Abstract
In genetically transformed plants, transgene silencing has been correlated with multiple and complex insertions of foreign DNA, e.g. T-DNA and vector backbone sequences. Occasionally, single-copy transgenes also suffer transgene silencing. We have compared integration patterns and T-DNA/plant DNA junctions in a collection of 37 single-copy T-DNA-transformed Arabidopsis lines, of which 13 displayed silencing. Vector sequences were found integrated in five lines, but only one of these displayed silencing. Truncated T-DNA copies, positioned in inverse orientation to an intact T-DNA copy, were discovered in three lines. The whole nptII gene with pnos promoter was present in the truncated copy of one such line in which heavy silencing has been observed. In the two other lines no silencing has been observed over five generations. Thus, vector sequences and short additional T-DNA sequences are not sufficient or necessary to induce transgene silencing. DNA methylation of selected restriction endonuclease sites could not be correlated with silencing. Our collection of T-DNA/plant DNA junctions has also been used to evaluate current models of T-DNA integration. Data for some of our lines are compatible with T-DNA integration in double-strand breaks, while for others initial invasion of plant DNA by the left or by the right T-DNA end seem important.
INTRODUCTION
Transformation of plants mediated by the microbe Agrobacterium tumefaciens is one of the most widely used means of generating transgenic plants. The bacterium is able to transfer a DNA segment (T-DNA) borne on a plasmid (Ti plasmid) that also contains the virulence (Vir) genes necessary for T-DNA transfer and integration into the genome of plant cells. The T-DNA is delineated by two imperfect direct repeats, the right and left borders. The VirD2 protein nicks at the two repeats, associates with the 5′ end of the right border and assists single-stranded T-DNA transfer to the plant nucleus and integration of T-DNA in the plant genome (reviewed in 1,2). The right border has been found to be essential for this process, while the left border defines the end of the T-DNA (3–5).
Ti plasmids have been modified into a number of binary vector systems, where the virulence genes are harboured on a helper plasmid and the T-DNA, in which foreign genes can be inserted, on another plasmid. The tumour-inducing genes found in the T-DNA of natural Ti plasmids have been removed and the binary vector system can therefore be used to generate plants with new traits of interest for research and commercial purposes. Models for T-DNA integration based on studies of junctions between T-DNA and plant DNA implicate the 3′ end in the invasion and anchoring of the single-stranded T-DNA into the plant DNA. The VirD2 protein, connected to the 5′ end of the T-DNA, may assist in the joining of this end to plant DNA. Nicked target DNA serves as primer for the synthesis of the complementary T-DNA strand. As a result of this process, double-stranded T-DNA is integrated and a short stretch of plant DNA is deleted at the target site (1,3–6). Some data, e.g. end-to-end fusions between two right T-DNA borders (7,8), indicate that after nuclear import, but before invasion into the plant DNA, the single strand is converted to a double strand. Different models have also been put forward to explain the finding of short sequences (fillers), often of unknown origin, at the junctions between plant and T-DNA (9,10).
A recurrent problem in the generation of genetically modified plants is that Agrobacterium T-DNA-mediated transformation, as well as other methods, often results in integration of more than one copy of foreign DNA, for instance as tandem or inverted repeats. Another complication is the transfer and integration of binary vector backbone (BVB) sequences (11–15). Several studies have been devoted to explain the mechanism of integration of two or more copies at the same site (7,8,16,17) and BVB transfer (13,15,18).
Complex integrations render the transferred genes susceptible to homology-dependent transgene silencing, either due to transcriptional silencing mechanisms, possibly involving ectopic DNA–DNA interactions and promoter methylation, or post-transcriptional silencing, thought to involve aberrant mRNAs leading to transcript degradation (19,20). It has been suggested that a general mechanism for scanning the genome for intrusive DNA is present in plants (21,22). Foreign sequences, like the T-DNA or BVB of prokaryotic origin, might be discovered due to differences in GC content compared to the GC content at the integration site in the plant genome. Transgene silencing is undesirable since it reduces the efficiency and reliability of transgene expression. In agricultural and research uses (e.g. efficient reverse genetics) one would therefore prefer single copy lines.
However, transgene silencing can occasionally also occur in lines with a single T-DNA copy (23–26). To investigate whether such T-DNAs have special features, we have cloned the flanking regions and analysed the integration events in a collection of transgenic lines which, based on Southern hybridisations, were single copy. The collection encompasses 13 lines showing silencing of the T-DNA-harboured nptII gene conferring kanamycin resistance (TS lines) and 24 control lines not displaying silencing (C lines).
