Abstract
We have sequenced the genome of Shigella flexneri serotype 2a, the most prevalent species and serotype that causes bacillary dysentery or shigellosis in man. The whole genome is composed of a 4 607 203 bp chromosome and a 221 618 bp virulence plasmid, designated pCP301. While the plasmid shows minor divergence from that sequenced in serotype 5a, striking characteristics of the chromosome have been revealed. The S.flexneri chromosome has, astonishingly, 314 IS elements, more than 7-fold over those possessed by its close relatives, the non-pathogenic K12 strain and enterohemorrhagic O157:H7 strain of Escherichia coli. There are 13 translocations and inversions compared with the E.coli sequences, all involve a segment larger than 5 kb, and most are associated with deletions or acquired DNA sequences, of which several are likely to be bacteriophage-transmitted pathogenicity islands. Furthermore, S.flexneri, resembling another human-restricted enteric pathogen, Salmonella typhi, also has hundreds of pseudogenes compared with the E.coli strains. All of these could be subjected to investigations towards novel preventative and treatment strategies against shigellosis.
INTRODUCTION
Shigella species are Gram-negative, non-sporulating, facultative anaerobes causing bacillary dysentery or shigellosis in man with estimated annual episodes of 160 million and 1.1 million deaths, most of which are children under 5 years old in developing countries (1). In China, more than 10 million cases are estimated per annum, of which 50–70% are caused by Shigella flexneri serotype 2a and most are associated with epidemic and pandemic shigellosis (2). Shigella are highly invasive in the colon and the rectum, and are able to proliferate in the host cell cytoplasm, triggering an inflammatory reaction. The clinical manifestations of Shigella infection vary from short-lasting watery diarrhea to acute inflammatory bowel disease characterized by fever, intestinal cramp and bloody diarrhea with mucopurulent feces (1). Since the current preventive and treatment strategies are found to be inadequate, the World Health Organization has placed an anti-Shigella vaccine as a priority (3).
Shigella was recognized as the etiologic agent for bacillary dysentery in the 1890s, and was adopted as a genus in the 1950s and subgrouped into four species: S.dysenteriae, S.flexneri, S.boydii and S.sonnei (4). However, a recent genetic study argues that Shigella emerged from multiple independent origins of Escherichia coli 35 000–270 000 years ago and may not constitute a genus (5). Genes on a virulence plasmid encode the primary virulence determinants, including the invasion plasmid antigens (Ipa) and their devoted Mxi-Spa type III secretion apparatus, but many chromosomal loci are also virulence required (6). Thus, the defining point for Shigella to evolve from E.coli must be the acquisition of the precursor of the current-day virulence plasmid carrying genes necessary for the bacteria to invade and access the host cell cytoplasm. This is a niche unique amongst the enteric pathogens with the exception of enteroinvasive E.coli that also possesses the virulence plasmid causing similar pathogenic characteristics (4). Subsequent evolution of the chromosome, however, enables the full expression of virulence. Hence, despite the fact that plasmid sequences from serotype 5a have become available (7,8), we felt it necessary to sequence the whole genome of S.flexneri 2a, the most prevalent species and serotype. Particularly, the expression of virulence depends on a complex regulation mechanism that involves dialog between the chromosome and the virulence plasmid (9), and a better understanding of this requires the availability of the whole genome sequence. Indeed, though the virulence plasmid from serotype 2a has minor divergence from that of serytype 5a, we have revealed the highly volatile and dynamic nature of the Shigella chromosome in comparison with the genomes of the non-pathogenic K12 strain and the enterohemorrhagic O157:H7 strain of E.coli (10,11). Furthermore, we have uncovered many chromosomal loci that potentially contribute to virulence in addition to those identified by the classic genetic studies (6).
MATERIALS AND METHODS
Shigella flexneri 2a strain and growth conditions
Shigella flexneri strain 301 (abbreviated Sf301), which we sequenced, was isolated from a patient with severe acute clinical manifestations of shigellosis in the Changping District, Beijing, in 1984, and has since been used as a reference strain for S.flexneri in China. The strain was routinely grown at 37°C overnight on tryptic soy agar containing 0.01% Congo red. Red colonies were inoculated into tryptic soy broth and grown to stationary phase at 37°C for isolating plasmid and chromosomal DNAs.
Shotgun sequencing and sequence assembly
The plasmid and the chromosomal libraries were separately constructed using pBluescript KS(–) (Strategene) as vectors. Approximately 48 000 clones were sequenced from both ends using the big-dye kit (ABI) and ABI377 or ABI3700 automated sequencers, giving rise to 10 times coverage of the genome.
Sequences were assembled initially using the phred/phrap program (12) when the sequence coverage was ∼4-fold over the estimated size of the genome. The program was run with optimized parameters and the quality score was set to ≥20. Further assembly was carried out repeatedly using the same program when more sequences were obtained. When 100 500 sequences were assembled into 318 contigs, the Consed program was used for sequence finishing (13). Gaps among contigs were closed either by primer walking on selected clones, which were identified by analysis on the forward and the reversed links between contigs using the perl/Tk algorithm, or by sequencing the DNA amplicons generated by polymerase chain reaction (PCR).
Prediction of open reading frames (ORFs) and identification of gene families
Glimmer 2.0, a program that searches for protein coding regions, was used to identify those ORFs possessing more than 30 consecutive codons (14). Overlapping and closely clustered ORFs were manually inspected. Predicted polypeptide sequences were used to search the non-redundant protein database with BLASTP, and the clusters of orthologous groups of proteins (COGs) database was used to identify families to which predicted proteins are related (15).
