Abstract
We seek to create useful biological diversity by exploiting the modular nature of genetic information. In this report we describe experiments that focus on the modular nature of plasmid cloning vectors. Bacterial plasmids are modular entities composed of origins of replication, selectable markers and other components. We describe a new ligation-independent cloning method that allows for rapid and seamless assembly of vectors from component modules. We further demonstrate that gene cloning can be accomplished simultaneously with assembly of a modular vector. This approach provides considerable flexibility as it allows for ‘menu driven’ cloning of genes into custom assembled modular vectors.
INTRODUCTION
We seek to automate the gene engineering process so that rapid assembly of modular genes is accomplished using standard laboratory robotics. To achieve this objective, the gene assembly process must be made as simple as possible. In particular, the number of enzymatic steps required for gene assembly should be kept to a minimum. In this report, we describe a method that can be used to assemble genes by simply mixing PCR products together and transforming Escherichia coli. Thus, cloning is accomplished without any enzymatic treatment of the PCR products.
We previously described a method called RNA-overhang cloning (ROC) that can be used for the seamless assembly of chimeric genes by directional ligation of PCR products (1). To perform an ROC experiment, PCR is conducted using chimeric DNA/RNA primers composed mostly of DNA, but including a 5′ region composed of one or more ribonucleotides. When PCR is performed using a polymerase that lacks reverse transcriptase activity (2), the result is double-stranded DNA molecules flanked by single-stranded RNA tails. Linking one or more PCR products together via these tails, followed by ligation of the PCR products to one another, creates chimeric genes.
In our original ROC experiments, we used tails of ≤4 nt to link PCR products to one another. Ligation-independent cloning (LIC) has been reported to require single-stranded tails of at least 12 nt for efficient linkage (3,4). We therefore modified our strategy and developed a system that could generate longer tails, made mostly of DNA. We demonstrated the utility of this method by using it to clone genes while simultaneously assembling the vectors into which the genes were cloned.
MATERIALS AND METHODS
Modular vectors
Table 1 provides the primer sequences and DNA template plasmids that were used for modular vector experiments. Three primers are shown for each vector module. The 2′-O-methyl residues are underlined. When plasmids were assembled from two components, the first two primers in each set were used in PCR. When plasmids were assembled from three components, the first and third primers in each set were used in PCR. Each 100 µl PCR contained 50 pmol of each primer, 1× Pfu buffer [10 mM (NH4)2SO4, 20 mM Tris pH 8.8, 2 mM MgSO4, 10 mM KCl, 0.1% Triton X-100 and 1 mg/ml bovine serum albumin], 1 mM additional MgSO4, 0.3 mM each dNTP, 5–10 ng of plasmid template and 1.25–1.85 U each of cloned Pfu and Pfu Turbo polymerases (Stratagene, La Jolla, CA). Chimeric RNA/DNA primers were purchased from Oligo’s Etc. (Willsonville, OR). A typical step program for PCR was as follows: one cycle of 95°C for 3 min and 52°C for 2 min, followed by 30 cycles of 95°C for 30 s, 52°C for 30 s and 72°C for 2 min.
Table 1. Vector module primers and templates.