Our data for 37 lines in total have allowed us to re-examine current models for T-DNA integration. We systematically found that plant/T-DNA junctions with perfect micro- homologies were without fillers. Such situations were found as often at the RB as the LB, suggesting that the ‘LB first’ model cannot account for all integration events. BVB sequences were found integrated upstream of both the LB and the RB and, in some cases, truncated T-DNA copies were found that had not been discovered by Southern hybridisation. However, BVB and extra T-DNA sequences were found in both TS lines and C lines. DNA methylation status of selected restriction sites was therefore investigated. Methylation was found in lines without silencing and vice versa. We conclude that BVB, complex integration patterns or DNA methylation are neither necessary nor sufficient for the induction of transgene silencing in single locus transgenes.
MATERIALS AND METHODS
Plant material
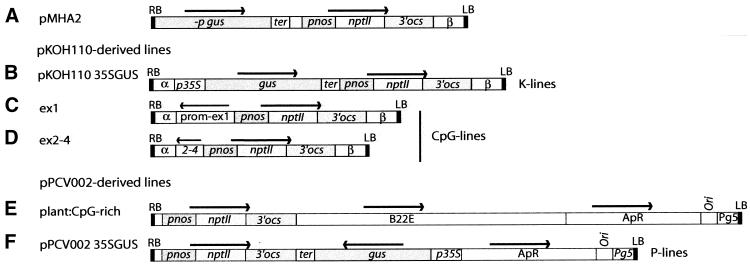
The 37 transgenic Arabidopsis lines used for this study were generated by Agrobacterium-mediated root transformation of Arabidopsis thaliana ecotype C24 as described (27) using the constructs pKOH110 35SGUS (24), pPCV002 35SGUS (24,28), ex1, ex2–4 (26), plant:CpG-rich with the barley gene B22E in the vector pPVC002 (26,29,30), and pMHA2 (31) (Fig. 1). For transformation with pPCV002-derived constructs the GV3101 strain with a nopaline helper vector was used (28), while the octopine-based helper plasmid pGV2260 in the C58C1 rifr strain was used for pKOH110/pMHA2 transformation (32).
Figure 1.
Vector constructs used in this study. (A) pMHA2; (B) pKOH110 35SGUS; (C) ex1; (D) ex2–4; (E) plant:CpG-rich; (F) pPCV002 35SGUS. RB, right border; LB, left border; gus, β-glucuronidase gene (udiA); –p gus, promoterless gus gene; prom–ex1 and 2–4, parts of the human CpG island comprising the promoter and exon 1, and exons 2–4 of the human MECL-1 gene, respectively; B22E, barley metallothionein-like gene; nptII, neomycin phosphotransferase gene; ApR, ampicillin resistance gene; pnos, nos promoter; 3′ ocs, ocs terminator; p35S, 35S promoter; ter, polyadenylation signal; α, β and Pg5 are natural T-DNA sequences. Horizontal arrows indicate direction of transcription.
Registration of nptII silencing
Segregation analyses of kanamycin-resistant and kanamycin-sensitive seedlings of the T2 generation and genomic Southern hybridisation data were used to estimate the number of T-DNA loci in each transgenic line (24). Seeds from up to 23 T2 sibling plants from each line for up to five generations were germinated on MS medium with 2% sucrose and 50 µg/ml kanamycin, to calculate the number of seedlings sensitive to kanamycin due to nptII silencing (24,26).
Analysis of genomic DNA
Genomic DNA was isolated from four to six rosette stage plants as described (33), with some modifications, including an RNase step (34). Southern blot and methylation analyses were performed as described previously (24,26).
Cloning of flanking regions
The regions flanking the right and left borders were cloned using two different methods, inverse PCR (IPCR) and genome walking. PCRs were performed with the Advantage2 polymerase mix (Clontech, Palo Alto, CA) or Elongase (BRL) enzyme.
