Abstract
The NMR structure of the 3′ stem–loop (3′SL) from human U4 snRNA was determined to gain insight into the structural basis for conservation of this stem–loop sequence from vertebrates. 3′SL sequences from human, rat, mouse and chicken U4 snRNA each consist of a 7 bp stem capped by a UACG tetraloop. No high resolution structure has previously been reported for a UACG tetraloop. The UACG tetraloop portion of the 3′SL was especially well defined by the NMR data, with a total of 92 NOE-derived restraints (about 15 per residue), including 48 inter-residue restraints (about 8 per residue) for the tetraloop and closing C-G base pair. Distance restraints were derived from NOESY spectra using MARDIGRAS with random error analysis. Refinement of the 20mer RNA hairpin structure was carried out using the programs DYANA and miniCarlo. In the UACG tetraloop, U and G formed a base pair stabilized by two hydrogen bonds, one between the 2′-hydroxyl proton of U and carbonyl oxygen of G, another between the imino proton of G and carbonyl oxygen O2 of U. In addition, the amino group of C formed a hydrogen bond with the phosphate oxygen of A. G adopted a syn orientation about the glycosidic bond, while the sugar puckers of A and C were either C2′-endo or flexible. The conformation of the UACG tetraloop was, overall, similar to that previously reported for UUCG tetraloops, another member of the UNCG class of tetraloops. The presence of an A, rather than a U, at the variable position, however, presents a distinct surface for interaction of the 3′SL tetraloop with either RNA or protein residues that may stabilize interactions important for active spliceosome formation. Such tertiary interactions may explain the conservation of the UACG tetraloop motif in 3′SL sequences from U4 snRNA in vertebrates.
INTRODUCTION
Pre-mRNA splicing is the essential process in eukaryotic cells that removes introns and ligates exons from transcribed genes (1). The process involves two transesterification reactions that are catalyzed by spliceosomes. Spliceosomes are macromolecular complexes constructed around splice junction sites of pre-mRNA from three RNA–protein particles, the U1 and U2 snRNPs and the U4/U6·U5 tri-snRNP (2). U1 and U2 snRNPs each contain a single snRNA while the U4/U6·U5 tri-snRNP consists of three discrete snRNAs, each with several proteins that bind the individual RNAs specifically (3). Assembly of the spliceosome occurs in the nucleus in a series of discrete steps (4). U1 snRNP initially binds the pre-mRNA at the 5′ splice site to form a commitment complex. U2 snRNP then binds at the 3′ splice site preceding formation of the lariat intermediate. Finally, the U4/U6·U5 tri-snRNP binds and a major structural rearrangement occurs, in which U4 snRNP is extruded, and the final steps of intron removal and exon ligation proceed (5).
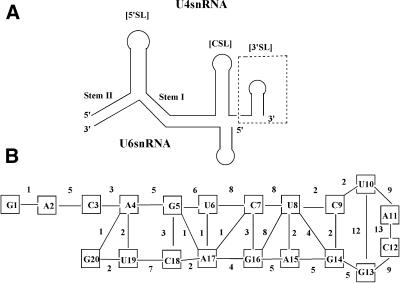
Although pre-mRNA splicing occurs in the nucleus, each of the snRNA components of the spliceosome, with the exception of U6 snRNA, are exported to the cytoplasm following transcription (6). In order to re-enter the nucleus, the 7mG cap of these snRNAs must be hypermethylated and the Sm (or common) proteins must form a complex that encircles the single-stranded Sm-binding site (7). The Sm-binding site in higher eukaryotes is flanked by two stem–loops, the central stem–loop (CSL) and the 3′ stem–loop (3′SL) (Fig. 1). The structural features of the Sm-binding site region that are important for Sm protein binding and assembly are the adenosines at the 5′-end of the single-stranded Sm-binding site and portions of the CSL (8,9). The function of the 3′SL from human U4 snRNA is not clear. The 3′SL does not occur in U4 snRNA from Saccharomyces cerevisae (10) and it may inhibit degradation of U4 snRNA from humans and other metazoans.
Figure 1.
(A) Depiction of the secondary structure of the U4/U6 snRNA complex. Interaction between the two snRNAs is stabilized by formation of two stem regions (I and II). The secondary structre of U4 snRNA in the U4/U6 snRNA complex consists of three stem–loops, the 5′SL, the [CSL] and the 3′SL. The NMR structure of the 3′SL (indicated in the dashed box) was determined in the present study. (B) Primary sequence and secondary structure of the 3′SL from human U4 snRNA. The numbers between the nucleotides indicate the number of inter-residue restraints used to determine the NMR structure.
Spliceosome assembly is a complex process involving numerous specific RNA–RNA, RNA–protein and protein– protein interactions (11). Information on snRNP structure and dynamics is essential in order to fully elucidate the mechanism of this intricate process. The secondary structures for the RNA components of the snRNPs have been known for many years (12) and, more recently, X-ray crystallography (13) and cryoelectron microscopy (14) have provided detailed structural information concerning RNA–protein and protein– protein complexes important for UsnRNP biogenesis and spliceosome assembly. Interestingly, the predicted secondary structures for 3′SL from U4 snRNA from human, rat, mouse and chicken are conserved, with each consisting of a 7 bp stem capped by a UACG tetraloop (15–18). Thus, the structure and flexibility of the UACG tetraloop in the context of the 3′SL is likely important in mediating RNA–RNA and/or RNA– protein interactions that are important for spliceosomal assembly and function in these organisms (19).