| Module | Template | Primers |
|---|---|---|
| Kanamycin (1231 bp) | pCR 2.1 | 5′-CTACCTAGCAAGCTUCTATCTGGACAAGGGAAAACG |
| 5′-GAGCTCACTTAGCAAGGCGAAAACTCTCAAGGATC | ||
| 5′-CTAGACAGTTCAGTCCTATCTGGACAAGGGAAAACG | ||
| Ampicillin (1420 bp) | pUC19 | 5′-CTACCTAGCAAGCTUCAGCCTGAATGGCGAATGG |
| 5′-GAGCTCACTTAGCAAGGTCATGAGATTATCAAAAAGG | ||
| 5′-CTAGACAGTTCAGTCCAGCCTGAATGGCGAATGG | ||
| Chloramphenicol (1083 bp) | pACYC184 | 5′-CTACCTAGCAAGCTUCCGAATAAATACCTGTGACGG |
| 5′-GAGCTCACTTAGCAAGCCTCAGGCATTTGAGAAGC | ||
| 5′-CTAGACAGTTCAGTCCCGAATAAATACCTGTGACGG | ||
| GFP (1068 bp) | pGFP | 5′-GACTGAACTGTCTAGAATGCAGCTGGCACGACAG |
| 5′-GATGACTTGACAGACGATTAGGGTGATGGTTCACG | ||
| LacZ (724 bp) | pBS II SK(–) | 5′-GACTGAACTGTCTAGAGCGCAACGCAATTAATGTG |
| 5′-GATGACTTGACAGACGATTAGGGTGATGGTTCACG | ||
| ColE1 Ori (859 bp) | pUC19 | 5′-TTGCTAAGTGAGCTCATCCCTTAACGTGAGTTTTCG |
| 5′-AAGCTTGCTAGGTAGGAGGCGGTTTGCGTATTGG | ||
| 5′-GTCTGTCAAGTCATCGAGGCGGTTTGCGTATTGG | ||
| p15 Ori (1104 bp) | pACYC184 | 5′-TTGCTAAGTGAGCTCCTACCGCATTAAAGCTTATCG |
| 5′-AAGCTTGCTAGGTAGGCGTCGGGTGATGCTGCC | ||
| 5′-GTCTGTCAAGTCATCGCGTCGGGTGATGCTGCC |
PCR products were gel purified from a 1% agarose gel using the QiaQuick gel extraction kit (Qiagen, Valencia, CA). Approximately 80 ng of each fragment were combined in a 20 µl reaction that included 2 µl of 10× USB ligation buffer (660 mM Tris–HCl pH 7.6, 66 mM MgCl2, 100 mM DTT and 660 µM ATP) (USB, Cleveland, OH). The reaction was heated to 65°C for 8 min and then cooled for 20 min to ∼35–40°C. Samples were centrifuged briefly and incubated for another 15 min at room temperature.
Annealing reactions were precipitated by adding 100 µl of 100% ethanol, incubating for 15 min at –80°C, followed by a 10 min centrifugation and 70 and 100% ethanol washes. Electrocompetent DH5α cells were transformed using a Bio-Rad E.coli pulser (Bio-Rad, Hercules, CA); 5 µl of each annealing reaction were combined with 40 µl of Electromax DH5α-E cells (Life Technologies, Rockville, MD).
Simplified PCR cloning
We combined 5 µl of the Kan PCR with 5 µl of the Ori PCR and added the sample to 100 µl of chemically competent E.coli cells. The mixture was incubated on ice for 15 min, followed by a 45 s heat shock at 42°C. Cells were incubated on ice for another 2 min before 800 µl of LB medium was added. Cells were incubated with shaking for 45 min at 37°C before plating.
Ligation-dependent cloning
For phosphorylation of primers 1 nmol of each primer was combined with 5 µl of 10 mM ATP, 5 µl of 10× PNK buffer (330 mM Tris–acetate pH 7.8, 660 mM potassium acetate, 100 mM magnesium acetate and 5 mM DTT), 2 µl (20 U) of T4 polynucleotide kinase (Epicentre, Madison, WI) and H2O to 50 µl. Reactions were incubated for 30 min at 37°C. Approximately 80 ng of each fragment were combined with 1.5 µl of USB ligase, 1.5 µl of 10× USB ligation buffer (660 mM Tris–HCl pH 7.6, 66 mM MgCl2, 100 mM DTT and 660 µM ATP) and 7.5 U T4 DNA ligase (USB) in a 15 µl reaction volume. Reactions were incubated for 2 h at room temperature, followed by 15 h at 14°C.
