt loop sequence for target binding. Figure 4A–D illustrates the thermal denaturation profiles of the four TMBs obtained under three conditions: in the absence of target (diamonds), in the presence of a match target (squares) and in the mixture containing a mismatch target (triangles). As expected, all three new TMBs can signal the presence of match nucleic acid targets by large fluorescence intensity change. They also exhibited an ability to discriminate against single-mismatch targets. Consistent with the findings previously reported for standard MBs (11), we found that the temperature adequate for carrying out single-mismatch discrimination was dependent on the GC content of the target sequence. When the target is AT-rich (as in TMB3 and TMB4), the TMBs can have a high level of performance in discrimination in the low temperature range. When GC content was sufficiently high (as in TMB5 and TMB6), the single-mismatch discrimination can be achieved in the high temperature range. For instance, the best temperature for TMB6 (its target is GC-rich with 67% GC content) was near 50°C while TMB3 (its target is AT-rich with 73% AT content) exhibited a large fold of discrimination even at 20°C.
Figure 4.
Four TMBs made with a common set of stem-1, stem-2 and stem-3. Four related TMBs, TMB3–6, are constructed with FDNA1 as the common fluorophore, QDNA1 as the common quencher, and individual LDNAs having the sequence arrangement of d(CCTGCCACGCTC CGCGGCGAGC-loop-GCTCGCCCTCGCAC CGTCCACC) (with FDNA1-binding sequence shown in bold, QDNA1-binding domain indicated in italic and self-complementary motifs underlined). The loop sequences are: TMB3, d(CACCAAATATGATAT); TMB4, d(CCGTATATCGAATGC); TMB5, d(CTACCGATGCTCAGA); TMB6, d(CATCGGCGCTAGGTC). Fluores cence intensity was measured for TMB3 (A), TMB4 (B), TMB5 (C) and TMB6 (D) as a function of temperature in the absence of a nucleic acid target (diamonds), in the presence of the match target (squares) and a mismatch target (triangles). The target sequences are: T3, d(ATATCA TA(C)TTTGGTG); T4, d(GCATTCGA(C)TATACGG); T5, d(TCTGAG CA(C)TCGGTAG); T6, d(GACCTAGC(A)GCCGATG). The single letter in the parenthesis in each target sequence is the single nucleotide change for the mismatch target at the position underlined. Each target also contains TACTCTT at the 5′ end and TTTGCTTT at the 3′ end.
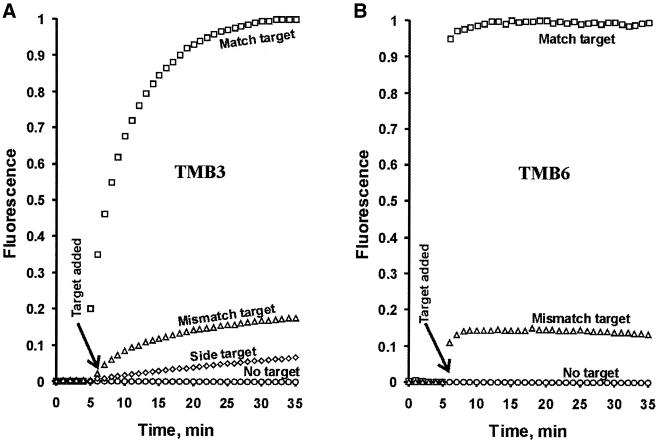
TMB3 (the AT-rich sequence) and TMB6 (the GC-rich sequence) were examined for the real-time signaling capability at a chosen temperature suitable for single-mismatch discrimination (22°C for TMB3 and 50°C for TMB6) and the results are shown in Figure 5A and B, respectively (fluorescence intensities were normalized; a ‘side target’ was also used for TMB3, see discussion below). Solutions made of each TMB were incubated at the designated temperature first for 5 min in the absence of any target, followed by the addition of water (i.e. no target; circles), the mismatch target (triangles) or the match target (squares), and the resultant mixtures were further incubated for 30 min more (the fluorescence intensity of each solution was monitored continuously before and after the target introduction). It is evident that TMBs can be used to effectively discriminate against targets differing only by a single nucleotide for both AT-rich and GC-rich targets. The performance levels of the TMBs were similar to those described previously for standard MBs with similar target sequences (1,4,11).
Figure 5.