Mobile elements and repetitive sequences were identified using pair-wise comparison. tRNA sequences were identified by the program tRNAscan-SE (16). Repetitive regions were defined as those that have at least 200 bp with the significance of e–10 by BLASTN against the Sf301 genome itself and known IS databases. Sequence annotation and graphs of the circular and linear genomic maps were prepared using a newly developed Perl-Script tool kit (available at ftp://ftp.chgb.org.cn/pub/).
Whole genomic comparison with E.coli K12 MG1655 (accession no. U00096) and O157 EDL933 (accession no. AE00517H) was performed using the GenomeComp program (J.Yang, J.Wang, Q.Jin, Y.Shen, Z.Yao and R.Chen, manuscript in preparation).
Accession of the genome sequence
The accession numbers for Sf301 chromosome and plasmid pCP301 are AE005674 and AF386526, respectively, in GenBank.
RESULTS AND DISCUSSION
General features of the genome
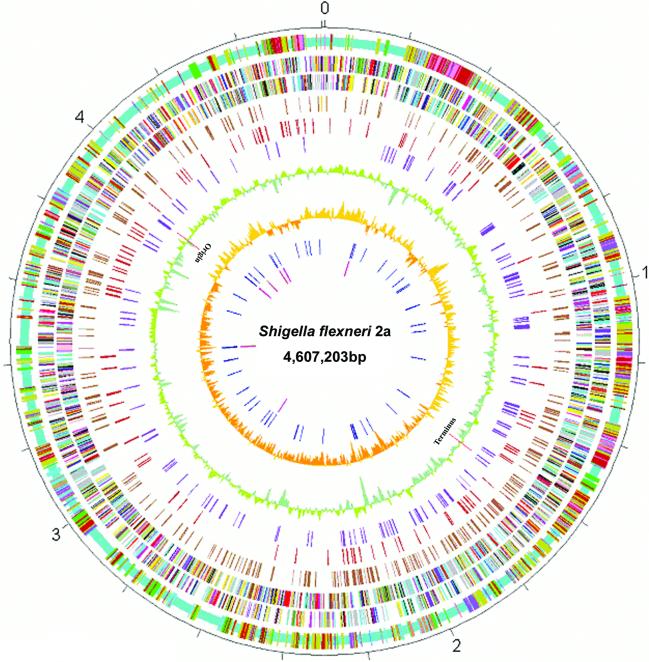
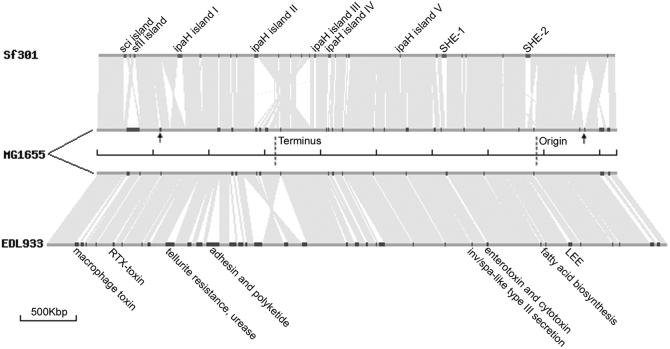
The primary features of the Sf301 genome are summarized in Table 1 and graphically viewed in Figures 1 and 2. The whole genome of Sf301 is composed of a 4 607 203 bp chromosome and a 221 618 bp virulence plasmid, designated pCP301. The chromosome shares a common ‘backbone’ sequence ∼3.9 Mb with those of E.coli K12 (MG1655) (10) and O157 (EDL933) (11), which is essentially collinear. However, the backbone sequence is interrupted by numerous segments of K12-, O157- and Shigella-specific DNA, designated ‘K-islands’ (KIs), ‘O-islands’ (OIs) and ‘S-islands’ (SIs), respectively (Fig. 1, circle 1). The co-linearity is also broken by numerous inversions and translocations compared with the E.coli sequences, 13 of which involve DNA segments >5 kb and are all bordered by IS elements and mostly associated with deletions or SIs (Fig. 2). All of these were confirmed by subsequent PCR sequencing of the junctions of each of the translocations and inversions. In the case of EDL933, there is only one inversion near the replication terminus with respect to K12 as noted previously (Fig. 2) (11). The dynamic gene shifts of the Sf301 chromosome are in contrast to the conserved genetic maps of E.coli K12, Salmonella typhimurium, other E.coli strains, other Salmonella spp., Klebsiella pneumoniae and many other enterics (17). However, there is no evidence for gene drift mediated by recombination between rRNA operons as observed in S.typhi (18) and in some Shigella strains (19). All the rRNA operons of Sf301 fall in approximately the same loci as those of E.coli (10,11). Natural selection that optimizes all promoters has operated to conserve genetic maps among enterics (17). A gradient of gene dosage generated from rapid chromosomal replication constrains genes to certain locations relative to the replication origin, and actively transcribed genes have a strong bias to be transcribed away from the origin, whereas weakly transcribed genes are evenly orientated (20). Genetic rearrangements alter the locations, orientations, and the coding strand (in the cases of inversions) with respect to the origin of replication, possibly changing the amount of transcription of many genes. This in turn may affect their dosage, and in some cases impair growth (21,22). Hence, the changed genetic map suggests that S.flexneri may have re-optimized its promoters to cope with selection pressures in the unique intracellular or ex vivo environments.