The UACG tetraloop is one of four possible UNCG tetraloop sequences, where N is U, C, G or A. RNA tetraloops fall into three main classes, with GNRA (R = A or G) and CUUG being the two main classes in addition to UNCG. Among UNCG tetraloops, the UUCG tetraloop has been studied the most extensively, with both NMR (20,21) and X-ray crystallography (22) studies previously reported. Previous spectroscopic studies of UACG tetraloops in RNA hairpins revealed similarities to UUCG tetraloops (23), but no high resolution structures of any UACG tetraloop are currently available. In this manuscript, we present the NMR structure of the 3′SL from human U4 snRNA. Overall, the structure is similar to those previously reported for the UUCG tetraloop, another member of the UNCG class of tetraloops (20–25). The presence of an A, rather than a U, at the variable position, however, presents a distinct surface for interaction of the 3′SL tetraloop with either RNA or protein residues that may stabilize interactions important for active spliceosome formation. Such tertiary interactions may explain the conservation of the UACG tetraloop motif in 3′SL sequences from U4 snRNA in vertebrates.
MATERIALS AND METHODS
RNA sample preparation
The 3′ stem–loop from human U4 snRNA (3′SL) was synthesized in vitro from a synthetic DNA template using T7 RNA polymerase (26). Samples of the 3′SL for NMR spectroscopy and UV hyperchromicity studies were purified from 20% denaturing polyacrylamide gels by electroelution, followed by desalting and dialysis. The samples were first dialyzed into 10 mM sodium phosphate buffer, pH 6.4, with 0.1 mM EDTA. Transfer of RNA samples into buffer containing higher sodium chloride concentrations was accomplished by dialysis. Preparation of samples containing magnesium chloride was also accomplished by dialysis, however, EDTA was not included in the dialysis buffer. All samples of the 3′SL were annealed by heating to 90°C, followed by cooling in ice-water. Sample concentrations were determined by UV absorbance at 260 nm using the extinction coefficient calculated for 3′SL using the Schepartz Laboratory Biopolymer Calculator (Yale University). All NMR samples were prepared with RNA concentrations between 1 and 2 mM. In all cases, unless otherwise specified, the RNA samples were dialyzed against 10 mM sodium phosphate, 50 mM sodium chloride, pH 6.4, with 0.1 mM EDTA.
UV melting studies
Optical absorbance melting studies were performed using a Cary 3E UV-Visible spectrophotometer with heating rates from 0.25 to 1°C/min. Typical RNA concentrations were 8 µM. Studies of the sodium and magnesium dependence of the melting temperature of the stem–loop were done using sodium chloride concentrations ranging from 0 to 200 mM and magnesium chloride concentrations ranging from 0 to 5 mM. To address the possibility of linear duplex formation in solution, melting temperatures were also determined at RNA concentrations of 2, 6 and 50 µM in sodium phosphate buffer and in buffer with 50 mM sodium chloride.
NMR spectroscopy
All NMR experiments were acquired using a Varian INOVA 600 NMR spectrometer at University of California at San Francisco (UCSF). TSP was used as an internal chemical shift reference. NMR data were processed using the NMRPipe suite of programs (27). NMR spectra were baseline corrected using Baseline (UCSF) and spectral annotations, assignments and peak integrations were completed using Sparky (28). Two-dimensional NMR data sets were typically collected using spectral widths of 6000 Hz in each dimension with 2048 complex data points in the directly detected dimension and 512 points in the indirectly collected dimension. NMR data were apodized in each dimension with either a phase-shifted sine bell or Gaussian function, followed by zero filling once in the indirect dimension prior to Fourier transformation. States’ method was used for quadrature detection in the indirectly detected dimension (29). NOESY spectra in D2O solution were recorded at 25, 30 and 35°C with mixing times of 80, 100, 200, 250, 300 and 400 ms. TOCSY and DQF COSY spectra were recorded at 25 and 30°C. The TOCSY experiments were acquired using a 50 ms mixing time with pre-saturation at low power applied during the evolution period. A natural abundance 13C HMQC spectrum was acquired at 30°C with spectral widths of 6000 and 7500 Hz in the 1H and 13C dimensions, respectively. The 13C frequency was centered at 85 p.p.m. A series of one-dimensional 1H NMR experiments were also recorded, which included temperature profiles of 3′SL in H2O and D2O solutions. The jump-and-return pulse sequence (30) was used for one-dimensional NMR experiments in H2O, while low power pre-saturation was used for experiments in D2O solution. One-dimensional NMR experiments were also used to determine T1 relaxation rates and measure linewidths for 1H resonances from samples of 3′SL over the concentration range 200 µM to 2 mM. SSNOESY spectra in H2O solution were recorded with spectral widths of 12 999 Hz in each dimension using the shaped pulse ‘S136g’, which had an excitation maximum at the imino region, for water suppression (31). Spectra were recorded at 10, 15 and 20°C with mixing times of 125, 150 and 250 ms.
Structure calculation
NMR data at 30°C in D2O and at 20°C in water were used for the structure calculation. Distance restraints involving non-exchangeable protons were calculated from integrated NOE intensities from two D2O NOESY data sets (80 and 400 ms) using MARDIGRAS (32), with the random error analysis procedure (33). For the distances involving exchangeable protons, only the upper bounds were used. Structure calculation was carried out with the help of the DYANA (34) and miniCarlo (35) programs, as described previously (36,37). The quality of refinement was assessed with Rx factors that directly compare simulated and experimental NOE intensities, calculated with CORMA (38). The structures were visualized using MidasPlus (39).
RESULTS
3′SL conformation
In order to determine if the RNA sequence corresponding to the 3′SL from human U4 snRNA formed a hairpin (rather than a linear duplex) under conditions suitable for determining the NMR structure, the RNA was synthesized using in vitro transcription and analyzed using gel electrophoresis, UV hyperchromicity and NMR spectroscopy. The sequence of the 3′SL is shown in Figure 1. Samples of the 3′SL were annealed prior to analysis by heating to 90°C followed by cooling in ice-water. The mobility of the RNA prepared by in vitro transcription through native polyacrylamide gels was consistent with formation of a single species having the molecular weight expected for the (unimolecular) hairpin. As described in greater detail in the following sections, thermal melting studies and NMR spectroscopy also verified that the 3′SL sequence adopted a hairpin conformation in solution at all concentrations studied.
