Abstract
Equilibrative nucleoside transporters (ENTs) are a recently characterized and poorly understood group of membrane proteins that are important in the uptake of endogenous nucleosides required for nucleic acid and nucleoside triphosphate synthesis. Despite their central importance in cellular metabolism and nucleoside analog chemotherapy, no human ENT gene has been described and nothing is known about gene structure and function. To gain insight into the ENT gene family, we used experimental and in silico comparative genomic approaches to identify ENT genes in three evolutionarily diverse organisms with completely (or almost completely) sequenced genomes, Homo sapiens, Caenorhabditis elegans and Drosophila melanogaster. We describe the chromosomal location, the predicted ENT gene structure and putative structural topologies of predicted ENT proteins derived from the open reading frames. Despite variations in genomic layout and limited ortholog protein sequence identity (≤27.45%), predicted topologies of ENT proteins are strikingly similar, suggesting an evolutionary conservation of a prototypic structure. In addition, a similar distribution of protein domains on exons is apparent in all three taxa. These data demonstrate that comparative sequence analyses should be combined with other approaches (such as genomic and proteomic analyses) to fully understand structure, function and evolution of protein families.
INTRODUCTION
The completion of the sequencing of several genomes has made available a wealth of data, which can provide insight into the molecular evolution and, possibly, functional relationships of protein families within eukaryotes (1,2). In addition, comparative genomics can facilitate identification of conserved sequences or motifs that may be indicative of nucleic acid and/or protein regions that are important in structure or regulation (3).
Membrane transport proteins are likely to have evolved shortly after the appearance of lipid biomembranes since they have a central role in cellular homeostasis. Nucleoside transporters (NTs) constitute a superfamily of integral membrane proteins that have been identified (based on sequence homology) in a wide variety of taxa (4,5).
NTs can be divided into two unique and non-homologous families based on mechanism of translocation of substrate, the equilibrative NTs (ENTs), which transport mainly by facilitated diffusion, and the concentrative NTs (CNTs), in which transport of a nucleoside is driven by co-transport of a cation down its concentration gradient. ENTs appear to be widely distributed in mammalian tissues and are likely to be primarily responsible for drug uptake of nucleoside analogs used clinically in humans. In addition, there are temporal, spatial and quantitative variations in ENT expression both within and between individuals (6), suggesting complex transcriptional regulation which, in a clinical setting, may influence drug efficacy. Although ENT proteins (and isoforms) have been reported in a variety of organisms (4), a comprehensive genomic analysis of the ENT family has not yet been conducted. Within metazoan eukaryotes with completely sequenced genomes, three human ENT proteins have been described (4). Of these, two, hENT1 and hENT2, have been functionally characterized (7,8). In Drosophila, two putative ENT proteins, which have not been functionally characterized, are reported and in Caenorhabditis elegans, six ENT proteins, three of which have been functionally characterized, are reported (4,9).
Despite their importance in chemotherapeutic drug uptake, the human ENT genes have not been fully characterized and none has yet been fully described in the literature. To date, only the chromosomal locations of the genes for hENT1 (10) and HNP36, a truncated hENT2-like protein (11) have been reported. Therefore, we undertook the first comprehensive analysis of the human ENT genes and describe here the complete gene structures of hENT1, hENT2 and hENT3 along with their precise chromosomal locations. In addition, we show that the hENT2 gene is alternatively spliced to produce a number of variants, including the HNP family of proteins.
As noted in humans, many other organisms possess multiple ENT proteins. Whether this is due to the presence of splice variants or different gene products is not known. To address this gap in our understanding of the ENT family, we have conducted a comprehensive and detailed genomic analysis of the ENT family in the model organisms with completely sequenced genomes, Drosophila melanogaster and C.elegans. We have identified and characterized all known and putative genes for ENT family members within these taxa. These studies lay the groundwork for future genetic analysis of ENTs in model organisms. In addition, protostomian invertebrates (such as D.melanogaster and C.elegans) possess adenosine receptor orthologs (1) and NTs have been implicated in the modulation of activity of adenosinergic signaling (12). Therefore, these organisms can be used as models to further study the relationship between transporters and receptors.
Our comparison of complete families of ENT genes from evolutionarily divergent organisms also provides insight into the general phylogenetic relationships among this protein family. Finally, these studies suggest a prototypic ENT structure that has been highly conserved during evolution despite considerable variation at the gene structure and protein sequence levels. Our findings demonstrate that low sequence identity can correlate with high structural similarity and suggest that multiple approaches (such as sequence alignments, analysis of gene structure and predicted protein structures) should be used to understand structure, function and relationships between orthologs in protein families.
See Supplementary Material for additional information.
MATERIALS AND METHODS
Identification and characterization of human ENT genes
A partial hENT1 gene sequence has already been deposited in GenBank (accession no. AF190884). However, this sequence lacks the upstream 5′-untranslated region (5′-UTR) and promoter. Therefore we cloned these regions of hENT1 by screening a λ DASH II genomic library (Stratagene, CA) using a PCR-generated probe corresponding to the 5′ end of the hENT1 open reading frame (ORF). Subcloning of positive inserts for further analysis was done using pBluescript II KS (+), (Stratagene, CA).
For the other genes, we used the DNA sequences of individual exons of hENT1 and scanned the completed human genome using BLASTN (http://www.ncbi.nlm.nih.gov/BLAST) to identify similar regions of DNA which were likely to contain hENT2 and hENT3. In addition, we used the cDNAs for hENT2 (accession no. AF034102) and hENT3 (accession no. AF326987) in a similar BLASTN search.
The chromosomal location for the hENT1 gene has been previously reported (10). Chromosomal locations for hENT2 and hENT3 were determined using NCBI Locus Link (http://www.ncbi.nlm.nih.gov/LocusLink).
Identification and characterization of D.melanogaster and C.elegans ENT genes
Putative ENT genes were identified using keyword searches (nucleoside, transporter, adenosine, etc.) of Flybase (http://www.flybase.org) and Wormbase (http://www.wormbase. org). In addition, all known NTs in GenBank were compared against the completed D.melanogaster and C.elegans genomes (Flybase and Wormbase) using BLASTN and BLASTP.
Identification of gene and predicted protein structure
For gene structure, exon/intron splice sites were identified based on typical splice donor–acceptor consensus sites (GT/AG) and we confirmed that the combined exons corresponded to the reported mRNAs and proteins. The predicted proteins were then analyzed using the transmembrane prediction program TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) to determine the putative topology. Amino acid identity (%) of all predicted protein sequences was determined using ClustalW (http://www.ebi.ac.uk/clustalw/).