Table 1. General features of the Sf301 genome compared with genomes of E.coli K12 and 0157, and the virulence plasmid, pWR501, from S.flexneri M90T 5a.
| Chromosome | Sf301 | MG1655a | EDL933b |
|---|---|---|---|
| Total length (bp) | 4 607 203 | 4 639 221 | 5 528 445 |
| No. of total ORFs | 4434 | 4289 | 5349 |
| Average length of ORFs (bp) | 891 | 954 | 905 |
| Percentage of coding sequence (%) | 80.4 | 87.8 | 87.1 |
| G + C content | |||
| Total genome (%) | 50.89 | 50.79 | 50.40 |
| Protein coding regions (%) | 51.95 | 51.85 | 51.51 |
| RNA genes (%) | 54.79 | 54.84 | 54.88 |
| Intergenic regions (%) | 46.07 | 42.28 | 42.76 |
| Ribosomal RNA | |||
| No. of 16S | 7 | 7 | 7 |
| No. of 23S | 7 | 7 | 7 |
| No. of 5S | 8 | 8 | 8 |
| No. of transfer RNA | 97 | 92 | 93 |
| No. of tmRNA | 1 | 1 | 1 |
| No. of non-classical RNA | 9 | 5 | 5 |
| Translocations and inversionsc | 13 | – | 1 |
| IS elements | 314 | 39 | 40 |
| Of which partial copies | 67 | 7 | 19 |
| Plasmid |
pCP301 |
pWR501d |
|
| Total length (bp) | 221 618 | 221 851 | |
| No. of total ORFs | 267 | 293 | |
| Average length of ORFs (bp) | 658 | 636 | |
| Percentage of coding sequence | 76.24 | 82.09 | |
| G + C content | |||
| Total (%) | 45.77 | 46.36 | |
| Coding regions (%) | 46.13 | 46.95 | |
| Intergenic regions (%) | 44.59 | 43.69 | |
| IS elements | 88 | 92 | |
| Of which partial copies | 62 | 69 |
Figure 1.
Circular map of Sf301 chromosome compared with those of E.coli K12 MG1655 and 0157 EDL933. Outer scale is marked in 200 kb. Circles range from 1 (outer circle) to 10 (inner circle). Circle 1, shared collinear backbone (light blue), and Shigella islands (SI) (cyan), K12 islands (KI) (green) and 0157 islands (OI) (tan); co-localized SI and KI (dark salmon); co-localized SI and OI (purple); co-localized KI and OI (dark blue); and co-localized SI, KI and OI (deep pink). Circles 2 and 3, ORFs encoded by leading and lagging strands with color code for functions: salmon, translation, ribosomal structure and biogenesis; light blue, transcription; cyan, DNA replication, recombination and repair; turquoise, cell division; deep pink, posttranslational modification, protein turnover and chaperones; olive drab, cell envelope biogenesis; purple, cell motility and secretion; forest green, inorganic ion transport and metabolism; magenta, signal transduction; red, energy production; sienna, carbohydrate transport and metabolism; yellow, amino acid transport; orange, nucleotide transport and metabolism; gold, co-enzyme transport and metabolism; dark blue, lipid metabolism; blue, secondary metabolites, transport and catabolism; gray, general function prediction only; black, function unclassified or unknown. Circle 4, distribution of pseudogenes. Circles 5 and 6, distribution of IS1 and other IS elements, respectively. Circle 7, G + C content with a window size of 10 kb. Circle 8, GC bias (G – C/G + C). Circle 9, distribution of tRNA genes. Circle 10, distribution of rrn operons. The replication origin and terminus are indicated.
Figure 2.
Schematic representation of translocations and inversions, and strain-specific islands (to scale). Chromosomes are represented by dark gray lines as indicated. Replication origin and terminus of MG1655 are indicated. The respective loci of Sf301 and EDL933 are at approximately the same positions. Regions in light gray indicate homologous sequences between paired chromosomes and triangular non-filling regions indicate the presence of inversions. Translocations are hardly visible because of the scale used. Regions in red, black and blue on the chromosomes represent SI, KI and OI, respectively. The known PAI among SIs and OIs are indicated. SIs with implications for a role in virulence are also indicated. Arrows indicate the KIs harboring ompT (left) and cadA (right) whose deletions are crucial for Shigella virulence.
The Shigella islands
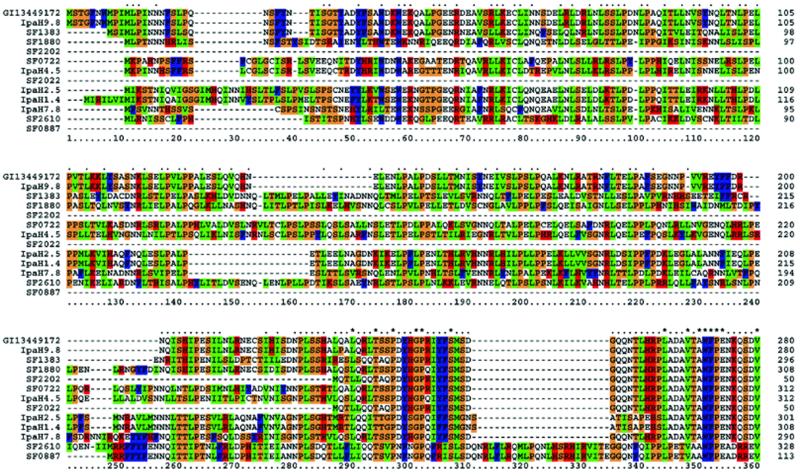
Sf301 has in total 64 SIs with sizes >1 kb, all of which are numbered and detailed in the ‘linear map 1’ (Supplementary Material). Among them, several, including the previously identified pathogenicity islands (PAIs) SHE-1 (23) and SHE-2 (24), have implications in virulence. Strikingly, there are seven ipaH genes, five of which are located in five large SIs, designated as ipaH islands 1–5 (Fig. 2). All five ipaH genes in the islands are next to the genes that potentially encode proteins sharing 73–76% identity with a 188 amino acid hypothetical protein of unknown function from Salmonella bacteriophage P27 (accession no. NP_543109). The majority of the remaining genes in the ipaH islands share homologies with genes of different phages including those identified in the genome of O157 EDL933. But the overall gene contents and organizations in all 5 ipaH islands have little similarity. This suggests that the chromosomal ipaH genes were originally linked with phage P27 and subsequently transmitted to S.flexneri by different phages. The plasmid, pCP301, carries five ipaH genes, termed ipaH9.8, ipaH7.8, ipaH4.5, ipaH2.5 and ipaH1.4, at approximately the same loci as those in pWR100 and pWR501 from serotype 5a (7,8). None of these is next to the genes of the phage P27 paralogs, suggesting that they came from different sources or, alternatively, were transmitted to the plasmid via different vehicles. The pWR501-borne ipaH7.8 is involved in the escape of Shigella from phagocytic vacuoles in the macrophages (25), but other ipaH genes have not been assigned a function. However, there is evidence that S.flexneri expresses more IpaH9.8 within host cells, and the proteins penetrate the host cell nuclei (26). This, and the fact that all IpaH proteins have a leucine-rich repeat region found in a diverse group of proteins from bacteria and eukaryotes (27), implies that IpaH might be involved in manipulating host gene expression. Alignment of all IpaH proteins indicates that they have identical C-terminal, but variable N-terminal, halves (Fig. 3). This suggests that they may interact with different host substrates, but exert similar functions.
