Abstract
Characterization of the thermodynamics of DNA– drug interactions is a very useful part in rational drug design. Isothermal titration calorimetry (ITC), differential scanning calorimetry (DSC) and UV melting experiments have been used to analyze the multivalent (intercalation plus minor groove) binding of the antitumor antibiotic chartreusin to DNA. Using DNA UV melting studies in the presence of the ligand and the binding enthalpy determined by ITC, we determined that the binding constant for the interaction was 3.6 × 105 M–1 at 20°C, in a solution containing 18 mM Na+. The DNA–drug interaction was enthalpy driven, with a ΔHb of –7.07 kcal/mol at 20°C. Binding enthalpies were determined by ITC in the 20–35°C range and used to calculate a binding-induced change in heat capacity (ΔCp) of –391 cal/mol K. We have obtained a detailed thermodynamic profile for the interaction of this multivalent drug, which makes possible a dissection of ΔGobs into the component free energy terms. The hydrophobic transfer of the chartreusin chromophore from the solution to the DNA intercalating site is the main contributor to the free energy of binding.
INTRODUCTION
Characterization of DNA–drug interactions requires not only knowledge of the structures of the complexes, but the free energy contributions to the interaction (1–5). The thermodynamic characterization of drug binding might be used, together with structural studies, in the development of new strategies to interfere with gene expression in living cells (6,7).
A thorough characterization of a sufficiently high number of DNA–drug complexes might be needed to understand the contributors that give rise to the observed affinity and the nature of the interactions (3). There are a considerable number of multivalent natural DNA-binding drugs that contain an intercalating chromophore and a carbohydrate or peptide moiety that binds in the minor groove. Both functional domains are considered to serve as DNA recognition elements. In several anthracycline antibiotics, and other DNA-binding drugs, the non-intercalating portion can form several bonds with DNA (1,2), thus contributing to the stability of the complex. Multivalent binding is not only important in terms of sequence specificity but also because the exact characteristics of a carbohydrate moiety can modify some activities, such as the DNA cleavage mediated by topoisomerase II (8). The detection and characterization of multivalent modes of binding may result in the formulation of better rules for rational drug design. Improved drug binding affinity and the ability to discriminate larger DNA sequences may allow us to target unique sites in the genome (6,9).
Chartreusin is an antitumor antibiotic (10,11) that contains the chromophore chartarin and an uncharged disaccharide moiety (Fig. 1), and possesses a broad spectrum of activity against various tumors (10). It is a well-characterized member of a wider group of coumarin-related compounds, which includes elsamicin A and the chrymutasins (12,13). Binding to DNA is believed to be central to the mechanism by which chartreusin exerts its antitumoral effects (11,14,15). The binding sites have been mapped in regions containing CpG steps (15). Moreover, chartreusin appears to be a strong inhibitor of topoisomerase II (16). It can be used as a model to study multivalent intercalation, which also involves interaction through the minor groove of DNA. Here, we present a thermodynamic characterization of the binding of chartreusin to DNA, which permitted a study of how the intercalating chromophore and uncharged disaccharide moieties contribute to the strength of binding.
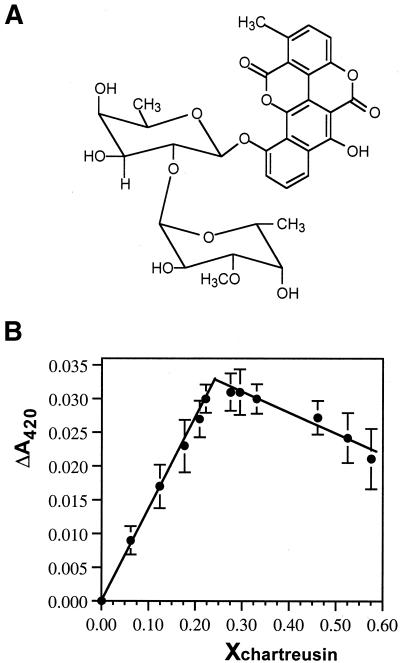
Figure 1.
(A) Structural formulae of chartreusin. (B) Continuous variation binding analysis (Job plot) for chartreusin binding to salmon testes DNA. The difference in absorbance at 420 nm as a function of the mole fraction of chartreusin is shown (mean ± SD, three independent experiments). The cross-over point was obtained from linear least squares fits to each data portion, with an inflection point at 0.244 mol fraction, which indicates a 1:3 stoichiometry of drug per DNA (bp).
Despite the pharmacological interest of chartreusin and some structurally related compounds (12), there is no experimental data on the conformation of the isolated drug or its complexes with DNA in the crystalline state. This shortcoming has been partially solved by a theoretical approach (17), which provided us with some details about the DNA– chartreusin interaction. Besides, previous binding data on chartreusin–DNA complexes were rather difficult to obtain and different results were obtained that were attributable to the formulations used to analyze the binding (11). In view of the modular nature of chartreusin, such knowledge of the multivalent binding can be used to improve the specificity of existing compounds and to design polyintercalators that should recognize longer DNA sequences.