Further evidence that samples of the 3′SL adopted a hairpin conformation was obtained from thermal melting studies. Studies of the sodium and magnesium dependence of the melting temperature of the stem–loop were done using sodium chloride concentrations ranging from 0 to 200 mM and magnesium chloride concentrations ranging from 0 to 5 mM. In the sodium chloride studies the buffer included 0.1 mM EDTA. The melting temperature (Tm) of the 3′SL was 74°C in 10 mM sodium phosphate buffer, pH 6.4. The Tm increased to 80°C in the presence of 50 mM sodium chloride and 84°C in 150 mM sodium chloride. Melting temperatures of 82, 85 and 86°C were obtained for magnesium chloride concentrations of 500 µM and 1 and 2 mM, respectively. The number of Mg2+ ions bound per molecule of hairpin was approximately 0.5, indicating the absence of a specific Mg2+-binding site in the 3′SL. In order to address the possibility of linear duplex formation in solution, melting temperatures were also determined at RNA concentrations of 2, 6 and 50 µM, in sodium phosphate buffer and in buffer with 50 mM sodium chloride. The results were within experimental uncertainty of one another, indicating that the Tm was independent of RNA concentration. The results indicated that the 3′SL adopted a hairpin conformation in solution and that Mg2+ was not required to stabilize the hairpin conformation. Although the hairpin was slightly stabilized by addition of sodium chloride above 50 mM concentration, this concentration of salt was sufficient to form a stable hairpin and was used in subsequent NMR studies.
3′SL NMR structure
Two-dimensional NMR studies of the 3′SL were conducted to determine the solution conformation of this RNA stem–loop. NMR spectra acquired on freshly annealed samples of 3′SL were consistent with formation of a single conformation for the hairpin. A second set of resonances was apparent in NMR spectra acquired after storing the sample for a few days at room temperature. These additional resonances were no longer apparent in NMR spectra acquired after the sample was re-annealed by heating to 90°C and cooling in ice-water. In order to determine if the second set of resonances that appeared in NMR spectra of the 3′SL after several days at room temperature were due to formation of a linear duplex structure, T1 relaxation rates and linewidths for 1H NMR resonances from both sets of resonances were compared. The T1 relaxation rates and linewidths for resolved 1H resonances from both sets of peaks were similar and were independent of RNA sample concentration between 200 µM and 2 mM. These results indicate that both sets of resonances were from unimolecular structures. Additional evidence that the two sets of resonances were both the result of unimolecular hairpin conformations was obtained by gel electrophoresis. NMR samples of 3′SL that displayed two sets of resonances displayed only a single band on native electrophoretic gels. The apparent molecular weight was consistent with the unimolecular hairpin conformation. These experiments established that the 3′SL can adopt either of two closely related, but distinct, conformations, depending on the annealing protocol and incubation of the sample. All of the results described here were obtained using a freshly annealed sample displaying a single set of resonances in NMR spectra.
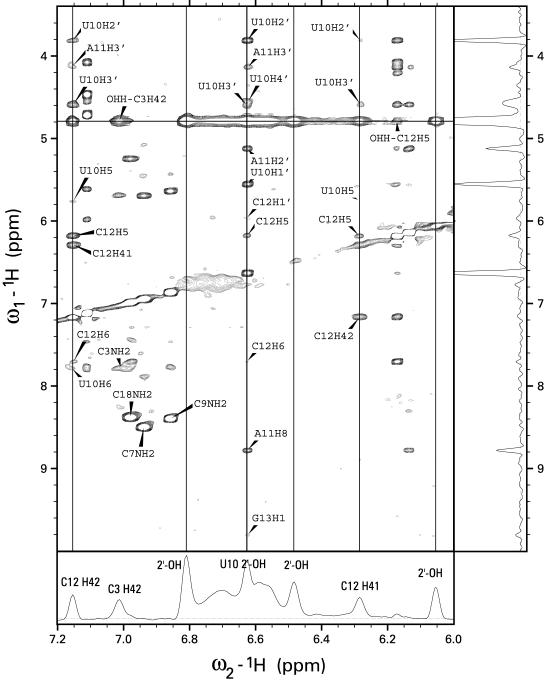
The 1H NMR assignments for 3′SL were completed using homonuclear NOESY and TOCSY experiments in D2O solution and NOESY experiments in H2O solution (Fig. 2), and then verified using a 13C HMQC spectrum. The nucleotides of the UACG tetraloop displayed excellent dispersion of the 1H resonances with a characteristic signature which facilitated nearly complete resonance assignments (23). The nucleotides in the stem region displayed poorer spectral dispersion and 1H resonance assignment for these residues was less complete. The stem region of the stem–loop displayed NMR spectral characteristics typical of an A-form RNA duplex. In particular, nucleotides in the stem region displayed stronger intra-residue than inter-residue H8/H6–H1′ crosspeaks in NOESY spectra acquired with an 80 ms mixing time, as well as characteristic sequential H2′–H1′ and very strong sequential H2′–H8/H6 cross-peaks. In a typical A-form duplex RNA, sequential H2′–H8/H6 distances are <2.5 Å, intra-residue H1′–H8/H6 distances are in the range 3.5–4.0 Å and sequential H1′–H8/H6 distances are ∼1 Å longer. All ribonucleotides in the stem, except C3, displayed 3JH1′–H2′ too small to be detected in DQF COSY experiments, thus intra-residue H1′–H2′correlations were determined from NOESY spectra with short mix times. Additional intra-residue assignments were determined from TOCSY spectra. The exchangeable 1H resonances were assigned from NOESY spectra in H2O solution.
Figure 2.
NOESY spectra of the 3′SL from human U4 snRNA in H2O buffer at 20°C acquired with a 250 ms mixing time. The bottom trace shows a slice through the water frequency with the exchange peaks labeled. The trace on the right shows a slice through the frequency from the 2′-OH of U10. The vertical lines are drawn through the frequencies of C12 H41 and H42 and the frequencies of three hydroxyl protons. Two broad, unlabeled exchange peaks in the bottom panel come from the unassigned amino groups of adenine or guanine. The NOE cross-peaks from U10 2′-OH establish the assignment and establish that the UACG tetraloop in 3′SL is stabilized by a hydrogen bond to O6 of G13. A similar hydrogen bond has been observed previously in the NMR structure of UUCG tetraloops. The pattern of hydrogen bonds that stabilize the UACG tetraloop is shown in Figure 4A.