Figure 3.
CLUSTALW amino acid sequence alignment of N-terminal halves of IpaH proteins identified in Sf301. IpaH9.8 of pWR501 (gi_13449172) serves as a reference on the top. The most homologous IpaH9.8 from pCP301 is placed in the second, and other IpaH family members are arranged in line with their homology to IpaH9.8. The consensus line displayed above the aligned sequences depicts identical amino acids as asterisks, with conserved residues shown as dots.
In ipaH island 2, four consecutive genes, similar to the Salmonella sitABCD and the Yersinea yfe operons (28), may encode proteins required for iron uptake. Since the Salmonella sitABCD can complement the growth in iron-restricted medium of an enterobactin-deficient E.coli, a role of the Salmonella sit-like (SSL) system in iron uptake is implicated. Iron uptake mechanisms have undergone complicated adjustments in S.flexneri. On the one hand, the enterobactin system is impaired due to the presence of stop codons in fepE, fhuE and entC genes (see Table 3), and on the other hand, the SSL system has been introduced, and additionally, SHE-2 encodes an aerobactin system (24). In some strains the E.coli fec enterobactin system is re-introduced along with so-called Shigella resistance locus PAI within a multiple resistance deletable element (MRDE) (29). However, MRDE is not present in Sf301.
Table 3. Pseudogenes with known functions identified in Sf301 genome.
| Pathway | Mutation | Description |
|---|---|---|
| Carbohydrate metabolism | ||
| araA | Stop codon | l-Arabinose isomerase; arabinose catabolism |
| ugd | Stop codon | UDP-glucose 6-dehydrogenase; colanic acid synthesis |
| fucK | Stop codon | l-Funulokinase, fucose catabolism |
| glcD | Stop codon | Glycolate oxidase subunit D |
| xylA | Stop codon | d-Xylose isomerase; d-xylose catabolism and d-glucose conversion |
| aceB | Stop codon | Malate synthetase A; glyoxylate bypass |
| dgoA | Stop codon | d-Galactonate hydro-lyase; galactonate catabolism |
| fdhFa | Stop codon | Formate dehydrogenase-H; anaerobic respiration |
| zwf | Stop codon | G6PD; oxidative branch of pentose phosphate pathway |
| Energy metabolism | ||
| cyoB | Stop codon | Cytochrome o ubiquinol oxidase subunit I; active under high oxygen growth conditions |
| cyoA | Truncation | Cytochrome o ubiquinol oxidase subunit II; as cuoB |
| acs | Stop codon | Acetyl-CoA synthetase; scavenging acetate |
| hyfB | Stop codon | Hydrogenase 4 subunit; anaerobic respiration |
| narZ | Stop codon | NRZ; anaerobic terminal electron acceptor |
| torA | Stop codon | Trimethylamine N-oxide reductase subunit; electron acceptor (anaerobic respiration) |
| torD | Insertion | Chaperone of TorA; preventing TorA degradation |
| Lipid metabolism | ||
| hcaD | Stop codon | Ferredoxin reductase; utilization of aromatic acids |
| Amino acid metabolism | ||
| speF | Stop codon | Ornithine decarboxylase isozyme; putrescine synthesis |
| speG | Frame shift | Spermidine acetyltransferase; polyamine synthesis |
| nadB | Stop codon | Quinolinate thynthetase B; pyridine synthesis |
| gabD | Stop codon | Succinate-semialdehyde dehydrogenase; aminobutyrate catabolism |
| mtgA | Frame shift | Peptidoglycan enzyme; cell wall formation |
| metA | Truncation | Homoserine transsuccinylase; methionine synthesis |
| cstC | Stop codon | Acetylornithine transaminase; arginine catabolism |
| Cofactors and vitamins | ||
| nfnB | Insertion | Dihydropteridine reductase; recycling the quinoid dihydrobiopterin cofactor by reducing it |
| lhr | Stop codon | ATP-dependent helicase, dispensable |
| lplA | Frame shift | Lipoate-protein ligase A; ligation of lipoyl to apoprotein |
| Complex lipids | ||
| gldA | Stop codon | Glycerol dehydrogenase; glycerol dissimilation |
| Complex carbohydrates | ||
| ycjM | Insertion | Putative polysaccharide hydrolase |
| otsA | Truncation | Trehalose-6-phosphate synthase; response to high osmolarity |
| aceK | Stop codon | Isocitrate dehydrogenase kinase/phosphatase; control flux between the TCA cycle and the glyoxylate bypass |
| Translation | ||
| prfB | Stop codon | Peptide chain release factor RF-2 |
| Transport | ||
| araF | Stop codon | l-Arabinose-binding periplasmic protein |
| cysW | Stop codon | Sulfate transport system permease W protein |
| yhdX | Truncation | Permease; putative amino acid ABC transporter |
| ugpC | Insertion | ATP-transporter; glycerol-3-phosphate uptake |
| rbsA | Insertion | ATP-biding component; d-ribose transport |
| rbsB | Stop codon | ABC transporter; d-ribose periplasmic binding protein |
| glvG | Frame shift | 6-Phospho-β-glucosidase; arbutin fermentation |
| ptsA | Stop codon | PEP-protein phosphotransferase system enzyme I |
| yphF | Stop codon | ABC transporter; periplasmic binding |
| Signal transduction | ||
| citB | Truncation | Regulator (paired with citR); citrate fermentation |
| kdpE | Stop codon | Regulator of the kdp operon; potassium transport |