Real-time detection using TMB3 and TMB6. The real-time signaling behaviors of TMB3 (A) and TMB6 (B) were assessed as follows: each solution was incubated at a constant temperature (22°C for TMB3 and 50°C for TMB6) for 5 min, followed by the addition of water (no target; circles), a match target (squares) or a mismatch target (triangles), and the continuing incubation at the same temperature for 30 min more. Fluorescent intensity of each solution was collected every minute during the 35 min incubation. The fluorescence intensity was normalized for each system as (Ft – F0) / (Fmax – F0) where Ft is the fluorescence reading of a DNA mixture at a given time, F0 is the initial reading for the sample containing no target, and Fmax is the reading at 35 min for the sample containing the match target. The sequences of TMB3 and TMB6 and their match and mismatch targets are listed in the legend of Figure 4. The data points in diamonds in (A) were obtained for TMB3 with the ‘side target’ ST-1, which has the following sequence d(AACACA AGGTGGCTCGCCGCGGAAAGCAAA) (the sequence underlined can form Watson–Crick base pairs with the LDNA3 segment covering the first 7-nt complementary sequence of the stem-1 and the nearby four nucleotides on each side).
Although a TMB is intended for the detection of a DNA target that can form specific Watson–Crick base pairs with the loop sequence of the LDNA (see Fig. 1), undesirable interactions that disrupt the formation of stem-1 can lead to false positive results. One possible scenario is that a DNA target might give rise to a false positive signal by binding to the LDNA segment consisting of one of the two complementary sequences of the stem-1 and its nearby nucleotides on each side. To assess the level of interference that might occur in this particular scenario, we used a special DNA target, side target 1 (ST-1), to test the false signaling possibility with TMB3. ST-1 contained a 15-nt sequence (the same length as the loop-binding sequences used as the targets throughout this study) intended to disrupt the stem-1 of TMB3 by forming Watson–Crick base pairs with the first seven nucleotides of the stem-1 (as the 5′ complementary sequence of the stem-1) as well as the eight nearby nucleotides (four on each side of the stem-1). Only a very weak signal was produced with the introduction of ST-1 (Fig. 5A, the data series in diamonds) and the fluorescence intensity was even lower than that seen with the mismatch target. Therefore, the interference caused by the hypothetical stem disruption was not very significant.
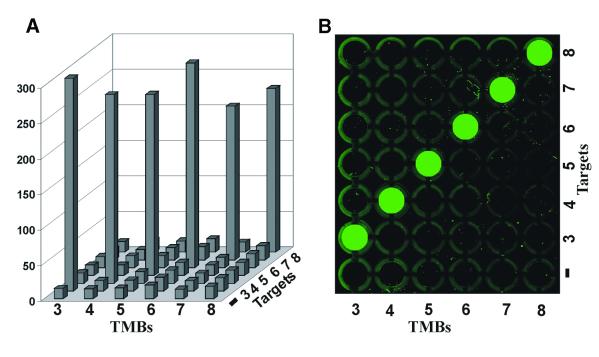
To further demonstrate the general utility of the common FDNA/QDNA pair, we conducted a simple array experiment for target sensing by fluorescence. In addition to TMB3–6, two new TMBs, TMB7 and TMB8, that again contained the common set of stem-1, stem-2 and stem-3 but different probing sequences, were included for the experiment. The fluorescence intensity of each TMB in the presence of each DNA target determined at 22°C is plotted in Figure 6A. Without exception, each TMB emits a very strong fluorescence in the presence of the match target, but only has very low background fluorescence in the presence of each of the unintended targets. For instance, TMB7 had a fluorescence intensity of 217 in the presence of T7, but only had fluorescence readings between 14 and 15 when the other five non-desirable targets were used (the background fluorescence at 13.5). The solutions used for Figure 6A were also placed in microplate wells and scanned to obtain the fluoroimage shown in Figure 6B. Only samples that contained the match target were able to give rise to detectable fluorescent signals. Each TMB was also examined for match target detection in the presence of all six targets (six-target mixture) as well as in the presence of only five unintended targets (five-target mixture), and was found to fluoresce at its maximal capability in the six-target mixture and only emit fluorescence at the background level in the five-target mixture (data not shown). These data clearly indicate the general applicability of FDNA and QDNA as universal probes in setting up parallel MBs for high throughput applications.
Figure 6.