Terminator-PCR (t-PCR) with a radiolabeled primer
One nanomole of a DNA primer called 5′Amp S/PE (5′-TGAGAGTGCACCATATGCG) was combined with 5 µl of [γ-32P]-labeled ATP (3000 Ci/mmol), 5 µl of 10× PNK buffer (330 mM Tris–acetate pH 7.8, 660 mM potassium acetate, 100 mM magnesium acetate and 5 mM DTT), 2 µl (20 U) of T4 polynucleotide kinase (Epicentre) and H2O to 50 µl. Reactions were incubated for 30 min at 37°C. A DNA sequencing reaction was performed using the Sequenase 2.0 DNA sequencing kit (USB) with the Amp/ColE1 vector as a template. Reactions contained 0.5 µl of [α-32P]-labeled ATP (800 Ci/mmol), 25 pmol primer and 1 µg DNA. This sequencing reaction was used as a size standard. Each 100 µl Pfu PCR contained 50 pmol of each primer, 1× cloned Pfu buffer [10 mM (NH4)2SO4, 20 mM Tris pH 8.8, 2 mM MgSO4, 10 mM KCl, 0.1% Triton X-100 and 0.1 mg/ml bovine serum albumin], 1 mM additional MgSO4, 0.3 mM each dNTP, 5–10 ng of plasmid template and 1.25 U of both cloned Pfu and Pfu Turbo polymerases (Stratagene). Pfu exo(–) reactions contained 2.5 U of Pfu exo(–) polymerase (Stratagene). Each 100 µl Taq PCR contained 50 pmol of each primer, 1× Taq buffer (10× reaction buffer without MgCl2: 500 mM KCl, 100 mM Tris–HCl pH 9.0 at 25°C and 1.0% Triton X-100), 1 mM MgCl2, 0.1 mM each dNTP and 5 U of Taq polymerase in storage buffer A (Promega, Madison, WI). The PCR cycling consisted of one cycle of 95°C for 3 min, 52°C for 2 min and 72°C for 15 s, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 15 s. Chimeric RNA/DNA primers were purchased from Oligo’s Etc.
RESULTS
Three ribonucleotide polymerase termination
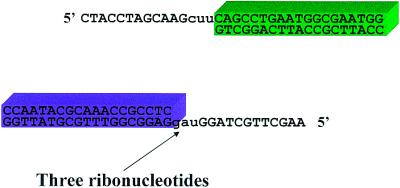
The experimental approach is described in Figure 1. We synthesized PCR primers that were composed of DNA, but included ribonucleotides at positions 13–15. We hypothesized that the ribonucleotides would cause Pfu to terminate polymerization, leaving a 15 nt overhang composed mostly of DNA. We tested this hypothesis by attempting to use these ‘terminator primers’ to create a functional E.coli plasmid by linking two PCR products together. One fragment contained the kanamycin resistance gene (Kan); the second contained a ColEl origin of replication (Ori). The tails flanking the Ori fragment were complementary to the tails flanking the Kan fragment so that the fragments could be combined via specific annealing of the tails.
Figure 1.
Termination of polymerization generates double-stranded DNA with specific single-stranded tails. Diagram of PCR products with tails generated by termination of polymerization. Deoxyribonucleotides are shown in upper case, ribonucleotides are shown in lower case.
The terminator primers were used to amplify the Ori and Kan fragments. The PCR products were combined, heated, and slow cooled to allow for annealing of the tails. DH5α electrocompetant E.coli was used for electroporation transformation. Approximately 80 ng of DNA was used in a transformation experiment; 40 000 colonies/µg DNA were obtained. We mapped 15 plasmids using SacI and HindIII. Fourteen plasmids exhibited the expected restriction map. Sequencing of a total of 18 overhang junctions demonstrated the high fidelity of the method since only four junctions contained point mutations.
Single ribonucleotide polymerase termination
To begin to define the requirements for termination of polymerization by ribonucleotides, we asked whether a single ribonucleotide is sufficient to create an overhang that can be used for LIC. PCR primers were generated that harbored a single ribonucleotide at position 15, relative to the 5′ terminus of the primer. Each of the four ribonucleotides (A, C, G and U) was tested individually for its ability to produce recombinants. Specifically, the Kan and Ori fragments were amplified with primers that contained a different single ribonucleotide at each of the four termination sites. The experiment produced 20 000 colonies/µg DNA. Sequence analysis of 10 junctions showed that eight were error free.