Therefore, we have analyzed the binding of chartreusin to DNA and thermodynamically characterized it. We use the data obtained from these analyses to dissect the components that act in drug binding to DNA (3), and compare the contribution of the intercalating and disaccharide portions with the interaction.
MATERIALS AND METHODS
Materials
Chartreusin was kindly provided by Dr W.C. Krueger (The Upjohn Co., Kalamazoo, MI). A molar extinction coefficient of 12 700 M–1 cm–1 at 420 nm was used to determine the free concentration of chartreusin and 7230 M–1 cm–1 for bound drug.
Salmon testes DNA (Sigma) was sonicated, phenol extracted twice and dialyzed against 10 mM cacodylate buffer (pH 7.0) containing 8 mM NaCl and 0.1 mM EDTA ([Na+] = 18 mM). DNA concentration, in base pairs, was determined spectrophotometrically by using a molar extinction coefficient of 12 824 M–1 cm–1 at 260 nm.
Continuous variation binding analysis
The stoichiometry for the binding of chartreusin to DNA was obtained using the method of continuous variation (18). The concentrations of chartreusin and DNA were each varied, while the sum of the concentrations was kept constant at 30 mM. Varying volumes of equally concentrated stock solutions of chartreusin and DNA were mixed to give a mole fraction of ligand ranging from 0 to 0.6. The difference in absorbance (ΔA420 nm) was plotted against the mole fraction of drug.
Isothermal titration calorimetry
Experiments were carried out at different temperatures (between 20 and 35°C) using a Microcal MCS-ITC calorimeter (Microcal Inc., Northampton, MA). The Origin software (Microcal) was used for data acquisition and analysis. In a typical experiment, 1.34 ml of a 0.75 mM (bp) DNA in 10 mM cacodylate buffer (pH 7.0) containing 8 mM NaCl and 0.1 mM EDTA ([Na+] = 18 mM) was titrated using a 50 µM chartreusin solution in the same buffer (15–25 injections of 10 µl each), using a 250 µl syringe rotating at 400 r.p.m. The injection time was 25 s and the delay between injections was 2.5 min. The peaks produced during titration were converted to heat output per injection by integration and correction for the cell volume and sample concentration.
Determination of the binding enthalpy by isothermal titration calorimetry
Binding enthalpies for chartreusin were determined from the heats of reaction obtained after each injection using a ‘model-free ITC’ protocol (19,20) to obtain multiple estimates of ΔH0 without any fitting bias. This protocol, which uses a high DNA concentration, ensures that all the titrated drug is effectively bound after each addition. The dilution heats, determined by injecting drug solution into the reaction cell loaded with buffer alone, were subtracted from the ΔH value determined for titration into DNA to render a corrected value for the binding-induced enthalpy change ΔHb (19).
Heat capacity measurements
Data obtained from ITC experiments were used to measure ΔCp, the change in heat capacity. ΔCp was measured as the temperature dependence of the binding enthalpy according to the relationship:
ΔCp = δ(ΔHb)/δT1
DNA UV melting studies
Profiles of absorbance at 260 nm versus temperature were measured between 25 and 98°C, at a heating rate of 0.5°C/min, in the presence of different concentrations of NaCl in 10 mM cacodylate buffer (pH 7.0) and saturating concentrations of chartreusin, using a Shimadzu UV-2101PC spectrophotometer equipped with a Shimadzu SPR-8 temperature controller.
Differential scanning calorimetry
Differential scanning calorimetry (DSC) measurements were performed using a Microcal MC-2 Scanning Calorimeter (Microcal Inc.). About 2 ml of a 0.56 mM DNA solution in 10 mM cacodylate buffer (pH 7.0) containing 8 mM NaCl and 0.1 mM EDTA ([Na+] = 18 mM) were used for DSC experiments. A heating rate of 0.5°C/min was used. DSC scans were corrected by subtraction of a buffer–buffer baseline, normalized to the concentration of DNA (in base pairs) and further baseline corrected. Data acquisition and analysis were performed with the graphics software Origin (Microcal Inc.).
Determination of binding constant by DNA UV melting analysis
The DNA melting temperatures (Tm) determined by UV melting in the presence of saturating amounts of chartreusin and in its absence (also determined by UV melting) were used to calculate the binding constant to salmon DNA at the DNA melting temperature (KTm), using the equation derived by Crothers (21):
1/Tm0 – 1/Tm = (R/nΔHwc) ln(1 + KTma)2
where Tm0 is the UV melting temperature of salmon testes DNA alone, Tm is the melting temperature in the presence of saturating amounts of the drug, ΔHwc is the enthalpy of DNA melting obtained by DSC, R is the gas constant, KTm is the drug binding constant at Tm, a is the free drug activity, which is estimated by one half of the total chartreusin concentration, and n is the size of the drug-binding site, determined by continuous variation analysis.