RT–PCR of D.melanogaster ENTs
Total RNA was isolated from adult D.melanogaster (Canton-S, both sexes) using Trizol reagent (Life Technologies). RNA was used to synthesize cDNA by reverse transcription using oligo(dT) primer from the Superscript Preamplification System (Life Technologies). The cDNA was then amplified by PCR with Herculase Enhanced DNA Polymerase (Stratagene, CA) as follows: one cycle of 94°C for 2 min; 30 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 1.5 min; and a final cycle of 68°C for 10 min. The primers for amplification of gene CG11045 were 5′-ctagtctagactgatacaacgcggcgata-3′ (forward) and 5′-ccggaattcggttaagacgaaaccaaatgg-3′ (reverse). The primers for CG11907 were 5′-ctagtctagaagtggcaatgccaggca-3′ (forward) and 5′-ccggaattctcaaagcatttgaacaaatacaaggctg-3′ (reverse). Forward primers contained an engineered XbaI site and reverse primers contained an EcoRI site. For CG11010 the forward primer used was 5′-ataagaatgcggccgcgatttgccagcggaaatc-3′ and the reverse primer used was 5′-cggggtaccttagatattttcgagcagtgcatctg-3′. The forward primer had a NotI engineered site while the reverse primer had a KpnI site.
RESULTS
Identification of genes
We have identified ENT and putative ENT genes within the human, D.melanogaster and C.elegans genomes. The chromosomal locations, accession numbers (genes and mRNAs), number of exons, predicted protein, transmembrane domain (TM) structures and function (if known) of all ENTs described in this paper are shown in Table 1.
Table 1. Chromosomal location, predicted gene and protein structures for human, Drosophila melanogaster and Caenorhabditis elegans ENTs.
| ENT | Chromosomal location | Gene (GenBank, Flybase or Wormbase) | No. of exons | mRNA or protein | TM domains (TMHMM 2.0 prediction) | Protein (amino acids) | Additional information (e.g. substrate preference, location) |
|---|---|---|---|---|---|---|---|
| hENT1 | 6p21.1–21.6 | AF495730 | 12 | U81375 gi:1845344 | 11 | 456 | Purines, pyrimidines |
| hENT2 | 11q12.1 | NT_008992.7 (contig no.) | 12 | AF034102 gi:2811136 | 11 | 456 | Nucleosides, nucleobases |
| HNP36 | 11q12.1 | NT_008992.7 (contig no.) | X86681 gi:951266 | 7 | 326 | Unknown | |
| hENT2 short variant | 11q12.1 | NT_008992.7 (contig no.) | AF401235 gi:15217146 | 6 | 301 | Unknown | |
| hENT3 | 10q22.1 | BK000392 (TPA section) | 5 | AF326987 gi:12656638 | 11 | 475 | Subcellular location? |
| DmENT1 | 2-21C1 | CG11907 gi:18466851 | 4 | AF217396a gi:8132774 | 11 | 476 | Unknown |
| DmENT2 | 2-26E2–E3 | CG11045 gi:18483578 | 1 | AAF52405 gi:7297138b | 11 | 458 | Unknown |
| DmENT3 | 3-69E4–E5 | CG11010 gi:10727955 | 3 (4)c | AAF49871c gi:7294530 | 10 (11)c | 586 (668)c | Unknown |
| CeENT1 | IV:5.13 | ZK809.4 | 5 | CAA92642 gi:3881800 | 11 | 461 (445)d | Purines, pyrimidines |
| CeENT2 | X:21.94 | K09A9.3 | 6 | CAB01882 gi:17568767 | 11 | 450 | Adenosine |
| CeENT3 | V:5.97 | K02E11.1 | 16 | CAB01223 gi:3878229 | 11 | 800 (729)d | Adenosine, uridine |
| CeENT4 | IV:10.84 | C47A4.2 | 8 | CAB62793 gi:6562351 | 11 | 418 (451)d | Unknown |
| CeENT5 | X:-7.27 | F16H11.3 | 7 | AAA98003 gi:1280131 | 11 | 434 | Unknown |
| CeENT6 | I:3.6 | F36H2.2 | 8 | CAB03075 gi:14530443 | 9 | 384 | 3′-Truncated based on ESTs |
aThere is an additional version of this mRNA submitted under accession no. gi:7296214 which contains an error in the predicted splice sites for the exons resulting in a shorter version of the protein.
bThis is a putative full-length mRNA based on the longest ORF derived from the gene product. However, identification of ESTs containing the 5′ region of this putative mRNA (5′-UTR and N-terminal sequence of the protein) and our own RT–PCR data suggest that this is an expressed mRNA.
cThis is a predicted protein sequence based on the gene. There is a longer version (668 amino acids) of the derived protein from this gene submitted under accession no. AAK92930, gi:15291323. This longer protein results from a longer ORF being determined based on a DNA clone isolated as part of the Drosophila Gene Collection 1 (1) and results from the presence of an additional exon being located within the second intron (leading to a total of four exons and a protein of 668 amino acids). However, the splice donor–acceptor sites for the additional intron (III) between the new exon (III) and exon IV do not conform to the usual GT-AG rule and therefore it is not clear whether this longer version of the mRNA exists. The mRNA used above for DmENT3 conforms to the splice donor–acceptor rule.
dProtein size in parentheses according to Hyde et al. (4).
To isolate the full hENT1 gene, including promoter, we screened approximately 8 × 105 independent plaque-forming units (p.f.u.) from a λ DASH II human genomic library and isolated six positive clones. One of these positive clones (4.1) was further analyzed and confirmed to contain an insert of ∼14 kb which included a region homologous to a sequence in GenBank (accession no. AL139392, 61.22 kb). Further analysis of this sequence (accession no. AL139392) revealed that it was also homologous to the partial hENT1 gene sequence (accession no. AF190884, 6.287 bp). However, our clone contains ∼10 kb of additional DNA, including the 5′-UTR and upstream regions. We have begun functional characterization of this region and confirmed it to be the promoter for hENT1 (13). The complete hENT1 gene, including the promoter and 3′-UTR, has been deposited in GenBank under accession no. AF495730.
The hENT2 mRNA (accession no. AF034102) was found to align to a draft sequence of chromosome 11 which correlates with a region of DNA previously identified to encode HNP36, a hENT2-like protein (11), which we have subsequently confirmed to be a splice variant of hENT2 (see below). This genomic sequence was compared to the mRNA to determine the gene location and intron/exon layout determined by finding consensus donor/acceptor splice sites. Approximately 4 kb of genomic DNA upstream of the putative transcriptional initiation site was included in the designation of this as the hENT2 gene and identified as the putative promoter.
The hENT3 gene was previously described as being located on chromosome 10 (4) based on sequence similarity to a clone (accession no. AL359384). However, this clone only contains a partial portion of the hENT3 gene, therefore we used the previously identified mRNA sequence (accession no. AF326987) and compared this with the complete draft sequence of the human genome. This resulted in identification of a sequenced contig (NT_008849.8) which contained the complete hENT3 gene (promoter, UTRs and coding region) consisting of ∼46 kb. Gene structure was determined using standard methods and the complete hENT3 gene (nucleotide sequence data) is available in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases under the accession no. TPA: BK000392.