Significantly, we were also able to generate the expected recombinant plasmid using a simplified protocol. The Kan and Ori PCR products were generated using terminator primers. We combined 5 µl of each PCR and directly transformed the DNA into chemically competent E.coli without a DNA purification step or an annealing step. This simple protocol produced 100 colonies/µg DNA. Although the cloning efficiency was low, this likely represents the simplest possible method for generating a recombinant plasmid from PCR products.
Ligation-dependent cloning with short overhangs
Ligation-dependent experiments were also performed in which the length of the overhang was shortened to 6, 3 or 1 nt. These experiments used chimeric primers consisting of a variable number of 5′ DNA nucleotides followed by a single ribonucleotide and a stretch of template binding DNA nucleotides. Thus, the overhangs included five, two or zero DNA nucleotides, respectively. All three of these primer configurations produce accurately ligated vectors, however, the yield of colonies was approximately 25-fold lower when a single ribonucleotide overhang was used (125 000 versus 5000 colonies/µg DNA). DNA sequence analysis revealed one case of a duplicated junction, apparently created by blunt-end ligation of PCR products, suggesting that the terminator nucleotide was being read through at some frequency. Blunt-ended fragments had not been detected in previous experiments, most likely due to the ligation-independent procedure that was used.
Single 2′-O-methyl ribonucleotide polymerase termination
In an attempt to improve the termination efficiency, we tested primers that harbored a single 2′-O-methyl ribonucleotide at position 15, rather than a single ribonucleotide. These chimeric 2′-O-methyl primers demonstrated greatly improved cloning efficiency. We obtained 500 000 colonies/µg DNA. Thus, cloning with primers containing single 2′-O-methyl ribonucleotides is 25-fold more efficient than cloning with primers that harbor single ribonucleotides. We tested each of the four possible 2′-O-methyl ribonucleotides and found that each of the four nucleotides functions to generate an overhang for terminator cloning.
Radiolabeled terminator-PCR experiments
In order to confirm the presence of the single-stranded overhangs and compare termination efficiencies with different types of chimeric primers and polymerases, we designed an experiment that used radiolabeled primers in t-PCRs. A 32P-labeled DNA reverse primer and a chimeric forward primer were used in a PCR to generate a 100 bp product. These reactions were denatured and resolved on an 8% polyacrylamide gel (Fig. 2). A DNA sequencing ladder was used as a size standard so that the lengths of the termination products could be measured with single nucleotide resolution. If the polymerase terminates at the terminator nucleotide(s) an 85 nt labeled product is expected; the 85 nt sequencing product is indicated by the lower arrow in Figure 2. Full read-through to the end of the chimeric primer would generate a 100 nt labeled product (indicated by the upper arrow in Fig. 2). The sequences of all chimeric primers were identical and were varied only with respect to the type of nucleotide(s) comprising the terminator positions (i.e. ribo, deoxy or 2′-O-methyl). The same 32P-labeled DNA reverse primer was used for all experiments.
Figure 2.
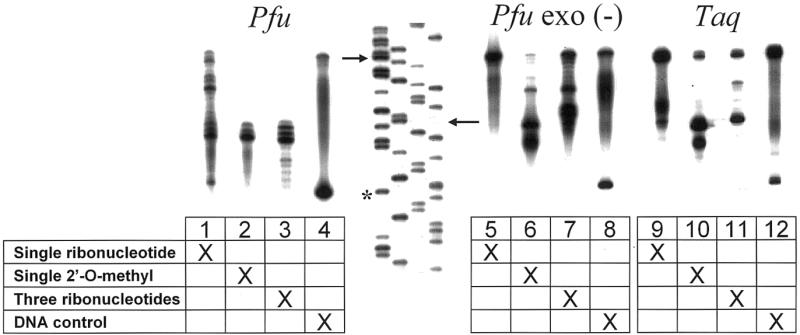
. Termination of polymerization by ribonucleotides and 2′-O-methyl ribonucleotides. PCR experiments were conducted with the following polymerases: Pfu (lanes 1–4), Pfu exo(–) (lanes 5–8) and Taq (lanes 9–12). The same 32P-labeled primer was used as a PCR primer in each experiment. Four different unlabeled primers were used. The four primers were identical except for the inclusion of one or three ribonucleotides or a single 2′-O-methyl ribonucleotide at a particular position (see Fig. 1). A sequencing ladder was used as a size standard so that termination products could be measured with single nucleotide resolution.