Results from a simple array experiment. Six TMBs, TMB3–8, that contained the same set of stem-1, stem-2 and stem-3 were used. Sequence information for TMB3–6 is given in Figure 4. TMB7 and TMB8 have the loop sequences of d(CATGCAGATCTACAC) and d(ATGGTAGAACAGAGA), respectively. Their target sequences are d(TACTCTTGTGTAGAT(G)CTGCATGTTTGCTTT) (T7) and d(TACTCTTTCTCTGTT(G)CTACCA TTTTGCTTT) (T8; the single letter in the parenthesis is the single nucleotide change for the mismatch target at the position underlined). (A) Fluorescence intensity of each TMB (each column) was recorded at 22°C in the absence of a nucleic acid target (first row), in the presence of T3 (second row), T4 (third row), T5 (fourth row), T6 (fifth row), T7 (sixth row) and T8 (seventh row). Each target DNA was directly mixed with a TMB stock solution and incubated for 1 h at room temperature before the fluorescence intensity of the solution was obtained. (B) A 200 µl aliquot of each of the above samples was placed in a well of 7 × 6 microplates and a fluoroimage was obtained using a Typhoon 9200 (Amersham Biosciences). The excitation was at 532 nm (green laser) and the emission was at 526 mm with a short-pass filter.
DISCUSSION
The results indicate that TMBs can have a high performance similar to the standard MBs and fluorescence signaling by TMBs is highly specific as a single base mutation within the target sequence can result in very significant signal reduction. The hairpin structures of TMBs appear to have somewhat decreased melting points as compared with related MBs with identical internal stem–loop sequences. This is likely caused by the difficulty of TMBs in forming the outside edge base pair in stem-1. This factor needs to be considered when designing TMBs with a desired melting point. After testing more than a dozen TMBs, we found that the melting points of TMBs can still be accurately predicted using the M-fold program if the periphery base pair of stem-1 is ignored. A convenient way to do this is to first design a stem–loop structure with a desired melting point and then to add a ‘fake’ base pair to the outside edge of the stem-1. The two bases in this dummy ‘base pair’ will of course not associate (or not fully associate) when the TMB is fully assembled, therefore their addition will not significantly affect the desired melting point.
Compared with the standard MBs, TMBs have the advantage of being easily adapted for applications that demand a great number of probes. With a single set of FDNA and QDNA and a series of standard LDNA oligodeoxyribonucleotides, a variety of TMBs can easily be assembled for detecting different nucleic acids. For these applications, the use of TMBs can be more cost-effective and also helps to eliminate the tedious procedures involved in synthesizing and purifying each double-labeled DNA probe.
TMBs also have the advantage of a greater flexibility in fluorophore changes. For example, a large number of nucleic acid samples can be probed easily with two or more fluorophores using the TMB approach. This is because the same LDNAs and QDNA can always be used and the additional cost to make new FDNAs labeled with different fluorophores is relatively small. Similarly, TMBs are also well suited for the construction of wavelength-shifting MBs (19) and for multiplex detections (4).
The TMBs have potential in the preparation of MB microarrays. Although DNA microarray technology is in commercial use and has yielded vast amounts of genetic and cellular information (20–25), current DNA array approaches require the labeling of nucleic acid targets with various fluorophores. Target labeling is not only time-consuming but it can change the level of targets originally present in a sample. With the use of MBs, there is no need to label nucleic acid targets. Since only unmodified oligodeoxyribonucleotides of TMBs need to be immobilized on the array surface, methods that are currently in use for coating microarrays with synthetic DNA oligodeoxyribonucleotides can be directly used to immobilize LDNAs. FDNA and QDNA can then be supplied as a universal stock solution that can be simply mixed with the sample of interest during the hybridization step. Fluorescence is generated during the hybridization and, thus, there is no need to label nucleic acid targets.
In summary, we have demonstrated that properly designed TMBs were as effective as standard MBs in signaling the presence of matching nucleic acid targets and in precisely discriminating targets that differ by a single nucleotide. We have also shown a single set of FDNA and QDNA can be used to construct multiple TMBs for detecting matching targets without false signaling. With the increased assembling flexibility, TMBs can be more cost-effective for applications that demand a large number of DNA probes and more compatible with surface immobilization. Therefore, the alternative MB we describe herein should broaden the utility of MBs as nucleic acid detection probes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the Li laboratory for helpful discussions. This work was supported by research grants from Canadian Institutes of Health Research, Natural Sciences and Engineering Research Council of Canada and Canadian Foundation for Innovation. Y.L. is a Canada Research Chair in Nucleic Acids Biochemistry.