| kdpD | Stop codon | Sensor of the kdpDE system; potassium transport |
| narQ | Stop codon | Nitrate/nitrite sensor protein; acts on NarL/NarP |
| arp | Stop codon | Regulator of acetyl CoA synthetase |
| malT | Stop codon | Positive regulator of mal operon |
| Cell motility | ||
| fliA | Frame shift | σ28 for flagellar operons |
| flgF | Stop codon | Cell-proximal portion of basal-body rod |
| flgK | Stop codon | Hook-filament junction protein 1 |
| flgL | Stop codon | Hook-filament junction protein |
| fliF | Stop codon | Basal-body MS-ring and collar protein |
| fliJ | Truncation | FliJ protein |
| flhA | Stop codon | Export of flagellar proteins |
| Unassigned enzymes | ||
| tesA | Stop codon | Acyl-CoA thioesterase I; hydrolyzes long chain acyl thioesters |
| pphA | Stop codon | Protein phosphatase 1; modulates phosphoproteins signaling protein misfolding |
| pphB | Stop codon | Removal of a phosphate group attached to serine or threonine residue; signaling protein misfolding through cpxRA system |
| Unassigned non-enzymes | ||
| yaaJ | Stop codon | Transport protein; sodium/alanine symporter |
| nfrA | Stop codon | Omp; bacteriophage N4 receptor |
| csgG | Stop codon | Transporter; curli assembly |
| csgA | Insertion | Curlin major subunit; coiled surface structures |
| fepE | Stop codon | Transporter; ferric enterobactin (enterochelin) |
| fhuE | Stop codon | Omp; receptor for ferric iron uptake |
| entC | Stop codon | Isochorismate synthase; enterobactin biosynthesis |
| hlyE | Stop codon | Hemolysin E; hemolytic to sheep blood |
| hslJ | Truncation | Heat shock protein HslJ |
| uidB | Truncation | Transporter; specific to α- and β-glucuronides |
| celD | Insertion | Negative regulator of cel operon (cryptic); ferment cellobiose, arbutin and salicin |
| molR | Insertion | Molybdate metabolism regulator, first fragment |
| molR_2 | Stop codon | Molybdate metabolism regulator, fragment 2 |
| cirA | Stop codon | Porin and receptor; colicin I uptake |
| focB | Frame shift | Formate transporter (formate channel 2) |
| emrA | Stop codon | Multidrug resistance secretion protein |
| ppdA | Frame shift | Prepilin peptidase dependent protein A |
| glcF | Frame shift | Glycolate oxidase iron–sulfur subunit; ferridoxin related |
| aer | Stop codon | Aerotaxis sensor receptor; transducing signals for aerotaxis |
| ompG | Truncation | Outer membrane protein; forms large channels |
| yaeG | Stop codon | Regulator of d-galactarate, d-glucarate and d-glycerate metabolism |
| nagD | Stop codon | N-Acetyleglucosamine metabolism |
| fimD | Insertion | Export and assembly of type 1 fimbriae |
afdhF has a stop codon (UAA) in addition to the stop codon UGA used for introducing selenocysteine.
Two other SIs are worthy of mention, the sci and the SfII islands (Fig. 2). The sci island is 22 789 bp in length and possesses a typical structure of PAI—inserted at an asp-tRNA and ends with an IS629 on the other side. It carries paralogs of the Salmonella sciCDEFF operon (accession no. AJ320483) of unknown function and of phage P22 and HK620, suggesting that it is possibly another phage-transmitted PAI. SfII has been demonstrated to be a lysogenic phage in which two genes, bgt encoding a bactoprenol glucosyl transferase and gtrII encoding a glucosyl transferase, are required for expression of the type II antigen (30). Thus, phage-mediated horizontal DNA transfer appears to be one of the major routes by which S.flexneri gains virulence determinants.
The IS elements
The IS elements identified in the Sf301 genome are listed in Table 2. In the chromosome, there are astonishingly 247 complete and 67 partial IS elements, which makes it the most IS-rich chromosome among enterics. The predominant species is IS1, followed by IS600, IS2 and IS4. They all are frequently associated with SIs, inversions and translocations, deletions and insertional gene inactivation (see ‘linear map 1’ in Supplementary Material). The IS elements are, therefore, probably the major cause of the dynamics of the Sf301 chromosome. Indeed, the presence of IS91 at two ends of MRDE (mentioned above) allows the precise acquisition and excision of the entire 99 kb segment (29). Furthermore, IS1 and other IS elements have also been shown to be able to mediate various genetic rearrangements (31,32), and IS1 in particular can cause inversions and deletions (32). It is plausible that the IS elements will mediate further evolution of the chromosome. Similarly, pCP301 has large numbers of IS elements, sharing similar composition with pWR501 from serotype 5a (Table 2), indicating that the virulence plasmids are also volatile and dynamic. One difference between pCP301 and pWR501 is that the former has two copies of iso-IS10R that may be transposed from the Sf301 chromosome (Table 2). But, it remains to be seen whether this IS element is present in the genome of serotype 5a. If not, it might be used as a marker for epidemiological studies.