The calculated apparent binding constant at Tm can be extrapolated to a reference temperature using the standard relationship:
δ[ln(Kobs)]/δ(1/T) = –(ΔHb/R)3
where Kobs is the DNA binding constant of the drug at the reference temperature T (Kelvin) and ΔHb the binding enthalpy, which we determined directly by ITC at the reference temperature.
The experimental Kobs, calculated at 20°C, and the binding enthalpy for chartreusin binding to DNA permitted the obtention of complete thermodynamic profiles. Free energies were obtained from the standard relation ΔG0 = –RT lnKobs and the entropy evaluated using the standard thermodynamic relationship ΔS0 = (ΔG0 – ΔH0)/T.
Analysis of the salt dependence of the binding constant
The effects of different NaCl concentrations upon the binding constant of chartreusin was calculated from the spectrophotometric UV melting profiles, as described above, and used to determine the ionic strength dependence of the equilibrium binding constants, at 20°C, according to the polyelectrolyte theory of Record et al. (22). The observed linear dependence is described by the relationship:
δ(log K)/δlog [Na+]) = –Zψ 4
where ψ is the fraction of sodium counterion associated per DNA phosphate (ψ = 0.88 for B-DNA) and Z the apparent charge of the drug.
Moreover, the Zψ value was used to evaluate the polyelectrolyte contribution to the free energy of binding to salmon testes DNA using the relationship (22):
ΔGpe = –ZψRT ln[Na+]5
and the ΔGpe obtained (Table 1) was used to calculate ΔGt, the ‘non-polyelectrolyte contribution to binding’ (23), using the following equation:
Table 1. Summary of measured binding enthalpy values for chartreusin at different temperatures and calculated heat capacity change (ΔCp).
| ΔHb(kcal/mol)a | ΔCp (cal/mol K)b | ||||
|---|---|---|---|---|---|
| 20°C | 25°C | 30°C | 35°C | ||
| Chartreusin | –7.1 ± 0.2 | –9.8 ± 0.7 | –11.4 ± 0.5 | –13.0 ± 0.3 | –391 ± 37 |
aValues are the mean ± SD of three independent titrations at each temperature, with 15–25 chartreusin injections per experiment.
bHeat capacity change calculated from the slope δ(ΔH)/δT obtained by linear least squares fit (r = 0.991).
ΔGobs = ΔGt + ΔGpe6
RESULTS AND DISCUSSION
The method of continuous variation was used to obtain the stoichiometry of the chartreusin–DNA interaction at 25 ± 1°C. The intersection of least squares fitted lines at X = 0.224 (Fig. 1B) corresponds to a site size of 3.1, about one chartreusin molecule per 3 bp DNA.
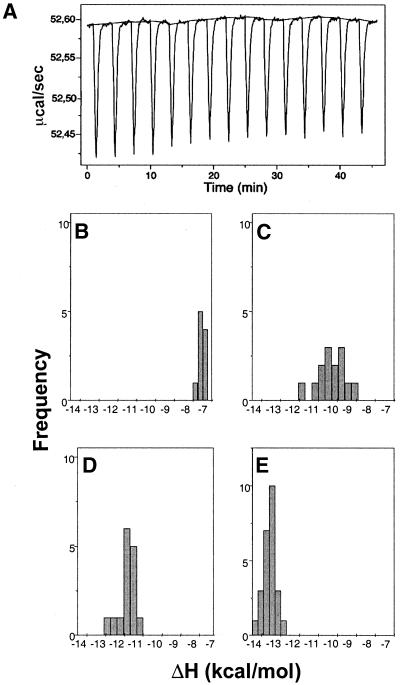
Figure 2A shows representative primary data from the calorimetric (ITC) titration of chartreusin into salmon testes DNA at 30°C. Figure 2B and C displays the distribution of binding enthalpy estimates obtained at four temperatures in the 20–35°C range. These distributions, which are essentially Gaussian, represent experimental error in the measurements rather than any site heterogeneity. Enthalpies were obtained from three independent titrations at each temperature and the different values averaged (Table 1).
Figure 2.
(A) Representative primary data from an isothermal titration calorimetry experiment, at 30°C, used to obtain multiple estimates of ΔH, in a titration of chartreusin on DNA, using a ‘model-free ITC’ protocol (20) in which a high DNA concentration ensures that all the added ligand is effectively bound. Panels (B)–(E) show the distribution of the DNA-binding enthalpy values for chartreusin, obtained as a function of temperature, at (B) 20°C, (C) 25°C, (D) 30°C and (E) 35°C. The distributions were calculated from three independent ITC analyses at the different temperatures, with 15–25 drug injections in each ITC experiment.