To identify any other putative isoforms, we also conducted exon scanning using hENT1 against the draft complete human genome and found regions of DNA that were confirmed to be hENT2 and hENT3 based on the predicted proteins derived from the genomic sequences, but no additional ENT-like genes. In addition, we scanned the human genome with complete sets of C.elegans and Drosophila ENTs. Extensive analyses using these sequences as queries did not reveal any additional novel human homologs (i.e. no BLAST hit with a P value < 10).
We identified putative ENTS in Drosophila using keyword (e.g. nucleoside, adenosine, transporter, etc.) searches within Flybase (http://www.flybase.org) and found five different putative NT genes. Additional analyses using all known ENTs to search the Drosophila genome produced no additional sequences. Based on similarity with known ENTs, three of the five sequences were determined to be putative Drosophila ENTs (while the remaining two sequences appear to be CNT homologs). Two of these ENT-like genes give rise to predicted proteins that have been previously identified as ENT family members (CG11907 and CG11045) (4). However, we identified an additional gene (CG11010) for which the predicted protein is a novel isoform that has not been previously described. We have designated these as follows: CG11907, DmENT1 gene; CG11045, DmENT2 gene and CG11010, DmENT3 gene (according to standard nomenclature for this field).
RT–PCR of adult fly RNA using primers against DmENT1 generated a product of ∼1.4 kb, while those for DmENT2 generated a product of 1.9 kb (data not shown). Both PCR products were subcloned into pGEM-T (Promega). Cloned products were sequenced in full at the Molecular Biology Core Facility (York University) and confirmed to be D.melanogaster ENTs (based on sequence homology). We were unable to amplify DmENT3 from RNA extracted from either adult or larval flies.
All C.elegans ENT genes were located as described for D.melanogaster, resulting in six different genes being identified (CeENT1–CeENT6). Exhaustive searches using other known ENTs did not result in any additional or novel isoforms being identified.
Chromosomal location of ENTs
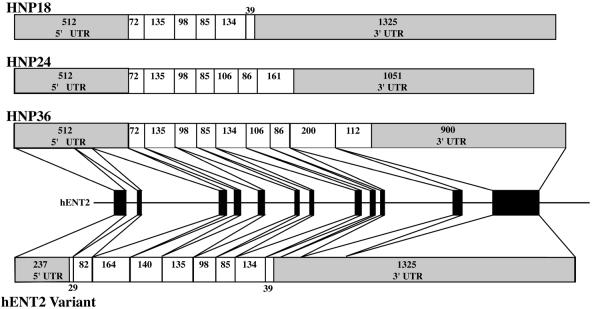
The chromosomal locations of all ENT genes are shown in Table 1. The three hENT genes are located on separate chromosomes. The hENT1 gene is one of 973 genes on chromosome 6 and its cytogenetic locus maps to 6p21.1–21.2 as previously described (10). The hENT2 gene, one of 1131 on chromosome 11, is located at 11q12.1. This location corresponds to the previous designation for HNP36 (11), confirming a single locus for this gene and indicating that the alternative forms of hENT2 (e.g. HNP36) are due to splice variants from a single gene. Two medical conditions of unknown origin have been linked to this region; however, it is not clear if genetic variations or mutations in hENT2 or HNP36 are responsible (11).
The chromosomal location of hENT3 is 10q22.1 and it is one of 1131 genes on this chromosome. Comparison of the chromosomal locations for hENT1, hENT2 and hENT3 with the OMIM database (http://www.ncbi.nlm.nih.gov/Omim/) did not provide any evidence of a disease phenotype currently being associated with these loci.
The three fly ENT genes are located on two chromosomes (Table 1). DmENT1 and DmENT2 are both found on chromosome 2 (2-21C1 and 2-26E3, respectively) within 6111 kb (DmENT1, 351 kb; DmENT2, 6462 kb) of each other, although they are oriented in opposite directions. DmENT3 is found on chromosome 3 (3-69E4–E5).
The C.elegans ENT genes are located on four different chromosomes I, IV, V and X. Although there are two chromosomes (IV and X) that each contain two different C.elegans ENT genes (CeENT1 and CeENT4 on IV, CeENT2 and CeENT5 on X), the locations of the genes are at a considerable distance from each other.
Gene structure of ENTs
Analysis of the gene structure for hENT1 and hENT2 revealed a similar layout of 12 exons (Fig. 1 and Table 2; hENT3, Supplementary Material). Exon sizes ranged from 79 to 1012 and intron sizes from 87 to 2048. The respective exon sizes for hENT1 and hENT2 were very similar while the comparable intron sizes varied considerably. We compared the exon layout and sizes of the hENT1 gene with the mENT1 gene (14), the only other characterized ENT gene. The overall layout of the genes and the exon sizes are identical between mENT1 and hENT1. The introns vary slightly in size. In contrast to hENT1, hENT2 produces a number of splice variants derived from varying translational strategies (Fig. 2; Supplementary Material). Briefly, the HNP family (11) has a translational start site located at the beginning of exon IV and then produces variants which stop half-way through either exon IX or exon XI or at the same location as the full-length hENT2. The other hENT2 variant (AF401235) has an identical start site to full-length hENT2 but the protein is truncated by a stop site half-way through exon IX, which is the same termination site as HNP18. These splice variants are predicted to produce proteins that are truncated at either the N- or C-terminus compared to full-length hENT2. However, all variants predict a protein that contains the central core domains encompassing approximately TMs 4–7. This region overlaps the predicted substrate translocation site (TM 3–6) (4). More recently, we have found that the most highly conserved region of the ENT protein in over 40 different family members from prokaryotes to humans is in TM 4–5 (5). Therefore, it is possible that these splice variants, which all contain TM 4–5, could give rise to functionally active proteins, although the physiological significance of the alternative forms remains unclear. Further detailed analysis of the transcriptional and translational regulation of hENT2 and the physiological relevance of the splice variants is required. In contrast to hENT1 and hENT2, hENT3 has a considerably different structure with only five exons and very large introns ranging in size from ∼4.3 to >21 kb (Fig. 1; Supplementary Material). The intronic region between exons I and II was scanned for potential additional transcriptional initiation sites and ORFs, but none were found. The putative polyadenylation signal for hENT1 (aataaa) is 597 bp downstream of the stop codon, for hENT2 (aataa) is 900 bp downstream of the stop codon and for hENT3 (aataaa) is 762 bp downstream of the stop codon. hENT1 and hENT2 appear to be similar, though smaller, to genes identified as part of the human genome sequence project where an ‘average’ gene is ∼49 kb in length, split into 12 exons producing transcripts of ∼2.4 kb with ∼30–40% of genes generating alternative splicing forms (L.-C.Tsui, personal communication).
Figure 1.