Table 2. IS elements identified in genomes of Sf301, MG1655 and EDL933, the virulence plasmid, and pWR501, from S.flexneri 5a.
| Name | Length (bp) | No. of ORFs | No. of intact elements | No. of partial elements | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sf301 | K12 | 0157 | pCP301 | pW501 | Sf301 | K12 | 0157 | pCP301 | pWR501 | |||
| IS1 | 768 | 2 | 108 | 6 | 2 | 2 | 3 | 9 | 0 | 0 | 1 | 1 |
| iso-IS1 | 803 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 5 | 5 |
| IS2 | 1331 | 2 | 30 | 6 | 1 | 1 | 2 | 5 | 1 | 0 | 2 | 2 |
| IS3 | 1258 | 2 | 5 | 5 | 0 | 0 | 0 | 3 | 0 | 2 | 7 | 8 |
| IS4 | 1428 | 2 | 18 | 1 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 2 |
| IS5 | 1198 | 1 | 0 | 10 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| iso-IS10Ra | 1329 | 1 | 13 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| IS21 | 2131 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 |
| IS91 | 1830 | 1 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 6 | 6 |
| IS100 | 1963 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 6 |
| IS150 | 1443 | 3 | 0 | 1 | 0 | 0 | 0 | 5 | 0 | 0 | 2 | 2 |
| IS186 | 1372 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IS600 | 1264 | 2 | 35 | 0 | 0 | 3 | 2 | 17 | 1 | 6 | 10 | 13 |
| IS629 | 1310 | 2 | 10 | 0 | 18 | 8 | 5 | 11 | 0 | 3 | 3 | 9 |
| IS630 | 1164 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 4 | 2 | 2 |
| IS911 | 1250 | 2 | 16 | 0 | 0 | 1 | 1 | 0 | 4 | 0 | 0 | 0 |
| IS1294 | 1714 | 1 | 0 | 0 | 0 | 1 | 2 | 3 | 0 | 0 | 7 | 4 |
| ISSfl1 | 929 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 3 |
| ISSfl2 | 1374 | 1 | 6 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | 0 |
| ISSfl3 | 1302 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| ISSfl4 | 2754 | 3 | 3 | 0 | 0 | 2 | 2 | 7 | 0 | 1 | 2 | 2 |
| Total | 247 | 32 | 21 | 26 | 23 | 67 | 7 | 19 | 62 | 69 | ||
aiso-IS10R is a homolog of IS10R identified in Sf301 in this study.
The Escherichia coli islands (KIs and OIs)
With the respect to the Sf301 chromosome, MG1655 and EDL933 possess two kinds of islands. One kind is formed owing to the deletions of the corresponding E.coli DNA segments from the Sf301 chromosome, which is hardly surprising given the dynamics of the genome. These include the so called ‘Black Hole’ harboring cadA responsible for converting lysine to cardverine that adversely affects virulence (33) and the kcp locus harboring ompT that inhibits the induction of guinea pig keratoconjunctivitis (34) (arrows in Fig. 2). It remains to be investigated how many such ‘Black Holes’ have deletions of genes that would otherwise inhibit full expression of virulence.
The other kind of island is apparently formed by laterally acquired DNA sequences, of which the large ones are evident in Figure 2 with the scales used. A FASTA query of these groups of OIs and KIs against the Sf301 genome reveals no significant homologous sequence, and a query of all the SIs against EDL933 and MG1655 genomes reveals no homologous sequence either. Thus, O157 and S.flexneri appear to have acquired their island DNA from different sources and have evolved from ancestral E.coli strains through unrelated paths. Furthermore, all the SIs, OIs and KIs have no duplicated copies, indicating that none of them is mobile.
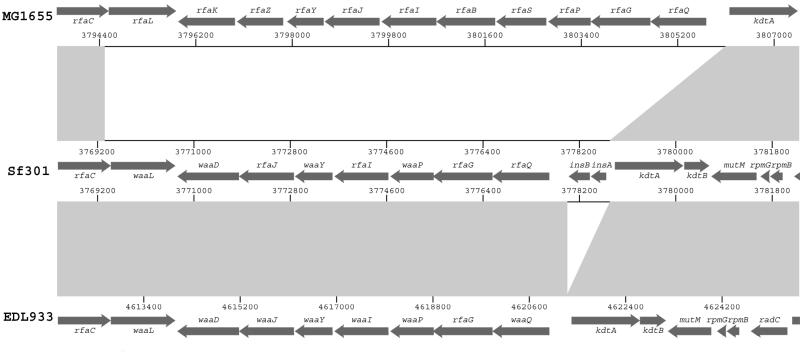
We must point out that we do not define sequences shared by paired strains (EDL933 or MG1655 with Sf301) as islands, though these may appear to be ‘islands’ with respect to the third genome. These sequences may reflect genetic properties of the ancestral E.coli strain that Sf301 evolved from. An example of these are the rfa/waa genes involving LPS biogenesis (Fig. 4). Sf301 and EDL933 have identical numbers of genes that share 99% identity in each case, whereas MG1655 has an equivalent functional operon with more genes and poor homology with the former (Fig. 4). Studies into this type of shared sequence may shed more light on strain diversity and evolution.