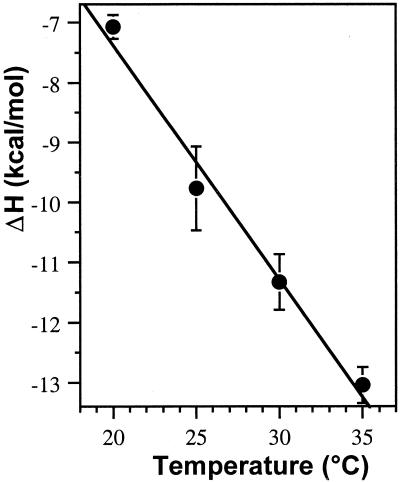
The temperature-dependent differences in the binding enthalpy of chartreusin were used to calculate changes in heat capacity (ΔCp) measured as the slope of a plot of ΔH versus temperature (Fig. 3 and Table 1). The observed ΔCp (–391 ± 37 cal/mol K) is clearly higher than that for other intercalators such as ethidium bromide (ΔCp = –139 cal/mol K) or the charged multivalent intercalator daunorubicin (ΔCp = –160 cal/mol K) (19), but it is close to the ΔCp of –375 cal/mol K determined for actinomycin D (19), which is also an uncharged multivalent intercalator (24).
Figure 3.
Temperature dependence of the binding enthalpy of chartreusin– DNA interactions. Data are means ± SD of three independent isothermal calorimetric titrations. The slope of the least squares fit of the data renders the heat capacity change, ΔCp, for chartreusin. See Table 1 for further details.
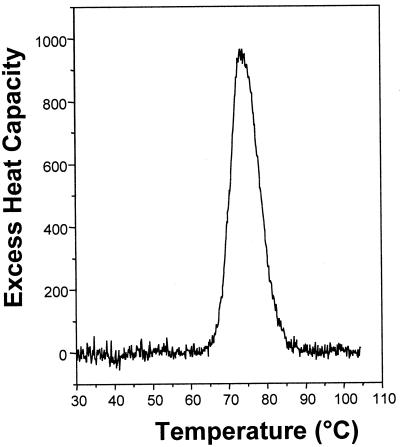
The Tm of DNA was 74.4°C (in 18 mM total Na+), while in the presence of saturating concentrations of chartreusin the melting temperature rose to 78.0°C. The enthalpy of the DNA melting, determined by differential scanning calorimetry, was ΔHwc = 8.8 ± 0.2 kcal/mol bp (Fig. 4). DNA UV melting analyses and the ΔHb measured by ITC were used to calculate the binding constant for the chartreusin–DNA interaction at 20°C in the presence of 18 mM final Na+ by application of equations 1 and 2 (Table 2). Using this method, we calculated Kobs without assuming that ΔHb was constant with temperature, as is usually adopted when the constant is derived from DSC studies only. Our ITC results demonstrate that this is not the case for the chartreusin binding to DNA (Fig. 3). Moreover, the relatively high concentration of DNA we used in the ‘model-free ITC’ protocol (20) ensured that all the added drug was bound to the nucleic acid, thereby there was no interference in the determination of ΔHb due to drug self-association (19,20).
Figure 4.
Differential scanning calorimetry curve of salmon testes DNA [excess heat capacity (cal/mol °C) plotted as a function of temperature]. Tm, 74.4 ± 0.1°C; ΔHwc, 8.8 ± 0.2 kcal/mol. The estimated error is the standard deviation of the mean (three experiments).
Table 2. UV melting temperatures of DNA and the DNA–chartreusin complex under different salt conditions showing the salt dependence of the binding constants at the melting temperature (KTm) and at 20°C (Kobs) for chartreusin binding to DNAa.
| [Na+] mM | Tm0 (°C) | Tm (°C) | KTm (M–1) | Kobs (M–1) |
|---|---|---|---|---|
| 8 | 69.0 | 72.8 | 5.4 × 104 | 4.0 × 105 |
| 18 | 74.4 | 78.0 | 4.9 × 104 | 3.6 × 105 |
| 30 | 80.0 | 83.5 | 4.5 × 104 | 3.3 × 105 |
| 60 | 85.8 | 89.0 | 4.2 × 104 | 3.1 × 105 |
| 120 | 92.6 | 96.0 | 3.8 × 104 | 2.8 × 105 |
KTm, the drug binding constant at Tm, and Kobs, the drug binding constant determined at 20°C, were calculated using equations 2 and 3 as described in the main text. For application of equation 3, the chartreusin binding enthalpy, ΔHb at 20°C (Table 1), was used.
aTm0 (°C) is the melting temperature of 20 µM DNA in the absence of drug. Tm (°C) is the melting temperature of 20 µM DNA in the presence of 10 µM chartreusin (saturating conditions).