The radiolabeled PCR experiments confirmed that there is a stop or pause induced by the terminator residues. PCRs using Pfu polymerase demonstrated termination with all versions of the terminator primers as expected. However, there was significant read-through with the single ribonucleotide primer (Fig. 2, lane 1). The experiment with Pfu exo(–) demonstrates that the 3′→5′ exonuclease activity of Pfu is important for termination since Pfu exo(–), which lacks 3′→5′ exonuclease activity, reads through the terminator with increased efficiency (lanes 5–7). PCRs using Taq polymerase showed significant termination by three RNA nucleotides and appeared to be strongly stopped by the 2′-O-methyl residue.
We expected termination to occur 1 nt prior to the terminator residue. Although Pfu terminated at that position, termination also occurred 1 or 2 nt prior to the expected position.
An unexpected early termination was observed adjacent to the template region of both the DNA control primer and the single ribonucleotide chimeric primer (Fig. 2, asterisk). This stop was not seen in the three ribonucleotide or 2′-O-methyl t-PCRs. The mechanism of this unexpected termination is unknown.
Modular vector assembly
We developed the t-PCR cloning method as a tool for creating useful biological diversity by linking together modular DNA elements in a ligation-independent manner. We tested this approach by using the method to clone genes while simultaneously assembling the vectors into which the genes were cloned.
The modular vector system works in the following way. Vector elements (i.e. drug resistance genes, replication origins or insert genes) are amplified separately with Pfu polymerase using 2′-O-methyl terminator primers. The overhangs on the amplified vector elements (modules) follow a specific set of rules that allow similar functional modules to be interchanged in a standardized orientation. Modules can be inserted or substituted into existing vectors or can be combined to construct new vectors.
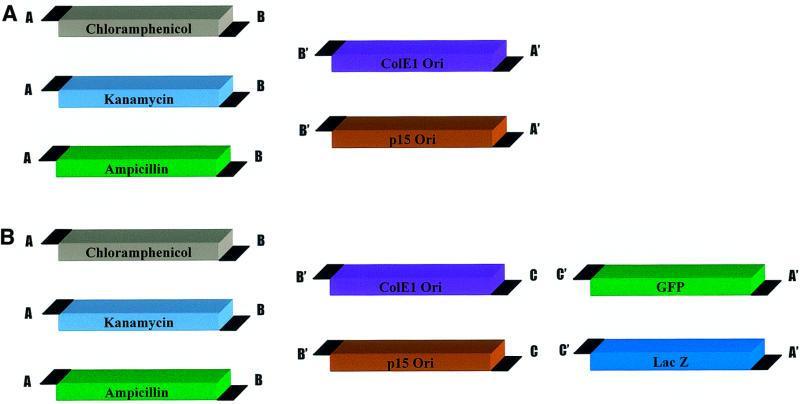
Combining drug resistance and origin of replication modules with insert modules produces cloned genes in customized vectors (Fig. 3). We constructed a set of vectors using two possible origins of replication, three possible drug resistance genes and two different marker genes. These fragments were combined in all possible combinations to generate a total of 18 distinct plasmids.
Figure 3.
Modular vector components. Identical overhang sequences are used for modules of analogous function (e.g. drug resistance genes), which make the modules readily interchangeable. (A) Components for combinatorial assembly of six modular vectors. (B) Components for expression cloning of two different genes into each of six modular vectors.