REFERENCES
- 1.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet G., Krichevsky,O. and Libchaber,A. (1998) Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl Acad. Sci. USA, 95, 8602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet G., Tyagi,S., Libchaber,A. and Kramer,F.R. (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA, 96, 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 5.Leone G., van Schijndel,H., van Gemen,B., Kramer,F.R. and Schoen,C.D. (1998) Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res., 26, 2150–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piatek A.S., Tyagi,S., Pol,A.C., Telenti,A., Miller,L.P., Kramer,F.R. and Alland,D. (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol., 16, 359–363. [DOI] [PubMed] [Google Scholar]
- 7.Vet J.A., Majithia,A.R., Marras,S.A.E., Tyagi,S., Dube,S., Poiesz,B.J. and Kramer,F.R. (1999) Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl Acad. Sci. USA, 96, 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostrikis L.G., Tyagi,S., Mhlanga,M.M., Ho,D.D. and Kramer,F.R. (1998) Spectral genotyping of human alleles. Science, 279, 1228–1229. [DOI] [PubMed] [Google Scholar]
- 9.Kostrikis L.G., Shin,S. and Ho,D.D. (1998) A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nature Med., 4, 350–353. [DOI] [PubMed] [Google Scholar]
- 10.Giesendorf B.A., Vet,J.A., Tyagi,S., Mensink,E.J., Trijbels,F.J. and Blom,H.J. (1998) Molecular beacons: a new approach for semi-automated mutation analysis. Clin. Chem., 44, 482–486. [PubMed] [Google Scholar]
- 11.Marras S.A.E., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal., 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 12.Matsuo T. (1998) In situ visualization of messenger RNA for basic fibroblast growth factor in living cells. Biochim. Biophys. Acta, 1379, 178–184. [DOI] [PubMed] [Google Scholar]
- 13.Sokol D.L., Zhang,X., Lu,P. and Gewirtz,A.M. (1998) Real time detection of DNA·RNA hybridization in living cells. Proc. Natl Acad. Sci. USA, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang X., Liu,X., Schuster,S. and Tan,W. (1999) Designing a novel molecular beacon for surface-immobilized DNA hybridization studies. J. Am. Chem. Soc., 121, 2921–2922. [Google Scholar]
- 15.Liu X. and Tan,W. (1999) A fiber-optic evanescent wave DNA biosensor based on novel molecular beacons. Anal. Chem., 71, 5054–5059. [DOI] [PubMed] [Google Scholar]
- 16.Steemers F.J., Ferguson,J.A. and Walt,D.R. (2000) Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. Nat. Biotechnol., 18, 91–94. [DOI] [PubMed] [Google Scholar]
- 17.Brown L.J., Cummins,J., Hamilton,A. and Brown,T. (2000) Molecular beacons attached to glass beads fluoresce upon hybridization to target DNA. Chem. Commun., 621–622. [Google Scholar]
- 18.Liu X., Farmerie,W., Schuster,S. and Tan,W. (2000) Molecular beacons for DNA biosensors with micrometer to submicrometer dimensions. Anal. Biochem., 283, 56–63. [DOI] [PubMed] [Google Scholar]
- 19.Tyagi S., Marras,S.A.E. and Kramer,F.R. (2000) Wavelength-shifting molecular beacons. Nat. Biotechnol., 18, 1191–1196. [DOI] [PubMed] [Google Scholar]
- 20.Chee M., Yang,R., Hubbell,E., Berno,A., Huang,X.C., Stern,D., Winkler,J., Lockhart,D.J., Morris,M.S. and Fodor,S.P. (1996) Accessing genetic information with high-density DNA arrays. Science, 274, 610–614. [DOI] [PubMed] [Google Scholar]
- 21.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 22.Ramsay G. (1998) DNA chips: state-of-the-art. Nat. Biotechnol., 16, 40–44. [DOI] [PubMed] [Google Scholar]
- 23.Whitecombe D., Newton,C.R. and Little,S. (1998) Advantages in approaches to DNA-based diagnostics. Curr. Opin. Biotechnol., 9, 602–608. [DOI] [PubMed] [Google Scholar]
- 24.Burns M.A., Johnson,B.N., Brahmasandra,S.N., Handique,K., Webster,J.R., Krishnan,M., Sammarco,T.S., Man,P.M., Jones,D., Heldsinger,D., Mastrangelo,C.H. and Burke,D.T. (1998) An integrated nanoliter DNA analysis device. Science, 282, 484–487. [DOI] [PubMed] [Google Scholar]
- 25.Case-Green S.C., Mir,K.U., Pritchard,C.E. and Southern,E.M. (1998) Analyzing genetic information with DNA arrays. Curr. Opin. Chem. Biol., 2, 404–410. [DOI] [PubMed] [Google Scholar]