Structure calculations for the 3′SL were carried out as previously described (36,37). An initial structure was modeled and energy minimized with miniCarlo. This structure was used in random error MARDIGRAS (RANDMARDI) (33) calculations, which were run 50 times for NOESY data sets collected with 80 and 400 ms mixing times with an overall correlation time of 2 ns for the 3′SL. The calculated inter-proton distance restraints were used for the restrained energy minimization calculations using the miniCarlo program. The resulting preliminary structure was used to re-evaluate the NOESY data sets. In particular, stereospecific assignments of H5′ and H5′′ protons were based on this preliminary structure. After that, the MARDIGRAS calculations were repeated once more. A total of 200 constraints (10 per residue) were determined from the NMR spectroscopic data and were used in the refinement protocol (Table 1). Of the 200 distance restraints, 140 occurred between non-exchangeable 1H resonances. Inter-proton distances calculated using MARDIGRAS were used to set the mean value of the distance restraint with the flat-well width of the restraint set to 0.67 Å about the mean value. Fifty-six of the distance restraints involved exchangeable 1H, and an upper limit of 6 Å was used for these restraints. Four of the restraints resulted from hydrogen bonding interactions observed in the NMR spectra. The loop portion of 3′SL, in particular, was very well defined by the NMR restraints (Fig. 1B). For the UACG tetraloop together with the closing pair (CG), 92 experimental restraints (15.3 per residue), including 48 inter-residue restraints (8 per residue) were obtained from the NMR data. This set of distance restraints was used in the final stage of the refinement. During the high temperature stages of the refinement in DYANA and miniCarlo, this set of distance restraints was supplemented by the Watson–Crick hydrogen bonds for the base pairs in the stem. In addition, during the DYANA calculations loose helical torsion angle restraints for the nucleotides in the stem were included. The refinement protocol started with torsion angle dynamics simulated annealing using the DYANA program, which produced 100 roughly folded structures. Fifty structures with the best score were further subjected to a Metropolis Monte Carlo simulated annealing protocol using the miniCarlo program. The 15 best structures were further restrained minimized with miniCarlo. The overlay of the 10 structures of lowest total energy (sum of conformational energy and restraint energy) is shown in Figure 3.
Table 1. Refinement statistics for 3′SLa.
| Number of distance restraints | ||
| Non-exchangeableb | 140 | |
| Exchangeable | 56 | |
| Hydrogen bondsc | 4 | |
| Total | 200 | |
| Per residue | 10.0 | |
| Conformational energy (kcal/mol)d | –161.0 ± 3.8 | |
| Residual distance deviation (Å)e | 0.10 ± 0.01 | |
| NOE-based Rx factor (×102) (80 ms data set) | ||
| Intra-residue | 4.99 ± 0.23 | |
| Inter-residue | 7.57 ± 0.33 | |
| Total | 6.02 ± 0.14 | |
| NOE-based Rx factor (×102) (400 ms data set) | ||
| Intra-residue | 5.69 ± 0.23 | |
| Inter-residue | 6.71 ± 0.31 | |
| Total | 6.16 ± 0.20 | |
| Number of restraints violated by >0.5 Åf | ||
| Stem residues | 2.2 | |
| Terminal residues | 2.4 | |
| Loop residues | 6.0 | |
| Pairwise atomic RMSD (Å)g | ||
| All residues | 2.08 ± 0.78 | |
| Residues 3–20 | 1.00 ± 0.44 | |
| Loop residues 9–14 | 0.65 ± 0.22 | |
| Stem residues 3–8 and 15–20 | 0.49 ± 0.15 |
aAverage values ± SD (where appropriate) calculated for the 10 final structures.
bQuantitative distance restraints calculated with MARDIGRAS, as described in the text, with an average flat-well width of 0.67 Å.
cIncludes both proton to heavy atom and heavy atom to heavy atom distances.
dAccording to the miniCarlo force field.
eAverage distance restraint deviation between the actual distance and the closest of the upper and lower bounds.
fAverage number per structure.
gIncluding all atoms.
Figure 3.
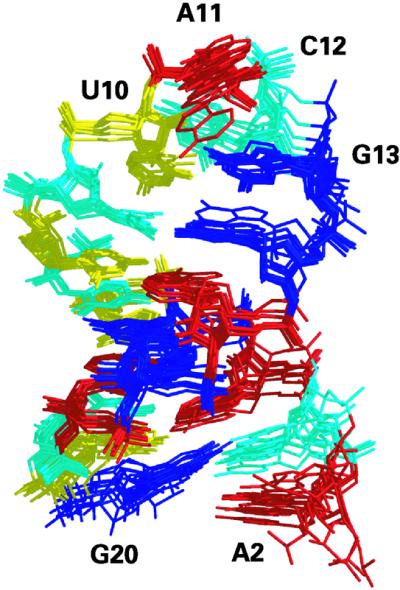
Overlay of the 10 structures of lowest total energy determined from the refinement protocol based on 200 distance restraints derived from NMR spectroscopic data. The stem region adopts an A-form helix while the UACG tetraloop is stabilized by a network of hydrogen bonds as well as base stacking. Adenines are colored in red, guanines in blue, uracils in yellow and cytosines in cyan. Coordinates of the 10 structures, distance restraints, integrated NOE intensities and proton chemical shifts have been deposited in the Protein Data Bank (accession number 1MFJ).