Six vectors were produced in two module assembly reactions that contained no insert (Fig. 3A). The GFP- and LacZ-containing vectors were assembled in three module assembly reactions (Fig. 3B), but were also assembled using two module reactions in order to compare cloning efficiencies between two and three module cloning. We found that two module assembly of GFP- or LacZ-containing vectors is approximately 6-fold more efficient than cloning of the same genes via a three module experiment (1400 versus 240 colonies/µg DNA). Unexpectedly, however, we found that the efficiency of two module assembly varies significantly depending on the design of the experiment. The two module assembly experiments that combined a resistance gene with an origin of replication (Fig. 3A) were typically 450-fold more efficient than the experiments that combined a marker gene (i.e. LacZ or GFP) with an entire amplified plasmid. The reason for this difference in efficiency remains to be determined.
We have successfully completed 30 modular assembly experiments. Twenty-five of the plasmids were produced on the first attempt. All recombinants were confirmed by restriction mapping. Approximately 80% of the plasmids that were mapped (90/116) exhibited the expected recombinant restriction map. Fifteen ligation junctions were sequenced and 14 junctions were free of point mutations.
DISCUSSION
In this report, we describe a LIC method (t-PCR) that allows for the creation of recombinant DNA molecules by simply mixing PCR products together and transforming E.coli. The method is sufficiently simple that we should now be able to fully automate the process of modular gene assembly.
We used the t-PCR method to construct a set of modular vectors. Furthermore, we demonstrated that gene cloning could be accomplished concomitant with modular vector assembly. The assembly of modular vectors follows certain design rules (5) that ensure that any module of a particular class (e.g. selectable markers) can be linked specifically to any module of another specific class (e.g. origins of replication). If this strategy is widely adopted, modular vector components will be universally interchangeable and the ability of individual researchers to rapidly generate novel vectors from modular components will increase dramatically. Furthermore, we envision the expansion of this approach to the assembly of other modular products such as modular proteins and cis-regulatory networks.
In addition to demonstrating the utility of the t-PCR method, we describe experiments designed to monitor the ability of certain polymerases to generate single-stranded tails in response to foreign residues in the template strand (Fig. 2). Our results suggest that two factors contribute to the generation of single-stranded tails: (i) a pause or termination of polymerization; (ii) the polymerase-associated 3′→5′ exonuclease activity of Pfu.
We describe experiments with Taq, Pfu and Pfu exo(–) and ribonucleotides or 2′-O-methyl ribonucleotides as terminator residues. We observe that three ribonucleotides terminate polymerization more efficiently than does a single ribonucleotide. Furthermore, a single 2′-O-methyl residue causes all three polymerases to terminate efficiently.
Others have mapped the precise termination points generated by termination of polymerization by various DNA polymerases in response to a foreign residue in the template strand. In the absence of exonuclease activity, termination generally occurs just prior to copying the foreign residue (6,7). Alternatively, termination sometimes occurs upon copying of the foreign residue (6,7). We find that both Pfu exo(–) and Taq terminate polymerization just before copying a 2′-O-methyl residue. Both enzymes appear to terminate after copying one or more of a series of three ribonucleotides. Both enzymes appear to read through a single ribonucleotide with high efficiency.
Why ribonucleotides or 2′-O-methyl ribonucleotides stimulate termination of polymerization is not obvious. One model is that the foreign residues are incompatible with the geometry of the catalytic site of the polymerase leading to a decreased rate of polymerization (8). Alternatively, Greagg and co-workers have suggested that certain archaeal DNA polymerases, such as Pfu, possess a ‘read-ahead’ function that allows the polymerase to detect deoxyuracil residues and terminate polymerization prior to copying those residues (9). The read-ahead function does not appear to be responsible for the termination we observe. First, the read-ahead function generates products that terminate 4–6 nt upstream of the position of the uracil in the DNA template. In contrast, we observe termination primarily 1, 2 or 3 nt upstream of a ribonucleotide or a 2′-O-methyl ribonucleotide. Furthermore, Greagg and co-workers observed similar termination patterns whether they used Pfu or Pfu exo(–) in their experiments. In contrast, we find that the exonuclease activity of Pfu is very important for the generation of single-stranded overhangs in response to ribonucleotides or 2′-O-methyl ribonucleotides in the template.