For IPCR, genomic DNA (100 ng) was digested with the appropriate restriction enzymes for 5–7 h. The digested DNA was cleaned using the QIAquick PCR Purification kit (Qiagen, Hilden, Germany) and eluted in 50 µl of distilled H2O. The DNA was ligated in a volume of 100 µl and 1 µl of this was used in the amplification reaction. The PCR was performed in two steps when the annealing temperature for the primers allowed that. Otherwise three steps were used. Denaturation was at 94°C for 1 min, followed by 30 cycles of 15 s at 94°C, 30 s at 52–53°C and 5 min at 68°C, followed by a final cycle of extension at 68°C for 5 min. When nested PCR was performed, 1 µl of a 1/100 dilution of the primary PCR was used.
In some cases genome walking (Clontech, Palo Alto, CA) was performed, using the adaptor of the Marathon cDNA amplification kit (Clontech, Palo Alto, CA). The adaptor was generated as described (35), using adaptor oligonucleotide 1 (5′-CTA ATA CGA CTC ACT ATA GGG CTC GAG CGG CCG CCC GGG CAG GT-3′) and adaptor oligonucleotide 2 (5′-PO4-ACC TGC CC-H2N-3′). The nested primers AP1 (5′-CCA TCC TAA TAC GAC TCA CTA TAG GGC-3′) and AP2 (5′-ACT CAC TAT AGG GCT CGA GCG GC-3′) were used in all the walks. Two-step PCRs were used. Denaturation was at 94°C for 1 min, followed by seven cycles of 94°C for 2 s and 72°C for 3 min and 32 cycles of 94°C for 2 s and 67°C for 3 min, followed by a final step of 67°C for 4 min. When one junction of a line had been cloned, the other junction was cloned by IPCR, genome walking or by PCR using one T-DNA-specific and one genomic primer.
Primer sequences and restriction enzymes used for the above purposes will be provided upon request.
Sequencing and data analysis
The resulting PCR products were either sequenced directly using gene-specific primers or preferably cloned into the pGEM-T easy vector (Promega, Madison, WI) and sequenced using the Universal primer (5′-GGA AAC AGC TAT GAC CAT G-3′) and M13 reverse primer (5′-GTA AAA CGA CGG CCA GT-3′). Manual sequencing was performed using the Sequenase V.2.0 sequencing kit (USB Corp., Cleveland, OH). Automated sequencing was performed on a MegaBACE500 using a DYEnamic ET Dye Terminator Cycle Sequencing Kit (Amersham Pharmacia Biotech, UK) or on an ABI PRISM using the Dynamics-ThermoSequenase radiolabelled terminator cycle sequencing kit (Amersham Pharmacia Biotech). Blast searches were performed against the Arabidopsis genome database. VecScreen was used to determine if the cloned fragments contained vector-derived sequences.
RESULTS
Six different constructs have been used to transform Arabidopsis (Fig. 1). The pMHA2 (31), pKOH11035S-GUS (K-lines; 24), ex1 and ex2–4 constructs (26) are all based on T-DNA from octopine strains and have identical T-DNA right and left borders. The three latter constructs harbour different inserts in the polylinker of the vector pKOH110, i.e. 35SGUS, or different parts of the CpG island associated with the human MECL-1 gene (ex1 and ex2–4, CpG lines) (26). The pMHA2 vector is similar to pKOH110 35SGUS, but harbours a promoter-less Gus gene (31). pPCV002 35SGUS (P lines) (24) and the plant:CpG-rich constructs (26), which harbour the CpG-rich barley gene B22E (Pl:CpG lines) (29,30), are based on the pPCV002 vector, which has a nopaline right border but an octopine left border (28).
The 13 lines showing nptII silencing belong to the K, P, CpG and Pl:CpG lines. The frequencies of silencing are given in Table 1. Six of these are novel single-copy lines displaying silencing identified in our laboratory; four K lines and two P lines with low (<10%) to medium (10–50%) overall frequency of silencing (Table 1). pMHA2, CpG and Pl:CpG lines have been included in the study as control lines (Table 2).
Table 1. Percent seedlings displaying silencing of the nptII gene in lines transformed with the constructs indicated.
| Construct | Line | Silencing (%) |
|---|---|---|
| pKOH 110 ex2–4a | 4 | 3.8–29.6 |
| 5 | 5.7b | |
| pPCV002 Pl:CpGa | 19 | 3.5–7.0 |
| 20 | 8.5–10 | |
| pKOH110 35SGUS | K11c | 0.7–4.2 |
| K13 | 100d | |
| K14 | 4.2–50 | |
| K15 | 2.7–15.2 | |
| K16 | 6.8d | |
| pPCV002 35SGUS | P4c | 1.7–100 |
| P6c | 28.1–50 | |
| P9 | 12.9–17.8d | |
| P10 | 3.0–10.7 |
The silencing level varies between siblings and generations, and the lowest and highest observed percentage silencing is therefore given for each line. For each line, 60–150 seeds from up to 23 siblings and up to five generations were tested.
aLines reported by Meza et al. (26).
bSilencing was only observed in the T5 generation of ex2–4 line 5.
cLines reported by Meza et al. (24).
dProgeny of one sibling only of lines K13 and K16, and two siblings of line P9, displayed silencing.