The experimental calculation of the binding constant and the binding enthalpy for chartreusin allowed us to obtain complete thermodynamic profiles evaluated at 20°C (Table 3). The favorable free energy of chartreusin binding to DNA (ΔG = –7.4 kcal/mol at 20°C) is derived from a large negative enthalpic contribution (ΔHb = –7.1 kcal/mol).
Table 3. Comparison of thermodynamic parameters for chartreusin binding to salmon testes DNAa.
| Kobs (M–1) | ΔGobs (kcal/mol) | –SK | ΔGpe (kcal/mol) | ΔGt (kcal/mol) | ΔHb (kcal/mol) | TΔS (cal/mol) | |
|---|---|---|---|---|---|---|---|
| Chartreusin | 3.6 × 105 | –7.4 | 0.13 | –0.3 | –7.1 | –7.1 (± 0.2) | 0.4 |
aKobs (M–1) is the binding constant for the interaction of the ligand with salmon testes DNA in 18 mM Na+ at 20°C. ΔGobs is the binding free energy calculated from ΔGobs = –RT lnK. SK is the slope of the plot of logKobs versus log[Na+] shown in Figure 5A. ΔGpe and ΔGt are the polyelectrolyte and non-polyelectrolyte contributions to the binding free energy (ΔGpe = SK RT ln[Na+]) evaluated at 18 mM Na+. ΔHb was determined by ITC at 20°C (see Table 1).
Previous attempts to determine DNA binding constants for chartreusin did not render reliable quantitative results (11), reflecting the difficulties associated with the experimental measurements (11,25). The uncertainties in the quantitative evaluation of the chartreusin binding constant could be attributable to problems in fitting the experimental spectroscopic data to mathematical models used in binding analysis (11). In our study, we have obtained a more direct estimate of the binding constant, without recourse to model-dependent curve fitting.
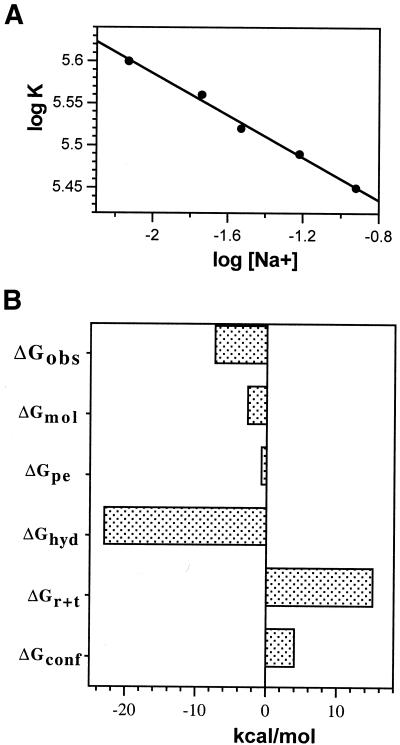
The DNA binding constant of chartreusin was also determined at different salt concentrations using the UV melting temperatures of drug–DNA complexes (Table 2). The binding constant for chartreusin changed only slightly with the different salt concentrations. Figure 5A shows the dependence of the binding constant on the salt concentration as determined by UV melting studies. These results indicated that chartreusin binding to DNA was accompanied by a rather low counterion release. The salt-dependent changes in binding constants were used, according to the polyelectrolyte theory (22), to calculate the ‘actual’ charge of a ligand and the salt dependence of the binding constant using equations 5 and 6. The slope of the double logarithmic Record’s plot in Figure 5A (SK values in Table 3) is equivalent to the number of counterions released upon drug binding. We found that only 0.13 counterions were released from DNA upon drug binding, which corresponds to a calculated apparent change (Z = 0.15) consistent with the absence of any net charge in chartreusin (Fig. 1A).
Figure 5.
(A) Salt dependence of chartreusin binding constants at 20°C. Data are presented according to Record’s theory (22). The linear least squares fit to the data yielded a slope of –0.13. From this value, a negligible positive charge of chartreusin is obtained (Z = 0.15), which is not at variance with the expected uncharged molecule at neutral pH. (B) A tentative parsing of the free energy (ΔGobs) of chartreusin binding to DNA at 20°C. The ΔGhyd presented in the figure corresponds to the lower estimate from the equation ΔGhyd = 80 (± 10) ΔCp (30). Details on all the estimated contributions to the free energy are discussed in the main text.
From the dependence of the binding constant on salt concentration, the observed binding free energy was partitioned in the non-polyelectrolyte (ΔGt) and polyelectrolyte (ΔGpe) components using equation 6 (Table 3). A ΔGt of –7.1 kcal/mol indicates that the non-electrolyte component of binding is fundamental in the stabilization of the chartreusin complex with DNA. As a comparison, we used equation 5 to calculate ΔGpe at 200 mM Na+ (ΔGpe = –0.12 kcal/mol). The polyelectrolyte contribution for chartreusin binding is close to the values determined, at this ionic strength, for the intercalation of the uncharged actinomycin or hydroxyrubicin molecules (23,24). Consequently, the slight salt dependence of chartreusin binding to DNA should arise from the ion release due to increased phosphate spacing that results from intercalation (24).