Excluding the dangling residues, G1 and A2, the 3′SL structure was reasonably well defined by the NMR restraints with an average pairwise RMSD of 1.0 Å (Table 1). The conformation of the 5′-terminal residue G1 (not shown in Fig. 3) was not defined by the NMR restraints, contributing to the large overall RMSD of 2.1 Å. In contrast, the dangling nucleotide A2, although also flexible, was stacked under C3 in most structures. Residues in the tetraloop and in the stem were better defined regionally than was the entire stem–loop, possibly resulting from flexibility at the step between the closing C-G base pair and the unusual U10-G13 base pair of the tetraloop. This step lacked the connectivities normally observed in NOESY spectra between sequential nucleotides of a right-handed helix. The structures were refined to a low overall residual distance violation of 0.1 Å and displayed low NOE-based Rx factors (Table 1). Despite the overall compliance with distance restraints derived from the NMR spectral data, some individual distance restraints were violated by >0.5 Å in some of the low energy structures. Most such distance violations occurred either for terminal nucleotides or for nucleotides in the tetraloop, and likely resulted from rapid interchange among multiple conformations (40). While the structure refinement data are consistent with the UACG tetraloop being flexible, an accurate accounting of the extent of flexibility in the UACG tetraloop is beyond the scope of this work.
UACG tetraloop structure
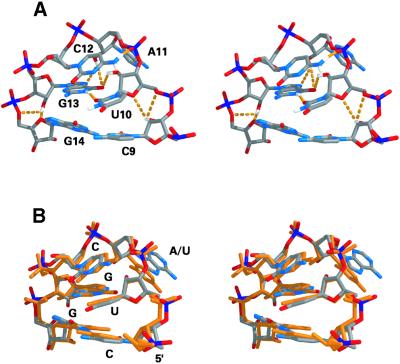
Overall, the structure of the UACG tetraloop in the 3′SL is similar to that reported previously for UUCG tetraloops in other RNA structures (21–25,41). The tetraloop is stabilized by base stacking and by a network of hydrogen bonding contacts (Fig. 4A; Table 2). G13 and U10 form a pair stabilized by two hydrogen bonds, one of which involves the 2′-OH of U10. The network of hydrogen bonds that stabilize the tetraloop is summarized in the following section. The imino proton of U10 is not involved in hydrogen bonding, rather it is exposed to solvent and does not show any cross-peaks in NOESY spectra, except for an exchange peak with water. Residue G13 adopts a syn configuration about the glycosidic bond and is stacked upon G14, while U10, the base pairing partner of G13, is stacked upon C9, the base pairing partner of G14. C12 is stacked upon the U10-G13 base pair. The sugar puckers for C12 and A11 are either C2′-endo or flexible, based on the intensities of H1′–H2′ cross-peaks in COSY spectra. The base of A11 points out into solution and both the N6 amino group and N1 are available for hydrogen bond formation with water, or with RNA or protein residues that may be involved in binding the UACG tetraloop structure. The superimposition of the UACG tetraloop with the UUCG tetraloop structure is shown in Figure 4B.
Figure 4.
Stereo depiction of (A) the NMR structure of the UACG tetraloop of 3′SL showing the network of hydrogen bonds that stabilize the tetraloop motif. The structures are colored according to atom types; hydrogen bonds are shown as orange dashed lines. (B) Superimposition of the UACG tetraloop structure from the present study with the NMR structure of the UUCG tetraloop determined previously (41). The two types of UNCG tetraloop are each stabilized by similar hydrogen bonding and base stacking arrangements. The UACG loop is colored according to atom types and the UUCG loop is colored in gold.
Table 2. Hydrogen bonds in the UACG tetraloopa.
| Hydrogen bond | Distance between heavy atoms (Å) | Distance between proton and heavy atom (Å) |
|---|---|---|
| G13 N1-H1…U10 O2 | 2.84 ± 0.01 | 1.88 ± 0.02 |
| U10 O2′-HO2′…G13 O6 | 3.02 ± 0.08 | 2.37 ± 0.04 |
| C12 N4-H42…A11 O2P | 2.89 ± 0.15 | 1.99 ± 0.17 |
| A11 O2′-HO2′…G13 N7 | 3.24 ± 0.12 | 2.60 ± 0.08 |
| A11 O2′-HO2′…G13 O6 | 3.88 ± 0.14 | 3.34 ± 0.20 |
| C9 O2′-HO2′…U10 O4′ | 3.45 ± 0.26 | 3.16 ± 0.49 |
| C9 O2′-HO2′…U10 O5′ | 3.41 ± 0.14 | 3.76 ± 0.18 |
| G13 O2′-HO2′…G14 O4′ | 3.54 ± 0.46 | 3.31 ± 0.86 |
| G13 O2′-HO2′…G14 O5′ | 3.83 ± 0.45 | 3.79 ± 0.36 |
aAverage values ± SD calculated for the 10 final structures.
The 2′-OH group of U10 is involved in numerous contacts within the UACG tetraloop structure. The U10 2′-OH 1H resonance was observed at 6.61 p.p.m., a value similar to that reported for the equivalent positions of UUCG tetraloops (21). The assignment was established based upon NOE cross-peaks from the 2′-OH of U10 to H1′, H2′, H3′ and H4′ of U10, H2′ and H3′ and H8 of A11, and H1′, H5 and H6 of C12 (Fig. 2). An NOE cross-peak between the imino 1H resonance of G13 and the 2′-OH 1H resonance of U10 was essential in establishing G13 O6 as the hydrogen bond acceptor for the 2′-OH of U10. The hydrogen bond between the 2′-OH of U10 and O6 of G13 is one of two that stabilized the G13-U10 base pair, with the second occurring between O2 of U10 and the imino hydrogen of G13. The imino hydrogen of G13 had NOESY cross-peaks with the following protons: U10, H1′, H2′ and 2′-OH; C12, H6, H1′ and H4′; G14, H8 and H1′. Cross-peaks with U10 H1′ and 2′-OH helped establish the acceptor of this hydrogen bond as the carbonyl oxygen O2 of U10. The presence of these hydrogen bonds is based on the NMR spectroscopic data, and their presence was used as restraints during the refinement procedure.