The observation that disruption of the Pfu exonuclease activity inhibits the formation of single-stranded overhangs suggests that neither a 2′-O-methyl residue nor three ribonucleotides create an absolute stop to polymerization. In the absence of the exonuclease activity, read-through is clearly seen. Given that read-through occurs, it is not surprising that the exonuclease activity stimulates the production of single-stranded overhangs. Copying of the foreign residue is a slow step. Once the polymerase has copied the foreign residue, removal of the newly incorporated nucleotides by the exonuclease would force the enzyme to attempt to accomplish this slow step yet again. Thus, the exonuclease activity is expected to enhance the production of single-stranded overhangs.
The presence of an active exonuclease site on the polymerase could also enhance formation of single-stranded overhangs by an alternative mechanism. It is known that the 3′ terminus that is created via polymerization sometimes switches into the exonuclease site of the enzyme (10). The presence of foreign residues on the template strand could stimulate this switch by either delaying polymerization (11) or disrupting duplex stability of a newly formed 3′ terminus that harbors the foreign residue (12). Thus, the presence of a foreign residue in the template strand could stimulate exonuclease activity in the vicinity of the foreign residue, even if the foreign residue is not actually copied by the enzyme (6). Alternatively, it is possible that the foreign residue is actually copied by the polymerase, but the newly created terminus is a poor substrate for further polymerization, resulting in enhanced transfer to the exonuclease site (6).
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by NIH grant AI48665.
REFERENCES
- 1.Coljee V.W., Murray,H.L., Donahue,W.F. and Jarrell,K.A. (2000) Seamless gene engineering using RNA- and DNA-overhang cloning. Nat. Biotechnol., 18, 789–791. [DOI] [PubMed] [Google Scholar]
- 2.Perler F.B., Kumar,S. and Kong,H. (1996) Thermostable DNA polymerases. Adv. Protein Chem., 48, 377–435. [DOI] [PubMed] [Google Scholar]
- 3.Aslandis C. and de Jong,P.J. (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res., 18, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aslandis C., de Jong,P.J. and Schmitz,G. (1994) Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products. PCR Methods Appl., 4, 172–177. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin C.Y. and Clark,K.B. (2000) Design Rules. MIT Press, Cambridge, MA.
- 6.Belguise-Valladier P., Maki,H., Sekiguchi,M. and Fuchs,R.P.P. (1994) Effects of single lesions on in vitro replication with DNA polymerase III holoenzyme. J. Mol. Biol., 236, 151–164. [DOI] [PubMed] [Google Scholar]
- 7.Khare V. and Eckert,K.A. (2001) The 3′→5′ exonuclease of T4 DNA polymerase removes premutagenic alkyl mispairs and contributes to futile cycling at O6-methylguanine lesions. J. Biol. Chem., 276, 24286–24292. [DOI] [PubMed] [Google Scholar]
- 8.Echols H. and Goodman,M.F. (1991) Fidelity mechanisms in DNA replication. Annu. Rev. Biochem., 60, 477–511. [DOI] [PubMed] [Google Scholar]
- 9.Greagg M.A., Fogg,M.J., Panayotou,G., Evans,S.J., Connolly,B.A. and Pearl,L.H. (1999) A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl Acad. Sci. USA, 96, 9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stocki S.A., Nonay,R.L. and Reha-Krantz,L.J. (1995) Dynamics of bacterial T4 DNA polymerase function: identification of amino acid residues that effect switching between polymerase and 3′→5′ exonuclease activities. J. Mol. Biol., 254, 15–28. [DOI] [PubMed] [Google Scholar]
- 11.Reha-Krantz L.J., Marquez,L.A., Elisseeva,E., Baker,R.P., Bloom,L.B., Dunford,H.B. and Goodman,M.F. (1998) The proofreading pathway of T4 DNA polymerase. J. Biol. Chem., 273, 22969–22976. [DOI] [PubMed] [Google Scholar]
- 12.Morales J.C. and Kool,E.T. (2000) Importance of terminal base pair hydrogen-bonding in the 3′-end proofreading by the Klenow fragment of DNA polymerase I. Biochemistry, 39, 2626–2632. [DOI] [PubMed] [Google Scholar]