Gene structure of the human ENTs. Filled black boxes represent exons and thin lines represent introns and flanking regions. Above each gene the spliced mRNA structure is shown. Clear boxes represent the ORF, grey boxes represent the UTR. Numbers inside the boxes represent the number of bases encoding each region.
Table 2. Gene structure, amino acid distribution and predicted domains of hENT1.
| Exons | Introns | Amino acids/exonb | Amino acids/domainc | Domainc | |||
|---|---|---|---|---|---|---|---|
| Exon | Size (bp) | Splice donor | Splice acceptor | Size (bp)a | |||
| 1 | 305 | 1–10 (10) | 1–12 | N-terminus (int.) | |||
| ACAG gtga | acag CTAC | 2048 (1836) | |||||
| 2 | 82 | 11–37 (27) | 13–35 | TM 1 | |||
| TCAG gtga | gcag TATT | 119 (131) | |||||
| 3 | 203 | 38–105 (68) | 36–75 | External loop (1) | |||
| 76–98 | TM 2 | ||||||
| AGAG gtga | gcag GATC | 116 (103) | |||||
| 4 | 140 | 106–152 (47) | 99–109 | Internal loop (1) | |||
| 110–132 | TM 3 | ||||||
| 133–146 | External loop (2) | ||||||
| AATT gtaa | ccag CATT | 300 (132) | |||||
| 5 | 135 | 153–197 (45) | 147–169 | TM 4 | |||
| 170–175 | Internal loop (2) | ||||||
| 176–195 | TM 5 | ||||||
| GCCA gtaa | ccag GTGG | 87 (108) | |||||
| 6 | 98 | 198–230 (33) | 196–204 | External loop (3) | |||
| 205–227 | TM 6 | ||||||
| CCTG gtga | atag GAAT | 145 (134) | |||||
| 7 | 79 | 231–256 (26) | 228–255 | Internal loop (3) | |||
| AAAG gtcc | gcag GAGA | 473 (609) | |||||
| 8 | 98 | 257–289 (33) | 256–289 | Internal loop (3) | |||
| AAAT gtac | ctag ATCT | 537 (614) | |||||
| 9 | 109 | 290–324 (35) | 290–312 | TM 7 | |||
| 313–326 | External loop (4) | ||||||
| TGGG gtga | ccag AACG | 237 (247) | |||||
| 10 | 86 | 325–353 (29) | 327–349 | TM 8 | |||
| GTGG gtaa | ggag CCTG | 379 (919) | |||||
| 11 | 200 | 354–420 (67) | 350–360 | Internal loop (4) | |||
| 361–380 | TM 9 | ||||||
| 381–394 | External loop (5) | ||||||
| 395–417 | TM 10 | ||||||
| CCAA gtga | ttag GAAA | 409 (310) | |||||
| 12 | 709 | 421–456 (36) | 418–429 | Internal loop (5) | |||
| 430–452 | TM 11 | ||||||
| 453–456 | C-terminus (ext.) | ||||||
aFigures in parentheses after hENT1 intron sizes are comparable mENT1 intron sizes.
bFigures in parentheses show number of amino acids encoded by an exon; in the case of amino acids split between exons, the amino acid is assigned to the exon carrying the majority of the codon.
cDomains and amino acids/domain are predicted based on analyses using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Figure 2.
Structure of splice variants derived from the hENT2 gene. Layout is as described for Figure 1. The HNP family consists of an extended 5′-UTR and a truncated N-terminal region of the ORF compared to hENT2. Each HNP family member has a different stop site resulting in variable C-terminals. For hENT2v, transcriptional initiation is identical to hENT2 but the ORF of the transcript is truncated at the same site as HNP18.
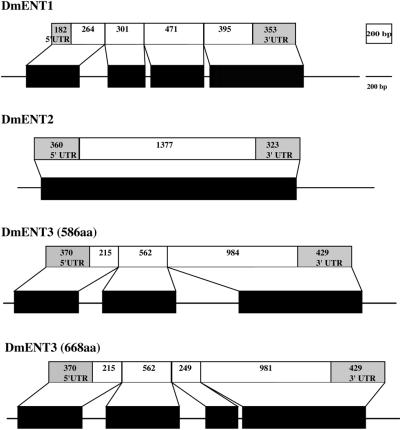
Drosophila ENTs are considerably simpler than human ENTs with fewer exons and relatively small (or non-existent) introns (Fig. 3). DmENT1 (CG11907) consists of four exons of approximately similar size (Table 3). DmENT2 is unusual in that initially it appears to be a pseudogene since it consists of a single exon (see Supplementary Material). However, there are no internal stop codons and no evidence of genomic poly(A) tracts. In addition, the gene is transcribed and mRNA is expressed, since we have successfully amplified this cDNA by RT–PCR from mRNA isolated from adult flies (data not shown). Moreover, the original GenBank submission for this mRNA (gi:8132774) states that it was isolated from an embryonic cDNA library. This apparently intronless gene could therefore be a retrogene. Since the predicted protein sequences of DmENT1 and DmENT2 are more similar to each other than DmENT3 (Fig. 4 and see Table 5), we speculate that these two genes may be the consequence of an ancient gene duplication event or possibly an incorporation into the genome of a processed DmENT1 transcript which has subsequently continued to evolve. DmENT3 consists of only three exons (Supplementary Material), although a larger protein has been predicted (Table 1) from a longer ORF derived from four putative exons. This longer ORF would result from the presence of an additional intron that does not conform to the usual splice donor–acceptor site rules.
Figure 3.
Gene structure of the Drosophila ENTs. Layout is as described for Figure 1.
Table 3. Gene structure, amino acid distribution and predicted domains of DmENT1.
| Exons | Introns | Amino acids/exon | Amino acids/domain | Domain | |||
|---|---|---|---|---|---|---|---|
| Exon | Size (bp) | Splice donor | Splice acceptor | Size (bp) | |||
| 1 | 446 | 1–88 (88) | 1–62 | N-terminus (int.) | |||
| 63–85 | TM 1 | ||||||
| AGAT gtaa | tcag TACT | 231 | |||||
| 2 | 301 | 89–188 (100) | 86–118 | External loop (1) | |||
| 119–141 | TM 2 | ||||||
| 142–147 | Internal loop (1) | ||||||
| 148–165 | TM 3 | ||||||
| 166–179 | External loop (2) | ||||||
| AATA gtgg | atag TATC | 53 | |||||
| 3 | 471 | 189–345 (157) | 180–202 | TM 4 | |||
| 203–208 | Internal loop (2) | ||||||
| 209–231 | TM 5 | ||||||
| 232–240 | External loop (3) | ||||||
| 241–263 | TM 6 | ||||||
| 264–309 | Internal loop (3) | ||||||
| 310–332 | TM 7 | ||||||
| 333–346 | External loop (4) | ||||||
| ACAG gtaa | acag ATGT | 54 | |||||
| 4 | 748 | 346–476 (131) | 347–369 | TM 8 | |||
| 370–381 | Internal loop (4) | ||||||
| 382–399 | TM 9 | ||||||
| 400–413 | External loop (5) | ||||||
| 414–436 | TM 10 | ||||||
| 437–448 | Internal loop (5) | ||||||
| 449–471 | TM 11 | ||||||
| 472–476 | C-terminus (ext.) | ||||||
Figure 4.