The amino proton, H42, of C12 is involved in formation of another strong hydrogen bond that stabilizes the UACG tetraloop. The H41 and H42 protons of the C12 amino group resonate at 6.29 and 7.16 p.p.m., respectively, and were distinguished from each other on the basis of relative NOE intensities with C12 H5. H41 and H42 display NOESY cross-peaks with each other, as well as with H2′, H3′ and H5 of U10, H5 and H6 of C12, and H3′ of A11 (Fig. 2). It is interesting that it is H42 (the downfield shifted proton) that is involved in hydrogen bond formation and not H41, which forms a hydrogen bond in Watson–Crick G-C base pairs. However, the acceptor of this hydrogen bond was not apparent based on the NMR spectroscopic data and, consequently, this information was not used during the structure refinement. Nevertheless, a very stable hydrogen bond between H42 of C12 and O2P of A11 was observed during the refinement protocol and was present in all of the low energy structures. A similar amino–phosphate hydrogen bond was observed in the UUCG tetraloop (21).
NMR spectroscopic data support the formation of three additional hydrogen bonds involving the 2′-OH of ribose sugars. Normally, the 2′-OH protons undergo rapid exchange with water and are not detected in 1H NMR spectra. For helical RNA, it has been shown that the preferred orientation of the hydroxyl groups prevent them from forming hydrogen bonds to the oxygens of the phosphodiester backbone, even at low temperatures (42). In contrast, when a hydroxyl group forms a strong hydrogen bond, as is the case with the U10 2′-OH, the 1H resonance is readily detected in 1H NMR spectra, and multiple cross-peaks involving this resonance are apparent in the NOESY spectra even though the exchange cross-peak with H2O is still strong (Fig. 2). In NOESY spectra of the 3′SL acquired in 95% H2O buffer, three exchange cross-peaks with water, at 6.81, 6.48 and 6.05 p.p.m., were observed in addition to that assigned to the 2′-OH of U10. The exchange cross-peaks with water were the only cross-peaks observed for these resonances in the NOESY spectra and it is likely that these resonances are due to hydroxyl groups that are almost fully exchanged during the mixing period of the NOESY experiment. A plausible interpretation of these resonances is that three hydroxyl groups participate in weak, transiently formed hydrogen bonds. Analysis of the refined structures revealed suitable candidates for these three hydroxyl groups: C9, A11 and G13. Based on distances in the low energy, refined structures, it is ambiguous if these hydroxyl groups are involved in true hydrogen bond formation or are just engaged in favorable electrostatic interactions, although the term ‘hydrogen bond’ will be used here for the sake of simplicity. All three hydroxyl groups formed bifurcated hydrogen bonds in the refined structures. One was a bifurcated hydrogen bond between the hydroxyl group of A11 and G13 N7 and O6. Two other hydrogen bonds formed between the 2′-OH of C9 and G13 with O4′ and O5′ of U10 and G14, respectively. Although these hydrogen bonds were consistent with the observation of exchange cross-peaks with water in the NOESY spectrum acquired in 95% H2O buffer, the lack of additional NOESY cross-peaks to confirm the assignments prohibited inclusion of these hydrogen bonds in the refinement protocol. It is interesting that a similar network of hydrogen bonds has been reported for another class of stable RNA tetraloops (GNRA; 43).
DISCUSSION
The structural basis for assembly and function of active spliceosomes remains an area of active research (1–4,11). In the present manuscript, we report the NMR structure of the 3′SL from human U4 snRNA (Fig. 3). Although the function of the 3′SL in U4 snRNA biogenesis, as well as in spliceosome assembly and function, is unclear, the length of the stem and the occurrence of the UACG tetraloop in 3′SL are conserved in U4 snRNA from human, rat, mouse and one form of U4 snRNA from chicken (15–18). Alternative stem lengths and tetraloop structures occur in the 3′SL from non-vertebrate organisms, such as Drosophila melanogaster (44), while in S.cerevisiae, the 3′SL is completely absent in U4 snRNA (10). Thus, it is likely that the 3′SL is important for formation of RNA–RNA and/or RNA–protein interactions that are important in either UsnRNP biogenesis or spliceosome assembly in many higher eukaryotes. In particular, the structure of a 3′SL consisting of a 7 bp stem with a UACG tetraloop is likely important in formation of active spliceosomes in mammals, including humans. The NMR structure presented in this manuscript provides insight into the structural features of this RNA stem–loop that contribute to spliceosome assembly.
The results from the present studies provide the first example of the structure of a UACG tetraloop in the context of an RNA hairpin. The UACG tetralooop is one member of the UNCG class of tetraloops. The best-studied member of the UNCG tetraloop class is the UUCG tetraloop, the structure of which has been previously determined using NMR spectroscopy and X-ray crystallography (20–25). The general structural features of generic U1U2C3G4 tetraloops include G4 adopting a syn conformation and forming a base pair with U1 that is stabilized by two hydrogen bonds. One hydrogen bond involves the imino hydrogen of G1 and O2 of U4. The second involves the 2′-OH of U1 and O6 of G4. U2 and C3 adopt C2′-endo sugar puckers. While C3 stacks upon the U1-G4 base pair, U2 neither efficiently stacks upon other nucleobases in the tetraloop nor does it form a base pair. All of these structural features characteristic of UUCG tetraloops were also apparent in the NMR structure of the UACG tetraloop from 3′SL of human U4 snRNA (Fig. 4). Our results are in agreement with previous comparative spectroscopic studies of UUCG and UACG tetraloops that indicated these sequences adopted very similar tetraloop structures in the context of short RNA hairpins (23). The NMR structure determined in this work for the UACG tetraloop from the 3′SL of human U4 snRNA suggests that the reason for conservation of the UACG tetraloop motif in 3′SL from U4 snRNA of higher eukaryotes is that the adenosine in the variable position is involved in tertiary contacts with other RNA or protein residues involved in spliceosome assembly. This is consistent with conservation of the 3′SL stem length in U4 snRNA in which the UACG tetraloop motif is conserved. The observation of co- conservation of tetraloop sequences and helix lengths in hairpins from rRNA has been suggested as evidence for the involvement of tetraloops in higher order interactions in rRNA (19). Alternatively, the UACG tetraloop has been shown to be slightly less thermodynamically stable than the UUCG tetraloop (45), and perhaps conservation of adenosine in the variable position of the UNCG tetraloop modulates hairpin stability and/or kinetics of U4 snRNA folding.