The binding reaction is driven by the enthalpy contribution, with a rather small entropy contribution (Table 3). The unfavorable entropic cost of DNA–drug complex formation seems to be partially compensated for by the release of counterions, as wells as the presence of the uncharged disaccharide moiety in the minor groove of DNA. The chartreusin disaccharide occupies the minor groove of the helix (17) providing some discrimination for the nucleotide on the 5′-side of the intercalation site (15). It may displace water molecules, providing a positive entropic term that is likely remedied by the negative entropy from the decreased flexibility of DNA. ΔHb became more negative as the temperature increased (Table 1), which was accompanied by changes in ΔS and an associated enthalpy–entropy compensation (26), thus rendering small changes in ΔG and Kobs (Tables 2 and 3). The peculiar disaccharide of chartreusin is not only important for the total strength of binding. Changes in the sugar of some useful antitumor drugs are considered to lead to alterations in their biological activity through mechanisms that are not always related to DNA binding (8,27).
In a previous paragraph, we analyzed the polyelectrolyte contributions to the binding free energies (ΔGpe). At least four other components are considered to parse (dissect) the drug binding energies of the binding free energy into contributions from the processes that lead to complex formation (2,3). Hence, the observed binding free energy (ΔGobs) would contain contributions from five terms:
ΔGobs = ΔGconf + ΔGr + t + ΔGpe + ΔGhyd + ΔGmol7
where ΔGconf is the free energy contribution from the conformational changes in DNA and drug upon complex formation, ΔGr + t is the free energy cost resulting from losses in rotational and translational degrees of freedom upon complex formation, ΔGhyd is the free energy for the hydrophobic transfer of a drug from solution into the DNA binding site and ΔGmol is the free energy contribution from DNA–drug interactions. We shall discuss the parsing of the free energy (ΔGobs) in the context of the structural characteristics of chartreusin and the binding to DNA, to help in the dissection of the free energy.
The formation of an intercalation complex would require the unstacking of adjacent bases and lengthening of the DNA helix. In general, the intercalation of a drug in DNA would involve an unfavorable ΔGconf term (Fig. 5B). The unwinding of the double helix after intercalation of elsamicin A, which bears the same chartarin chromophore as chartreusin, has been measured experimentally (28). However, the effect of binding the disaccharide moiety of chartreusin in the minor groove is less obvious. A negligible ΔGconf (ΔGconf = 0) has been contemplated in Hoechst 33258 binding into the minor groove of DNA (29). The values for ΔGconf and ΔGr + t used in Figure 5B may be considered extreme (19,20). The magnitude used for ΔGr + t in equation 7 was taken from those in the bibliography about intercalating reactions (3,5).
The ΔHb estimated by ITC at different temperatures (Fig. 3 and Table 1) were used to experimentally determine ΔCp for chartreusin, which can be used to determine ΔGhyd using the relationship ΔGhyd = 80 ΔCp, with an ∼25% error, providing insights into the hydrophobic contribution to binding (30). The experimental measurement of the heat capacity change for chartreusin shows that the free energy contribution for the hydrophobic transfer process is large and favorable (negative), a consequence of the several hydration contributions involved in the formation of an intercalative complex. Molecular dynamics calculations have been used to study specific drug–DNA interactions and the role of the solvent water molecules in the drug–DNA complex of the chartarin-containing molecule elsamicin A (17). These investigations showed that the chromophore shared by elsamicin A and chartreusin is largely buried inside the DNA intercalation site and the solvent-accessible surface is rather small (17). Accessibility to solvent has been correlated with the magnitude of ΔCp and the hydrophobic component of binding (19). From a quantitative point of view, the largely buried chartarin chromophore should render high ΔCp values, as experimentally observed (Table 1). Since the computed calculations were on the intercalated chromophore, we cannot use them to analyze the participation of the disaccharide moiety in the hydrophobic component of chartreusin binding to DNA.
In general, it is considered that the somewhat large contribution of the hydrophobic transfer (ΔGhyd) might suffice to overwhelm other unfavorable contributions from changes in conformation and the entropic cost of forming a bimolecular complex (3,5), thus achieving the ΔGobs and Kobs observed. We found that considering ΔGhyd to be about –23 kcal/mol [which is the lower limit of the estimate derived from the relation ΔGhyd = 80 (± 10) ΔCp] was committing to obtaining a negative value for ΔGmol, thus accounting for the different non-covalent interactions that are formed in a drug–DNA complex (Fig. 5B). Using a ΔGhyd of –31 kcal/mol would generate the cumbersome participation of positive (unfavorable) interactions in a multivalent intercalation. In the parsing of free energy (Fig. 5B) ΔGmol is assigned following an approach in which the sum of the other contributions discussed above is considered. The resulting value is subtracted from the experimental ΔGobs (19). Therefore, ΔGmol is the contribution of all molecular interactions not considered in the ΔGhyd and ΔGpe terms, such as, for example, van der Waals interactions.