Alignment of three D.melanogaster ENT predicted proteins using Clustal X (v.1.8). Black shaded amino acids are conserved between proteins, grey shaded amino acids are similar (conservative substitutions). Numbered bars below sequences represent putative transmembrane domains.
Table 5. Percent amino acid identity between predicted ENT proteins from three taxa: human, Drosophila melanogaster and Caenorhabditis elegans.
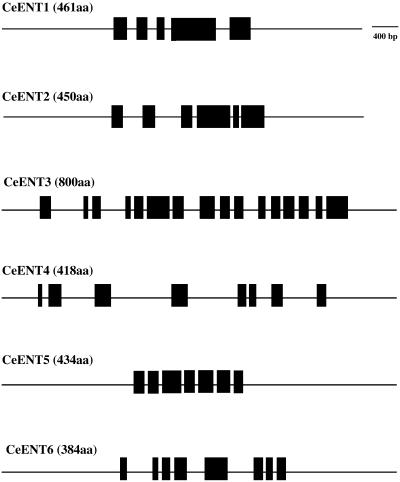
Caenorhabditis elegans genes have a diversity of structure ranging from five to 16 exons (Fig. 5). While most C.elegans ENT genes have few obvious structural similarities, the sizes of exons 1, 2 and 3 of CeENT1 and CeENT2 are identical (or very close; see Table 4 and Supplementary Material). Exon 4 in CeENT1 is identical to exons 4 and 5 combined of CeENT2. Moreover, CeENT1 exon 5 and CeENT2 exon 6 are identical. Overall, this similarity in gene structure suggests that CeENT1 and CeENT2 result from a relatively recent gene duplication event. This speculation is supported by the very high primary sequence similarity of these two gene products (Table 5). The most complex C.elegans ENT gene, CeENT3, appears to be derived from a gene fusion event since the first eight exons encode a non-transporter-like protein which then leads into a typical ENT-type protein. Sequence comparison of this novel N-terminal portion of CeENT3 with all sequences in GenBank did not reveal homology to a known protein so the origin of this part of the protein is unclear. However, CeENT3 is apparently expressed and has been functionally characterized (9), suggesting that it is an active ENT. Other noted structural variations include the larger introns in CeENT4. Data for the gene structure of CeENT2 are shown in Table 4. All other CeENT gene structures can be found in the Supplementary Material accompanying this paper.
Figure 5.
Gene structure of the C.elegans ENTs. Layout is as described in Figure 1. For clarity, predicted mRNA structures are not shown.
Table 4. Gene structure, amino acid distribution and predicted domains of CeENT2.
| Exons | Introns | Amino acids/exon | Amino acids/domain | Domain | |||
|---|---|---|---|---|---|---|---|
| Exon | Size (bp) | Splice donor | Splice acceptor | Size (bp) | |||
| 1 | 177 | 1–59 (59) | 1–33 | N-terminus (int.) | |||
| 34–56 | TM 1 | ||||||
| TGAC gtga | ccag TACT | 315 | |||||
| 2 | 166 | 60–114 (55) | 57–90 | External loop (1) | |||
| 91–113 | TM 2 | ||||||
| TCAA gtaa | tcag GGGA | 407 | |||||
| 3 | 94 | 115–209 (94) | 114–119 | Internal loop (1) | |||
| 120–142 | TM 3 | ||||||
| 143–146 | External loop (2) | ||||||
| 147–169 | TM 4 | ||||||
| 170–181 | Internal loop (2) | ||||||
| 182–204 | TM 5 | ||||||
| ACAT gtga | ccag GGCT | 87 | |||||
| 4 | 512 | 210–381 (171) | 205–218 | External loop (3) | |||
| 219–238 | TM 6 | ||||||
| 239–277 | Internal loop (3) | ||||||
| 278–300 | TM 7 | ||||||
| 301–319 | External loop (4) | ||||||
| 320–342 | TM 8 | ||||||
| 343–354 | Internal loop (4) | ||||||
| 355–372 | TM 9 | ||||||
| 373–386 | External loop (5) | ||||||
| CCAC gtga | ttag AGAA | 50 | |||||
| 5 | 86 | 382–410 (28) | 387–409 | TM 10 | |||
| ATGG gtaa | acag CCCG | 44 | |||||
| 6 | 314 | 411–450 (39) | 410–421 | Internal loop (5) | |||
| 422–444 | TM 11 | ||||||
| 445–450 | C-terminus (ext.) | ||||||
Exon/protein domain distribution and predicted protein structure
A domain is defined as an independent structural unit which can be found alone or in conjunction with other domains or repeats. The evolution of protein domains (and their conservation within genomes) is being increasingly studied as sequence data build up. Domains are often evolutionarily related and evolution of these proteins is likely to ensure conservation of ‘active’ domains (15). Even though the precise structure of a domain is not always known, it is still possible to define the boundaries, in many cases, from sequence data alone. Transporters and many other membrane proteins are made up of structurally defined domains such as TMs, substrate-binding domains and ‘helical hairpins’ (two TMs plus an external loop) (16,17). In addition to structural domains, functional domains (which may or may not correlate with structure) can also be predicted based on functional assays. To determine if we could recognize structural (and possibly functional) domains in the ENTs, we used the transmembrane prediction analysis program TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) to define domains in the predicted proteins. We then determined if there was any correlation with gene structure.
For the human ENTs, we found the distribution of the protein on the exons to be similar for hENT1 and hENT2 with between 9 and 67 amino acids per exon (Table 2, Supplementary Material). There was a marked correlation between the structural domains predicted by TMHMM 2.0 and exons. For example, exon 2 of hENT1 encodes amino acids 11–37 of the hENT1 protein, which correspond closely with the first predicted transmembrane region (TM 1, amino acids 13–35). In contrast, the exons of the hENT3 gene encoded between 28 and 208 amino acids, with the final exon (V) encompassing almost half (208 of 475) of the protein (Supplementary Material). The only domain which is split between two exons is the large intracellular loop (between TMs 6 and 7) and this structural arrangement is present in all three human ENTs.