The observation of two distinct conformations for the 3′SL from human U4 snRNA raises the possibility that this stem–loop may be involved in a ‘conformational switch’ that regulates spliceosome function in vivo. The NMR structure presented here (Fig. 3) was obtained from analysis of NMR spectra collected using only freshly annealed samples of the 3′SL. Although the NMR structure of the alternative hairpin conformation for the 3′SL that occurred following equilibration of the sample at ambient temperature for several days was not determined, the differences apparent in the NMR spectra for the two conformations are consistent with the structural differences occurring mainly in the stem region, while the UACG tetraloop structure was conserved. The maintenance of UACG tetraloop structure is consistent with thermodynamic studies and molecular dynamics simulations that indicate that this structural motif is quite stable (23–25).
In summary, we have determined the NMR structure of the 3′SL from human U4 snRNA.
A functional significance for the 3′SL of U4 snRNA, other than conferring stability, has not been established. The conservation of stem length and the UACG tetraloop motif in 3′SL from U4 snRNA of vertebrates suggests, however, that this stem–loop is involved in tertiary interactions important for the formation of active spliceosomes. The NMR structure of the UACG tetraloop clearly indicates that the adenosine of the tetraloop is available for hydrogen bonding or other interactions important for spliceosome assembly without any structural rearrangement. These studies provide the first high resolution structural data for the UACG tetraloop motif and provide an insight into the basis of conservation of this stem–loop structure in vertebrates.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Drs Anwer Mujeeb and Zhihua Du for helpful discussions. The authors wish to acknowledge the use of start-up funds from WFUSM (W.H.G.), NIH P30 CA12197 (W.H.G.), NIH R01 GM 39247 (T.L.J.), P41 RR01081 (T.L.J.) and NIH RO1 GM 42223 (L.A.M.).
REFERENCES
- 1.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- 2.Will C. and Lührmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- 3.Urlaub H., Hartmuth,K., Kostka,S., Grelle,G. and Lührmann,R. (2000) A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). J. Biol. Chem., 275, 41458–41468. [DOI] [PubMed] [Google Scholar]
- 4.Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 52, 315–326. [DOI] [PubMed] [Google Scholar]
- 5.Maroney P.A., Romfo,C.M. and Nilsen,T.W. (2000) Functional recognition of the 5′ splice site by U4/U6·U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell, 6, 317–328. [DOI] [PubMed] [Google Scholar]
- 6.Kuersten S., Ohno,M. and Mattaj,I.W. (2001) Nucleocytoplasmic transport: ran, beta and beyond. Trends Cell Biol., 11, 497–503. [DOI] [PubMed] [Google Scholar]
- 7.Plessel G., Lührmann,R. and Kastner,B. (1997) Electron microscopy of assembly intermediates of the snRNP core: morphological similarities of the RNA-free (E.F.G.) protein heteromer and the intact snRNP core. J. Mol. Biol., 265, 87–94. [DOI] [PubMed] [Google Scholar]
- 8.Hartmuth K., Raker,V.A., Huber,J., Branlant,C. and Lührmann,R. (1999) An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J. Mol. Biol., 285, 133–147. [DOI] [PubMed] [Google Scholar]
- 9.Cui W. and Gmeiner,W.H. (2002) Effect of 5-FU substitution and mutation on Sm protein binding to human U4 snRNA. Nucl. Nucl., 21, 139–155. [DOI] [PubMed] [Google Scholar]
- 10.Bordonne R., Banroques,J., Abelson,J. and Guthrie,C. (1990) Domains of yeast U4 spliceosomal RNA required for PRP4 protein binding, anRNP-snRNP interactions and pre-mRNA splicing in vivo. Genes Dev., 4, 1185–1196. [DOI] [PubMed] [Google Scholar]
- 11.Hastings M.L. and Krainer,A.R. (2001) Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol., 13, 302–309. [DOI] [PubMed] [Google Scholar]
- 12.Wolff T. and Bindereif,A. (1992) Reconstituted mammalian U4/U6 snRNP complements splicing: a mutational analysis. EMBO J., 11, 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kambach C., Walke,S. and Nagai,K. (1999) Structure and assembly of the spliceosomal small ribonucleoprotein particles. Curr. Opin. Struct. Biol., 9, 222–230. [DOI] [PubMed] [Google Scholar]
- 14.Stark H., Dube,P., Lührmann,R. and Kastner,B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature, 409, 539–542. [DOI] [PubMed] [Google Scholar]
- 15.Krol A., Branlant,C., Lazar,E., Gallinaro,H. and Jacob,M. (1981) Primary and secondary structures of chicken, rat and man U4 RNAs. Homologies with U1 and U5 RNAs. Nucleic Acids Res., 9, 2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy R., Henning,D. and Busch,H. (1981) The primary nucleotide sequence of U4 RNA. J. Biol. Chem., 256, 3532–3538. [PubMed] [Google Scholar]
- 17.Kato N. and Harada,F. (1981) Nucleotide sequence of 5.7S RNA of mouse cells. Biochem. Biophys. Res. Commun., 99, 1477–1485. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman M.L., Korf,G.M., McNamara,K.J. and Stumph,W.E. (1986) Structural and functional analysis of chicken U4 small nuclear RNA genes. Mol. Cell. Biol., 6, 3910–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedenstierna K.O., Siefert,J.L., Fox,G.E. and Murgola,E.J. (2000) Co-conservation of rRNA tetraloop sequences and helix length suggests involvement of the tetraloops in higher-order interactions. Biochimie, 82, 221–227. [DOI] [PubMed] [Google Scholar]