Although a parsing exercise has recognized limitations (19,20), it was worth performing because chartreusin is an example of a multivalent molecule consisting of an intercalating chromophore bound to a disaccharide moiety. Therefore, the data obtained may be useful in the rational design of polyintercalating antitumor drugs containing a saccharide or other moieties lying in the minor groove. Sugar moieties have been shown to be an essential component of some antibiotics for their topoisomerase poisoning activity and antitumor efficacy (27) and might play a key role in the high activity of chartreusin against topoisomerase II (16). It would be interesting to understand whether an uncharged carbohydrate moiety in multivalent intercalators is compatible with strong binding to DNA, as observed here, while the presence, or absence, of protonatable amino groups in the carbohydrate moiety could also participate in the ability of a polyfunctional drug to overcome its processing by multidrug resistance systems (31).
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants PB98-0469 and BMC-2000-0898 from DGESIC (Spain), and the support of the Centre de Referencia en Biotecnologia of the Generalitat de Catalunya.
REFERENCES
- 1.Marky L.A., Snyder,J.G., Remeta,D.P. and Breslauer,K.J. (1983) Thermodynamics of drug-DNA interactions. J. Biomol. Struct. Dyn., 1, 487–507. [DOI] [PubMed] [Google Scholar]
- 2.Chaires J.B., Satyanarayana,S., Suh,D., Fokt,I., Przewloka,T. and Priebe,W. (1996) Parsing the free energy of anthracycline antibiotic binding to DNA. Biochemistry, 35, 2047–2053. [DOI] [PubMed] [Google Scholar]
- 3.Chaires J.B. (1997) Energetics of drug-DNA interactions. Biopolymers, 44, 201–215. [DOI] [PubMed] [Google Scholar]
- 4.Lane A.N. and Jenkins,T.C. (2000) Thermodynamics of nucleic acids and their interactions with ligands. Q. Rev. Biophys., 33, 255–306. [DOI] [PubMed] [Google Scholar]
- 5.Haq I. (2002) Thermodynamics of drug–DNA interactions. Arch. Biochem. Biophys., 403, 1–15. [DOI] [PubMed] [Google Scholar]
- 6.Gottesfeld J.M., Turner,J.M. and Dervan,P.B. (2000) Chemical approaches to control gene expression. Gene Expr., 9, 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Priebe W., Fokt,I., Przewloka,T., Chaires,J.B., Portugal,J. and Trent,J.O. (2001) Exploiting anthracycline scaffold for designing DNA-targeting agents. Methods Enzymol., 340, 529–555. [DOI] [PubMed] [Google Scholar]
- 8.Capranico G., Supino,R., Binaschi,M., Capolongo,L., Grandi,M., Suarato,A. and Zunino,F. (1994) Influence of structural modifications at the 3′ and 4′ positions of doxorubicin on the drug ability to trap topoisomerase II and to overcome multidrug resistance. Mol. Pharmacol., 45, 908–915. [PubMed] [Google Scholar]
- 9.Mansilla S. and Portugal,J. (2002) Occurrence of DNA sequences specifically recognized by drugs in human promoters. J. Biomol. Struct. Dyn., 19, 669–679. [DOI] [PubMed] [Google Scholar]
- 10.McGovren J.P., Neil,G.L., Crampton,S.L., Robinson,M.I. and Douros,J.D. (1977) Antitumor activity and preliminary drug disposition studies on chartreusin (NSC 5159). Cancer Res., 37, 1666–1672. [PubMed] [Google Scholar]
- 11.Krueger W.C., Pschigoda,L.M. and Moscowitz,A. (1986) The binding of the antitumor antibiotic chartreusin to poly(dA-dT).poly(dA-dT), poly(dG-dC).poly(dG-dC), calf thymus DNA, transfer RNA and ribosomal RNA. J. Antibiot., 39, 1298–1303. [DOI] [PubMed] [Google Scholar]
- 12.Konishi J., Sugawara,K., Kofu,F., Nishiyama,Y., Tomita,K., Miyaki,T. and Kawagushi,H. (1986) Elsamicins, new antitumor antibiotics related to chartreusin. J. Antibiot., 39, 784–791. [DOI] [PubMed] [Google Scholar]