The predicted protein structures of hENT1 and hENT2 derived from the gene sequence correlate exactly with the previously published data for these proteins (7,8). The predicted hENT3 protein sequence has three amino acids which are different from the previously published sequence (4), due to differences of four nucleotides in the genomic sequence (TPA: BK000392) compared to the mRNA sequence (AF326987). In the gene, the presence of a cytidine rather than a thymine (in the reported mRNA sequence) results in the presence of a serine at position 159 rather than a phenylalanine. A serine at this position would be identical to the mENT3 sequence (AAF326986) and also hENT2 and mENT2. In contrast, mENT1 and hENT1 possess an aspartic acid at this location. In addition, a guanine rather than an adenine (in the reported mRNA) results in a valine at position 240 rather than the isoleucine in the reported mRNA (AF326987). The change from a valine to an isoleucine is a conservative substitution and unlikely to have any effect on protein function. Finally, the genomic sequence predicts an isoleucine at position 323 (as a consequence of the presence of an adenine rather than guanine in the genomic sequence) rather than the valine in the reported mRNA. The presence of an isoleucine (rather than a valine) at this position is also consistent with the mENT3 sequence. Whether these variations are due to sequencing errors or underlying variability in the sequence within the population is not clear. The rest of the protein sequence is identical between the genomic DNA and the reported protein.
The Drosophila ENTs are similarly encoded by exons that correlate relatively closely to structural domains (Table 3, Supplementary Material), although more domains are encoded by fewer exons. (DmENT2 cannot be analyzed in this way due to its single exonic structure.) The DmENT1 and DmENT2 proteins are predicted to have 11 TMs and are more similar to each other than to DmENT3 (Fig. 4 and Table 5). Structurally, they are also predicted to be similar to the human ENTs despite the low sequence identity at the amino acid level (Table 5). This suggests that DmENT1 and DmENT2 have the prototypic ENT structure (Fig. 6). The structural topology of DmENT3 is less clear.
Figure 6.
Predicted topologies for members of the human, D.melanogaster and C.elegans ENT families. The analysis is based on TMHMM 2.0 as described in Materials and Methods. Red bars and peaks represent putative transmembrane domains. Blue lines represent putative intracellular portions of the protein; pink lines represent putative extracellular portions of the protein.
In general, the C.elegans ENT proteins are distributed onto exons in a structurally defined manner (Table 4 and Supplementary Material). Similar to DmENT1 and DmENT2, the N-terminal tail and TM 1 are found in exon 1 for CeENT1, CeENT2, CeENT5 and CeENT6. This domain is also located on one exon for CeENT3, although preceded by the exons encoding the non-transporter protein sequence. Except for the N-terminal region of the C.elegans ENTs and the overall exon/domain conservation, there appears to be relatively little similarity in overall protein/exon structure among the six C.elegans ENTs such that the domains are distributed among different exons for the different genes. Despite this variation in domain/exon distribution between C.elegans ENTs and the overall low sequence similarity at the protein level with both Drosophila and human ENTs (Table 5), the predicted topologies of the C.elegans ENTs are remarkably similar to those previously described (Fig. 6, Supplementary Material). In general, the C.elegans ENT proteins are predicted to have between nine and 11 TMs, with CeENT2, CeENT4 and CeENT5 in particular showing similar topology to the ENT1 and ENT2 proteins from humans and Drosophila. CeENT3 also shows an overall similar topology with the exception of an extended intracellular N-terminal region as a consequence of the non-transporter like protein sequence. CeENT6 appears to be truncated and the full-length protein may conform to the general ENT structure.
The striking similarity in predicted topology of the invertebrate and vertebrate proteins combined with the functional characterization that has occurred for some C.elegans ENTs and human ENTs suggests that a prototypic eukaryotic ENT model can be developed which consists of an integral membrane protein with an intracellular N-terminus, 11 TMs, a central intracellular loop (between TMs 6 and 7) and an extracellular C-terminus. This ENT prototypic structure has been highly conserved during evolution despite considerable changes in primary amino acid sequence.
DISCUSSION
We report in this paper the first complete and comprehensive analysis of the ENT gene family in three evolutionarily diverse taxa, Homo sapiens, D.melanogaster and C.elegans.
The presence of multiple ENTs appears to be a characteristic of most (but not all) eukaryotic organisms studied to date (5). The generation of gene families encoding similar proteins can occur by large-scale events such as duplication of the entire genome (polyploidization), as has been proposed for the teleost lineage (18), and by smaller regional duplications, such as unequal crossing over (19). In the case of the ENTs, polyploidization of an ancestral genome containing an ancestral ENT would produce two homologs. This event, followed by taxa-specific duplications, would be the most parsimonious explanation for our findings. Caenorhabditis elegans and Drosophila are proposed to have core proteomes of approximately the same size (8000–9000 proteins) (1). However, much of the genome of worms and flies consists of duplicated genes (1) and both the C.elegans and Drosophila genomes have been shown to contain regions of active local duplication (20), although worms appear to have twice as many local duplications compared to flies (1). These findings concur with the presence of twice as many ENTs in C.elegans. In both C.elegans and Drosophila, most of the gene duplications (70%) are on the same chromosomal element (1), as appears to be the case for DmENT1 and DmENT2 and some of the C.elegans ENTs.
Compared to the invertebrates, the human genome consists of many more genes and this increase has been proposed to be due to rapid and recent local gene duplications (21). The structural arrangement of hENT1 and hENT2 would support the concept that these genes are paralogs and have arisen relatively recently, as a consequence of a duplication event in the mammalian (rather than primate) evolutionary lineage, since the ENT1 and ENT2 forms are also present in other mammals. Both hENT1 and mENT1 (14) show considerable similarity in genomic organization, although there are differences in sizes of introns. This is consistent with the concept of more rapid evolution of non-coding regions of the gene (19). The ENT1/2 gene duplication and relocation must have occurred at least prior to the split between rodents and primates ∼100 000 000 years ago (22). It is also possible that this occurred earlier in the pre-mammalian vertebrate lineage. However, clarification of the evolutionary history of the vertebrate ENT1 and ENT2 isoforms awaits identification and analysis of ENTs from other mammals and from non-mammalian vertebrates.
While the genomic organization of hENT1 and hENT2 points to a recent duplication, the origin of hENT3 and its evolutionary relationship to the other human ENTs is not clear. It has a very different gene structure and the presence of a very large intron between exons I and II is unusual. Although a mouse ENT3 protein has been reported, ENT3 genes in other species have not yet been fully described. There is no clear evidence for specific ENT orthologs to hENT1, hENT2 and/or hENT3 in invertebrates based on genomic organization (i.e. DmENT1 is not the ortholog of hENT1). This lack of genomic conservation between vertebrates and invertebrates contrasts somewhat with findings for some other membrane proteins where gene structure is conserved (23).
Similar to human ENTs, local gene duplication events appear to have given rise to multiple paralogous ENT family members in C.elegans (most strikingly CeENT1 and CeENT2, where similar gene structure is still evident) and possibly in D.melanogaster (DmENT1 and DmENT2). Similarly, other organisms such as Arabidopsis and the parasitic protozoa (4,24) have undergone taxa-specific gene duplication events to generate additional paralagous ENTs, suggesting that this is the primary reason for multiple (i.e. greater than two) ENT isoforms in the eukaryotes.