Figure 4.
Comparison of the rfa/waa region (to scale). Arrows indicate predicted ORFs in both strands. Regions in gray indicate identical sequences among strains and the non-filling areas indicate sequences with non or low homology.
Pseudogenes
Apart from deletions of corresponding E.coli DNA segments, the formation of pseudogenes through introduction of stop codons, frame shifts, truncations and insertions in the coding regions appears to also play a major part in losing unwanted genes in S.flexneri. Pseudogenes with known functions according to the E.coli protein database are listed in Table 3. Answers to many of the phenotypic characteristics of Shigella, such as the loss of motility and utilization of lactose, maltose and xylose, etc., can be found here. It is noted that 90% of these pseudogenes are intact in O157 EDL933. To this end, S.flexneri resembles S.typhi, another enteric pathogen restricted to humans. The presence of large numbers of pseudogenes has been postulated to be one of the main reasons that S.typhi evolved from the rest of the Salmonella species to become a solely human pathogen (35). Likewise, the originally closely linked O157 and Shigella have evolved in diverse directions. Strain O157 became a successful pathogen with broad host range mainly by acquiring DNA (Table 1 and Fig. 2), whereas Shigella also became a successful pathogen but restricted to humans only, by acquiring, as well as losing, DNA.
The virulence plasmid pCP301
Like previously sequenced virulence plasmids (pWR100 and pWR501) from serotype 5a strains (7,8), pCP301 is a mosaic of potential virulence-related genes, IS elements, maintenance genes and functionally unknown ORFs. All the previously identified virulence genes are present in pCP301. These include the primary invasion genes ipa and mxi-spa (encoding the invasion plasmid antigens and the type III secretion system, respectively), virG/IcsA (required for polymerizing host actin to provide propelling force for intra- and inter-cellular spread) and virF (necessary for regulating virulence gene expression). The replication origin (R100-like) ori and G site (single-strand initiation site) in pCP301 are identical to those of pWR501 and pWR100. pCP301 also has maintenance genes, repA, copA and copB, for replication; parA and parB for partitioning; and ccdA and ccdB for post-segregation killing. The noticeable difference between pCP301 and the plasmids from serotype 5a is the presence of more IS-related DNA in pCP301, making its size close to pWR501 (221 851 bp) which is larger than pWR100 because of a Tn501 (8360 bp) insertion (8). So, both Shigella serotypes most likely acquired the ancestral virulence plasmid from the same source. One other minor divergence is that the ipa-mxi-spa loci in pWR501 and pCP301 are in the same orientation, whereas in pMYSH6000, the virulence plasmid from another 2a strain, they are in inverse orders (36). This indicates that the divergence of the plasmids does not necessarily correlate with serotypes. A detailed comparison of pCP301 with pWR501 is available in the Supplementary Material (‘linear map 2’).
CONCLUSION
Comparison of the S.flexneri genome with that of E.coli supports the previous genetic study (5) that S.flexneri is closely related to E.coli and may turn out to belong to the same genus. The global gene content (Table 1) and alignments (Fig. 2) indicate that S.flexneri is more closely related to the non-pathogenic K12 strain MG1655 rather than the pathogenic O157 strain EDL933. This is in agreement with the suggestions that O157 and K12 last shared an ancestor ∼4.5 million years ago (37), whereas Shigella evolved from multiple E.coli strains much later, correlating with the appearance of early man in the paleolithic (5). All these studies call strongly to reclassify Shigella species as members of E.coli.
To meet the demand of its unique pathogenic lifestyle, the S.flexneri chromosome has evolved distinctive characteristics after acquisition of the large virulence plasmid. Most importantly, there are several potential bacteriophage-transmitted PAIs, many translocations, inversions and deletions of the corresponding E.coli DNA segments, and numerous pseudogenes. These findings provide an invaluable genetic basis for future studies into understanding bacterial evolution, as well as pathogenicity, and the development of novel preventive and treatment strategies against shigellosis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Paul Langford, Janine Bosse and Alick Stephens for critical reading of the manuscript, and the following for technical support: Shu Wang, Tianjing Cai, Yingying Xi, Xinyu Tan, Yanrui Jiang, Shitao Zhuang, Xinfeng Zhou, Li Rong, Tao Lu, Wei Liu and Lihong Chen from National Center of Human Genome Research, Beijing, and Shuiyun Lan, Yueqing Zhang and Minjie Chen from Fudan University for their expert technical assistance. This project was supported by the State Key Basic Research Program (grant no. G1999054103) and High Technology Project (grant no. Z19-02-05-01) from the Ministry of Science and Technology of China.