- 20.Cheong C., Varani,G. and Tinoco,I.,Jr (1990) Solution structure of an unusually stable RNA hairpin 5′GGAC(UUCG)GUCC. Nature, 346, 680–682. [DOI] [PubMed] [Google Scholar]
- 21.Allain F.H. and Varani,G. (1995) Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol., 250, 333–353. [DOI] [PubMed] [Google Scholar]
- 22.Ennifar E., Nikulin,A., Tischenko,S., Serganov,A., Nevskaya,N., Garber,M., Ehresmann,C., Nikonov,S. and Dunas,P. (2000) The crystal structure of UUCG tetraloop. J. Mol. Biol., 304, 35–42. [DOI] [PubMed] [Google Scholar]
- 23.Abdelkafi M., Ghomi,M., Turpin,P.Y., Baumruk,V., Herve du Penhoat,C., Lampire,O., Bouchemal-Chibani,N., Goyer,P., Namane,A., Gouyette,C., Huynh-Dinh,T. and Bednarova,L. (1997) Common structural features of UUCG and UACG tetraloops in very short hairpins determined by UV absorption, Raman, IR and NMR spectroscopies. J. Biomol. Struct. Dyn., 14, 579–593. [DOI] [PubMed] [Google Scholar]
- 24.Baumruk V., Gouyette,C., Huynh-Dinh,T., Sun,J.S. and Ghomi,M. (2001) Comparison between CUUG and UUCG tetraloops: thermodynamic stability and structural features analyzed by UV absorption and vibrational spectroscopy. Nucleic Acids Res., 29, 4089–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams D.J., Boots,J.L. and Hall,K.B. (2001) Thermodynamics of 2′-ribose substitutions in UUCG tetraloops. RNA, 7, 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 27.Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 28.Goddard T.D. and Kneller,D.G. (1998) SPARKY v. 3.0. University of California, San Francisco, CA.
- 29.States D.J., Haberkorn,R.A. and Ruben,D.J. (1982) A two dimensional nuclear Overhauser experiment with pure absorption phase in four quandrants J. Magn. Reson., 48, 286–292. [Google Scholar]
- 30.Hore P.J. (1983) Solvent suppression in Fourier transform nuclear magnetic resonance. J. Magn. Reson., 55, 283–300. [Google Scholar]
- 31.Smallcombe S.H. (1993) Solvent suppression with symmetrically-shifted pulses. J. Am. Chem. Soc., 115, 4776–4785. [Google Scholar]
- 32.Borgias B.A. and James,T.L. (1990) MARDIGRAS – a procedure for matrix analysis of relaxation for discerning geometry of an aqueous structure. J. Magn. Reson., 87, 475–487. [Google Scholar]
- 33.Liu H., Spielmann,H.P., Ulyanov,N.B., Wemmer,D.E. and James,T.L. (1995) Interproton distance bounds from 2D-NOE intensities: effect of experimental noise and peak integration errors. J. Biomol. NMR., 6, 390–402. [DOI] [PubMed] [Google Scholar]
- 34.Güntert P., Mumenthaler,C. and Wüthrich,K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol., 273, 283–298. [DOI] [PubMed] [Google Scholar]
- 35.Zhurkin V.B., Ulyanov,N.B., Gorin,A.A. and Jernigan,R.L. (1991) Static and statistical bending of DNA evaluated by Monte Carlo simulations. Proc. Natl Acad. Sci. USA, 88, 7046–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulyanov N.B., Ivanov,V.I., Minyat,E.E., Khomyakov,E.B., Petrova,M.V., Lesiak,K. and James,T.L. (1998) A pseudosquare knot structure of DNA in solution. Biochemistry, 37, 12715–12726. [DOI] [PubMed] [Google Scholar]
- 37.Ulyanov N.B., Bauer,W.R. and James,T.L. (2002) High-resolution NMR structure of an AT-rich DNA sequence. J. Biomol. NMR, 22, 265–280. [DOI] [PubMed] [Google Scholar]
- 38.Keepers J.W. and James,T.L. (1984) A theoretical study of distance determinations from NMR. Two-dimensional nuclear Overhauser effect spectra. J. Magn. Reson., 57, 404–426. [Google Scholar]
- 39.Ferrin T.E., Huang,C.C., Jarvis,L.E. and Langridge,R. (1988) The MIDAS display system. J. Mol. Graphics, 6, 13–27. [Google Scholar]
- 40.Ulyanov N.B., Schmitz,U., Kumar,A. and James,T.L. (1995) Probability assessment of conformational ensembles: sugar repuckering in a DNA duplex in solution. Biophys. J., 68, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colmenarejo G. and Tinoco,I.,Jr (1999) Structure and thermodynamics of metal binding in the P5 helix of a group I intron ribozyme. J. Mol. Biol., 290, 119–135. [DOI] [PubMed] [Google Scholar]
- 42.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) The orientation and dynamics of the C2′-OH and hydration of RNA and DNA.RNA hybrids. Nucleic Acids Res., 26, 3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jucker F.M., Heus,H.A., Yip,P.F., Moors,E.H.M. and Pardi,A. (1996) A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol., 264, 968–980. [DOI] [PubMed] [Google Scholar]
- 44.Saba J.A., Busch,H., Wright,D. and Reddy,R. (1986) Isolation and characterization of two putative full-length Drosophila U4 small nuclear RNA genes. J. Biol. Chem., 261, 8750–8753. [PubMed] [Google Scholar]
- 45.Abdelkafi M., Luelliot,N., Baumruk,V., Bednarova,L., Turpin,P.Y., Namani,A., Gouyette,C., Huynh-Dinh,T. and Bednarova,L. (1998) Structural features of the UCCG and UGCG tetraloops in very short hairpins as evidenced by optical spectroscopy. Biochemistry, 37, 7878–7884. [DOI] [PubMed] [Google Scholar]