Following gene duplication, subsequent diversification can occur, as appears to have happened with many ENTs (e.g. hENT1 and hENT2, CeENT1 and CeENT2, DmENT1 and DmENT2). Gene duplication (regardless of mechanism) is an important and powerful evolutionary mechanism that can give rise to families of structurally and often functionally related genes (19). Newly duplicated genes can undergo sequence divergence while the progenitor maintains the original function, resulting in related genes with slightly different functions or substrate preferences. While many of the identified ENTs remain to be functionally characterized, some homologs have already been shown to have different substrate preferences, as seen with hENT1/2 and CeENT1/2 (4,9).
Although gene structure and protein similarity suggest that hENT1 and hENT2 are relatively recent gene duplications, they are located in different chromosomes. For ENTs located on the same chromosome (DmENT1 and DmENT2 on chromosome 2, CeENT1 and CeENT4 on chromosome IV and CeENT2 and CeENT5 on chromosome X), there is considerable genetic distance between homologs. This contrasts with recent observations that membrane proteins are often found in tandem clusters within genomes (25) and may represent one of the genomic differences between transporters with high numbers of genes (e.g. ABC transporters) and those encoded by few genes, such as the ENTs.
Despite various duplication events, overall there are relatively few ENT genes compared to other transporter families such as the GLUTs, where there are 12 genes in humans (26), or the ABC transporters, where there are numerous genes in both humans and Drosophila (27). The underlying reasons for such differences are not clear, although the human ENT family appears to use an alternative mechanism for the generation of protein diversity by the production of splice variants (seen in hENT2). Since all of the hENT2 splice variants contain the apparent functional ENT core of TM 3–6 (4,28), they are potentially active transporters, although their physiological relevance is unknown. This ‘combinatorial’ approach to increasing protein diversity appears to be a hallmark of the human genome (1,29) and splice variants for other membrane proteins (the GABAB receptor) have been previously described (23).
A requirement of the ‘combinatorial’ approach to generating protein diversity is the presence of functional units within protein that can be used in various arrangements. In some proteins, the presence of functional units is reflected in the overall gene organization. For instance, in the hemocyanin proteins, individual functional units are encoded by separate exons and this arrangement is evolutionarily conserved (30). In the absence of crystallographic data to show three-dimensional structures (and therefore predict functional units), protein domains can be identified based on predicted topologies which may correlate or contribute to functionally important regions of the protein. While gene structure has not been clearly conserved (in terms of number of exons/introns, etc.) for the ENTs and functional units are unknown, there is some evidence that domains correlate with exons in all phyla. Individual domains tend to be encoded by separate exons (hENT1 and hENT2) or are grouped together on an exon and are rarely split by intron regions (e.g. hENT3 and invertebrate ENTs). Whether these structural domains correlate with function is not clear. The substrate translocation domain is proposed to be within TM 3–6 and a number of amino acids have been implicated in other functions (4,28,31). Future work to identify ENT functional units, such as recognition and discrimination domains (i.e. purine or pyrimidine) and translocation domains, is required.
An increased understanding of the structure and function of human ENTs is critical in determining their role in nucleoside analog drug uptake, efficacy and resistance. Despite the widespread use of nucleoside analogs in chemotherapy, nothing is known about the genetic regulation of any human ENT. Differential tissue expression of hENT1 and hENT2 has been previously described (6,7), suggesting spatial transcriptional regulation. In addition, there are striking differences between expression levels of ENTs in different individuals in both normal and matched tumor tissue (6), suggesting additional temporal regulation. These observations highlight the need for an improved understanding of the transcriptional regulation of human ENTs correlated with an awareness of their central role in determining the clinical efficacy of nucleoside analog chemotherapeutic drugs. In this paper, we have identified the complete gene structure, including putative promoter regions, of all reported human ENTs. Our work lays the foundation for future analysis of transcriptional regulation of these clinically important proteins. Promoter studies on these genes are in their infancy (13) and the reasons for tissue and individual specific and highly variable expression are not yet clear.
We have found striking similarities and differences between human, Drosophila and C.elegans with respect to their ENT genes. There is the possibility that an additional novel human ENT exists (possibly of very low sequence similarity which has not been detected by our analyses to date), highlighting the need for caution when using significant sequence similarity as a defining feature of comparative genomic studies. Identification of all members of the ENT family within these evolutionarily diverse organisms demonstrates the need and lays the groundwork for more detailed phylogenetic analyses using other organisms which may be better representatives of evolutionary history. Within a single organism, identification and characterization of the entire ENT protein family provides a framework on which to build additional studies on the impact of transcriptional and post-transcriptional processing events on total ENT functional diversity in each proteome.
In conclusion, the gene structures and primary amino acid sequences of the ENTs described here are relatively poorly conserved between taxa. However, the predicted topologies are remarkably conserved, suggesting that sequence similarity comparisons alone can be insufficient in determining evolutionary, structural and/or functional relationships between proteins both within and between taxa. Despite the extensive diversity in sequence of the ENTs or putative ENTs described in this paper, they are clearly unified by common structural motifs. Our data highlight the need for a variety of approaches to be used in improving our understanding of this group of proteins. Identification (by functional or bioinformatics methods) of additional ENT members in diverse organisms and, in particular, crystallographic analyses (to obtain three-dimensional data) are essential in future studies of the ENTs.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Natural Science and Engineering Research Council (NSERC, 203397-98) and National Cancer Institutes (NCIC, 011070) of Canada to I.R.C.
DDBJ/EMBL/GenBank accession nos AF495730, BK000392
REFERENCES
- 1.Rubin G., Yandell,M.D., Wortmann,J.R., Gabor Miklos,G.L., Nelson,C.R., Hariharan,I.K. Fortini,M.E., Li,P.W., Apweiler,R., Fleischmann,W. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maglich J.M., Sluder,A., Guan,X., Shi,Y., McKee,D.D., Carrick,K., Kamdar,K., Willson,T.M. and Moore,J.T. (2001) Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol., 2, 0029.1–0029.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane R.P., Cutforth,T., Young,J., Athanasiou,M., Friedman,C., Rowen,L., Evans,G., Axel,R., Hood,L. and Trask,B.J. (2001) Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc. Natl Acad. Sci. USA, 98, 7390–7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyde R.J., Cass,C.E., Young,J.D. and Baldwin,S.A. (2001) The ENT family of eukaryote nucleoside and nucleobase transporters: recent advances in the investigation of structure/function relationships and the identification of novel isoforms. Mol. Membr. Biol., 18, 53–63. [PubMed] [Google Scholar]
- 5.Acimovic Y and Coe,I.R. (2002) Molecular evolution of the equilibrative nucleoside transporter family: identification of novel family members in prokaryotes and eukaryotes. Mol. Biol. Evol., in press. [DOI] [PubMed] [Google Scholar]
- 6.Pennycooke M., Chaudary,N., Shuralyova,I., Zhang,Y. and Coe,I.R. (2001) Differential expression of human nucleoside transporters in normal and tumor tissue. Biochem. Biophys. Res. Commun., 280, 951–959. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths M., Beaumont,N., Yao,S.Y.M., Sundaram,M., Boumah,C.E., Davies,A., Kwong,F.Y.P., Coe,I.R., Cass,C.E., Young,J.D. and Baldwin,S.A. (1997) Cloning of a human nucleoside transporter implicated in the cellular uptake of adenosine and chemotherapeutic drugs. Nature Med., 3, 89–93. [DOI] [PubMed] [Google Scholar]
- 8.Crawford C.R., Patel,D.H., Naeve,C. and Belt,J.A. (1998) Cloning of the human equilibrative, nitrobenzylmercaptopurine riboside (NBMPR)-insensitive nucleoside transporter ei by functional expression in a transport-deficient cell line. J. Biol. Chem., 273, 5288–5293. [DOI] [PubMed] [Google Scholar]
- 9.Appleford P.J., Griffiths. M., Chomey,E.G., Yao,S.Y.M., MacGregor,D., Isaac,R.E., Hope,I.A., Coates,D., Cass,C.E., Young,J.D. and Baldwin,S.A. (2001) Caenorhabditis elegans as a model organism in the study of nucleoside transporters. In Abstracts of the International C.elegans Meeting, abstract 952. www.wormbase.org.