DDBJ/EMBL/GenBank accession nos AE005674, AF386526
REFERENCES
- 1.Sansonetti P.J. (2001) Microbes and microbial toxins: paradigms for microbial–mucosal interactions III. Shigellosis: from symptoms to molecular pathogenesis. Am. J. Physiol. Gastrointest. Liver Physiol., 280, G319–G323. [DOI] [PubMed] [Google Scholar]
- 2.Mei Y., Liu,H. and Xu,J. (1989) Cloning and application of genus specific DNA probes for Shigella. Chinese J. Epidemiol., 10, 167–170. [Google Scholar]
- 3.Sansonetti P.J. (1998) Vaccines against enteric infections. Slaying the Hydra at once or head by head? Nature Med., 4 (Suppl.), 499–500. [DOI] [PubMed] [Google Scholar]
- 4.Hale T.L. (1991) Genetic basis of virulence in Shigella species. Microbiol. Rev., 55, 206–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pupo G.M., Lan,R. and Reeves,P.R. (2000) Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl Acad. Sci. USA, 97, 10567–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sansonetti P.J., Hale,T.L., Dammin,G.J., Kapfer,C., Collins,H.H.,Jr and Formal,S.B. (1983) Alternations in the pathogenicity of Escherichia coli K12 after transfer of plasmid and chromosomal genes from Shigella flexneri. Infect. Immun., 39, 1392–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchrieser C., Glaser,P., Rusniok,C., Nedjari,H., D’Hauteville,H., Kunst,F., Sansonetti,P. and Parsot,C. (2000) The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri.Mol. Microbiol., 38, 760–771. [DOI] [PubMed] [Google Scholar]
- 8.Venkatesan M.M., Goldberg,M.B., Rose,D.J., Grotbeck,E.J., Burland,V. and Blattner,F.R. (2001) Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun., 69, 3271–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman C.J., McKenna,S. and Beloin,C. (2001) Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Int. J. Med. Microbiol., 290, 89–96. [DOI] [PubMed] [Google Scholar]
- 10.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 11.Perna E.S., Plunkett,G.,III, Burland,V., Mau,B., Glasner,J.D., Rose,D.J., Mayhew,G.F., Evans,P.S., Gregor,J., Kirkpatrick,H.A. et al. (2001) Genomic sequence of enterohaemorrhagic Escherichia coli 0157:H7. Nature, 409, 529–533. [DOI] [PubMed] [Google Scholar]
- 12.Ewing B., Hillier,L., Wendi,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 13.Gordon D., Abajian,C. and Green,P. (1998) Consed: a graphical tool for sequence finishing. Genome Res., 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 14.Salzberg S.L., Delcher,A.L., Kasif,S. and White,O. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res., 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth J.R., Benson,N., Galitski,T., Haack,K., Lawrence,J.G. and Miesel,L. (1996) Rearrangements of the bacterial chromosome: formation and applications. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella, 2nd Edn. ASM Press, Washington DC, pp. 2256–2276.
- 18.Liu S.-L. and Sanderson,K.E. (1995) Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl Acad. Sci. USA, 92, 1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu S., Setianingrum,E., Zhao,L., Li,Z., Xu,H., Kawamura,Y. and Ezaki,T. (2000) I-CeuI fragment analysis of the Shigella species: evidence for large-scale chromosome rearrangement in S. dysenteriae and S. flexneri. FEMS Microbiol. Lett., 182, 93–98. [DOI] [PubMed] [Google Scholar]
- 20.Brewer B.J. (1990) Replication and the transcriptional organization of the Escherichia coli chromosome. In Drlica,K. and Riley,M. (ed.), The Bacterial Chromosome. ASM Press, Washington DC, pp. 61–83.
- 21.Hill C.W. and Gray,J.A. (1988) Effect of chromosomal inversion on cell fitness in Escherichia coli K12. Genetics, 119, 771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebollo J.E., Francois,V. and Louarn,J.M. (1988) Detection of possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc. Natl Acad. Sci. USA, 85, 9391–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajakumar K., Sasakawa,C. and Adler,B. (1997) Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect. Immun., 65, 4606–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss J.E., Cardozo,T.J., Zychlinsky,A. and Groisman,E.A. (1999) The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol., 33, 74–83. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Prada C.M., Hoover,D.L., Tall,B.D., Hartman,A.B., Kopelowitz,J. and Venkatesan,M.M. (2000) Shigella flexneri IpaH7.8 facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect. Immun., 68, 3608–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyotome T., Suzuki,T., Kuwae,A., Nonaka,T., Fukuda,H., Imajoh-Ohmi,S., Toyofuku,T., Hori,M. and Sasakawa,C. (2001) Shigella protein IpaH9.8 is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem., 276, 32071–32079. [DOI] [PubMed] [Google Scholar]
- 27.Buchanan S.G. and Gay,N.J. (1996) Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol., 65, 1–44. [DOI] [PubMed] [Google Scholar]
- 28.Shou D., Hardt,W.-D. and Galan,J.E. (1999) Salmonella typhimurium encodes a putative transport system within the centisome 63 pathogenicity island. Infect. Immun., 67, 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luck S.N., Turner,S.A., Rajakumar,K., Sakellaris,H. and Adler,B. (2001) Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun., 69, 6012–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mavris M., Manning,P.A. and Morona,R. (1997) Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol. Microbiol., 26, 939–950. [DOI] [PubMed] [Google Scholar]
- 31.Turlan C. and Chandler,M. (1995) IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J., 14, 5410–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider D., Duperchy,E., Coursange,E., Lenski,R.E. and Blot,M. (2000) Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics, 156, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurelli A.T., Fernandez,R.E., Bloch,C.A., Rode,C.K. and Fasano,A. (1998) ‘Black holes’ and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 3943–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakata N., Tobe,T., Fukuda,I., Suzuki,T., Komatsu,K., Yoshikawa,M. and Sasakawa,C. (1993) The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol. Microbiol., 9, 459–468. [DOI] [PubMed] [Google Scholar]
- 35.Parkhill J., Dougan,G., James,K.D., Thomson,N.R., Pickard,D., Wain,J., Churcher,C., Mungall,K.L., Bentley,S.D., Holden,M.T.G. et al. (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature, 413, 848–852. [DOI] [PubMed] [Google Scholar]
- 36.Sasakawa C., Makino,S., Kamata,K. and Yoshikawa,M. (1986) Isolation, characterization, and mapping of Tn5 insertions into the 140-megadalton invasion plasmid defective in the mouse Sereny test in Shigella flexneri 2a. Infect. Immun., 54, 32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid S.D., Herbeline,C.J., Bumbaugh,A.C., Selander,R.K. and Whittman,T.S. (2000) Parallel evolution of virulence in pathogenic Escherichia coli. Nature, 406, 64–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.