- 13.Uchida H., Nakakita,Y., Enoki,N., Abe,N., Nakamura,T. and Munekata,M. (1994) Chrymutasins: novel-aglycone antitumor antibiotics from a mutant of Streptomyces chartreusis. II. Characterization and structural elucidation. J. Antibiot., 47, 655–667. [DOI] [PubMed] [Google Scholar]
- 14.Uramoto M., Kusano,T., Nishio,K., Isono,K., Shishido,K. and Ando,T. (1983) Specific binding of chartreusin, an antitumor antibiotic, to DNA. FEBS Lett., 153, 325–328. [DOI] [PubMed] [Google Scholar]
- 15.Salas X. and Portugal,J. (1991) Map of chartreusin and elsamicin binding sites on DNA. FEBS Lett., 292, 223–228. [DOI] [PubMed] [Google Scholar]
- 16.Lorico A. and Long,B.H. (1993) Biochemical characterisation of elsamicin and other coumarin-related antitumour agents as potent inhibitors of human topoisomerase II. Eur. J. Cancer, 29, 1985–1991. [DOI] [PubMed] [Google Scholar]
- 17.Alhambra C., Luque,F.J., Portugal,J. and Orozco,M. (1995) Molecular dynamics study of the binding of elsamicin A to DNA. Eur. J. Biochem., 230, 555–566. [DOI] [PubMed] [Google Scholar]
- 18.Jenkins T.C. (1997) Optical absorbance and fluorescence techniques for measuring DNA-drug interactions. Methods Mol. Biol., 90, 195–218. [DOI] [PubMed] [Google Scholar]
- 19.Ren J., Jenkins,T.C. and Chaires,J.B. (2000) Energetics of DNA intercalation reactions. Biochemistry, 39, 8439–8447. [DOI] [PubMed] [Google Scholar]
- 20.Haq I., Jenkins,T.C., Chowdhry,B.Z., Ren,J. and Chaires,J.B. (2000) Parsing free energies of drug-DNA interactions. Methods Enzymol., 323, 373–405. [DOI] [PubMed] [Google Scholar]
- 21.Crothers D.M. (1971) Statistical thermodynamics of nucleic acid melting transitions with coupled binding equilibria. Biopolymers, 10, 2147–2160. [DOI] [PubMed] [Google Scholar]
- 22.Record M.T., Anderson,C.F. and Lohman,T.M. (1978) Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening and ion effects on water activity. Q. Rev. Biophys., 11, 103–178. [DOI] [PubMed] [Google Scholar]
- 23.Chaires J.B. (1996) Dissecting the free energy of drug binding to DNA. Anticancer Drug Des., 11, 569–580. [PubMed] [Google Scholar]
- 24.Wilson W.D. and Tanious,F.A. (1994) Kinetic analysis of drug-nucleic acid binding modes: absolute rates and effects of salt concentration. In Neidle,S. and Waring,M. (eds), Molecular Aspects of Anticancer Drug–DNA Interactions. MacMillan, London, Vol. 2, pp. 243–269.
- 25.Jiménez-García E. and Portugal,J. (1992) Elsamicin A can convert the Z-form of poly[d(G-C)] and poly[(G-m5C)] back to B-form DNA. Biochemistry, 31, 11641–11646. [DOI] [PubMed] [Google Scholar]
- 26.Breslauer K.J., Remeta,D.P., Chou,W.Y., Ferrante,R., Curry,J., Zaunczkowski,D., Snyder,J.G. and Marky,L.A. (1987) Enthalpy-entropy compensations in drug-DNA binding studies. Proc. Natl Acad. Sci. USA, 84, 8922–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zunino F., Pratesi,G. and Perego,P. (2001) Role of the sugar moiety in the pharmacologicalactivity of anthracyclines: development of a novel series of disaccharide analogs. Biochem. Pharmacol., 61, 933–938. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez-Campos A., Azorín,F. and Portugal,J. (1996) Influence of elsamicin A on the activity of mammalian topoisomerase I. Biochemistry, 35, 11177–11182. [DOI] [PubMed] [Google Scholar]
- 29.Haq I., Ladbury,J.E., Chowdhry,B.Z., Jenkins,T.C. and Chaires,J.B. (1997) Specific binding of Hoechst 33258 to the d(CGCAAATTTGCG)2 duplex: calorimetric and spectroscopic studies. J. Mol. Biol., 271, 244–257. [DOI] [PubMed] [Google Scholar]
- 30.Record M.T. Jr, Ha,J.H. and Fisher,M.A. (1991) Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol., 208, 291–343. [DOI] [PubMed] [Google Scholar]
- 31.Frezard F., Pereira-Maia,E., Quidu,P., Priebe,W. and Garnier-Suillerot,A. (2001) P-glycoprotein preferentially effluxes anthracyclines containing free basic versus charged amine. Eur. J. Biochem., 268, 1561–1567. [PubMed] [Google Scholar]