- 10.Coe I.R., Griffiths,M., Young,J.D., Baldwin,S.A. and Cass,C.E. (1997) Assignment of the human equilibrative nucleoside transporter (hENT1) to 6p21.1–21.2. Genomics, 45, 459–460. [DOI] [PubMed] [Google Scholar]
- 11.Williams J.B., Rexer,B., Sirripurapu,S., John,S., Goldstein,R., Phillips,J.A., Haley,L.L., Sait,S.N.J., Shows,T.B., Smith,C.M. and Gerhard,D.S. (1997) The human HNP36 gene is localized to chromosome 11q13 and produces alternative transcripts that are not mutated in multiple endocrine neoplasia, type 1 (MEN 1) syndrome. Genomics, 42, 325–330. [DOI] [PubMed] [Google Scholar]
- 12.Szkotak A.J., Ng,A.M.L., Sawicka,J., Baldwin,S.A., Man,S.F.P., Cass,C.E., Young,J.D. and Duszyk,M. (2001) Regulation of K+ current in human airway epithelial cells by exogenous and autocrine adenosine. Am. J. Physiol. Cell Physiol., 281, C1991–C2002. [DOI] [PubMed] [Google Scholar]
- 13.Abdulla P., Sankar,N. and Coe,I.R. (2002) Isolation and characterization of the human equilibrative nucleoside transporter genes, hENT1, hENT2 and hENT3. Proc. AACR, 43, abstract 506. Cadmus Journal Services, Linthicum, MD. [Google Scholar]
- 14.Choi D.-P., Handa,M., Young,H., Gordon,A.S., Diamond,I. and Messing,R.O. (2000) Genomic organization and expression of the mouse equilibrative, nitrobenzyl-sensitive nucleoside transporter 1 (ENT1) gene. Biochem. Biophys. Res. Commun., 277, 200–208. [DOI] [PubMed] [Google Scholar]
- 15.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 16.Gafvelin G. and von Heijne,G. (1994) Topological “frustration” in multispanning E. coli inner membrane proteins. Cell, 77, 401–412. [DOI] [PubMed] [Google Scholar]
- 17.Gafvelin G., Sakaaguchi,M., Andersson,H. and von Heijne,G. (1997) Topological rules for membrane protein assembly in eukaryotic cells. J. Biol. Chem., 272, 6119–6127. [DOI] [PubMed] [Google Scholar]
- 18.Taylor J.S. and Brinkmann,H. (2001) 2R or not 2R? Trends Genet., 17, 488–489. [DOI] [PubMed] [Google Scholar]
- 19.Graur D. and Li,W.-H. (2000) Fundamentals of Molecular Evolution, 2nd Edn. Sinauer Associates, Sunderland, MA.
- 20.Friedman R. and Hughes,A.L. (2001) Gene duplication and the structure of eukaryotic genomes. Genome Res., 11, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman R. and Hughes,A.L. (2001) Pattern and timing of gene duplication in animal genomes. Genome Res., 11, 1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S. and Hedges,S.B. (1998) A molecular timescale for vertebrate evolution. Nature, 392, 917–920. [DOI] [PubMed] [Google Scholar]
- 23.Martin S.C., Russek,S.J. and Farb,D.H. (2001) Human GABABR genomic structure: evidence for splice variants in GABABR1 but not GABABR2. Gene, 278, 63–79. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez M.A., Tryon,R., Green,J., Boor,I. and Landfear,S.M. (2002) Six related nucleoside/nucleobase transporters from Trypanosoma brucei exhibit distinct biochemical function. J. Biol. Chem., 277, 21499–21504. [DOI] [PubMed] [Google Scholar]
- 25.Kihara D. and Kaneshisa,M. (2000) Tandem clusters of membrane proteins in complete genome sequences. Genome Res., 10, 731–743. [DOI] [PubMed] [Google Scholar]
- 26.Joost H.G. and Thorens,B. (2001) The extended GLUT-family of sugar/polyol transport facilitators: nomenclature, sequence characteristics and potential function of its novel members. Mol. Membr. Biol., 18, 247–256. [DOI] [PubMed] [Google Scholar]
- 27.Dean M., Rzhetsky A. and Allikmets,R. (2001) The human ATP-binding cassette (ABC) transporter superfamily. Genome Res., 11, 1156–1166. [DOI] [PubMed] [Google Scholar]
- 28.SenGupta D.J., Lum,P.Y., Lai,Y., Shubochkina,E., Bakken,A.H., Schneider,G. and Unadkat,J.D. (2002) A single glycine mutation in the equilibrative nucleoside transporter gene, hENT1, alters nucleoside transport activity and sensitivity to nitrobenzylthioinosine. Biochemistry, 41, 1512–1519. [DOI] [PubMed] [Google Scholar]
- 29.Croft L., Schandorff,S., Clarek,F., Burrage,K., Arctander,P. and Mattick,J.S. (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nature Genet., 24, 340–341. [DOI] [PubMed] [Google Scholar]
- 30.Lieb B., Altenhein,B., Markl,J., Vincent,A., van Olden,E., van Holde,K.E. and Miller,K.I. (2001) Structures of two molluscan hemocyanin genes: significance for gene evolution. Proc. Natl Acad. Sci. USA, 98, 4546–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visser F., Vickers,M.F., Ng,A.M.L., Baldwin,S.A., Young,J.D. and Cass,C.E. (2002) Mutation of residue 33 of human equilibrative nucleoside transporters alters sensitivity to inhibition of transport by dilazep and dipyridamole. J. Biol. Chem., 277, 395–401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.