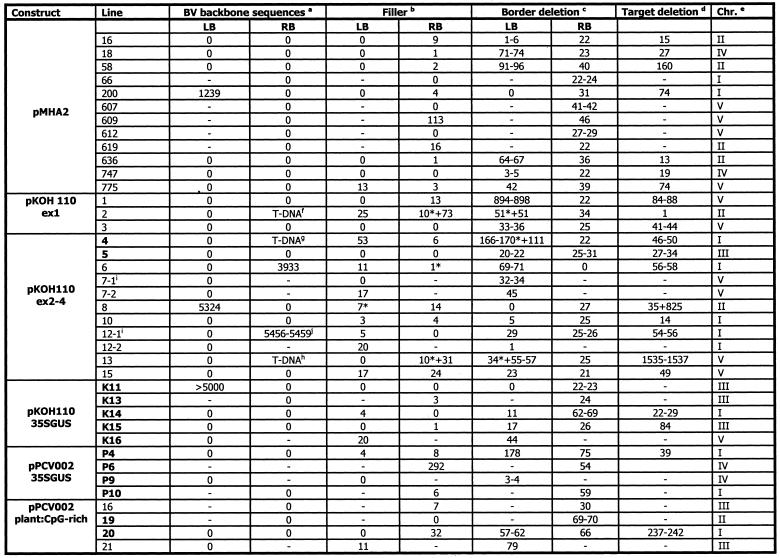
Table 2. Target and T-DNA characteristics and integration of BVB or additional T-DNA sequences of lines presented in this study.
Lines given in bold display nptII silencing. –, not determined. *Data for truncated T-DNA copies.
aNumber of base pairs upstream of the borders derived from the backbone sequence of the T-DNA vector.
bNumber of base pairs at the T-DNA/plant DNA junction that are neither a continuation of the T-DNA nor the plant-DNA.
cDeletion of the border counted from the first base in the border repeat. In cases of micro-homology between target and T-DNA, a range is given for the border and target deletion.
dNumber of base pairs deleted at the target upon integration of the T-DNA. In cases of micro-homology between target and T-DNA, a range is given for the border and target deletion.
eChromosome into which the T-DNA has integrated.
f,g,hTruncated T-DNA sequences found upstream of RB (see Fig. 2).
iCloning indicated the presence of an additional T-DNA fragment in close proximity to another T-DNA copy.
jThe BVB sequence has micro-homology at both ends.
The number of inserted T-DNA copies was initially determined by Southern hybridisation. Normally, two enzymes with a single digestion site in the T-DNA and probes hybridising on each side of the restriction endonuclease sites were used (24,26). The analysis indicated that all lines contained a single T-DNA locus comprised of a single T-DNA insert with the exception of the single locus ex2–4 line 5, which has two tandem repeated T-DNAs (24,26). IPCR or genome walking was used to isolate the T-DNA/plant DNA junction sites (see Materials and Methods). Sequenced flanking fragments were used for BLAST searches against the Arabidopsis genome databases (data not shown). T-DNA integrations were found spread on all chromosomes in independent loci (Table 2).
Genomic deletions and border deletions do not distinguish TS lines from C lines, but RB deletions differ with the vectors used
Both flanking sequences were isolated from 22 lines and the T-DNA integration sites were investigated for the presence of genomic deletions (Table 2). No striking differences were seen between the TS and C lines. The majority of the deletions are <75 bp, with an average of 36 bp. The smallest deletion was 1 bp. In four cases, deletions of >100 bp were found, the largest of 1537 bp. Normally, the deletion represented a continuous stretch of genomic DNA (Fig. 2A and Table 2). A somewhat more complex pattern was observed in only one line (ex2–4 line 8), where a deletion of 35 bp at the integration site was followed by 60 bp of genomic DNA preceding a second deletion of 825 bp (Fig. 2B and Table 2).
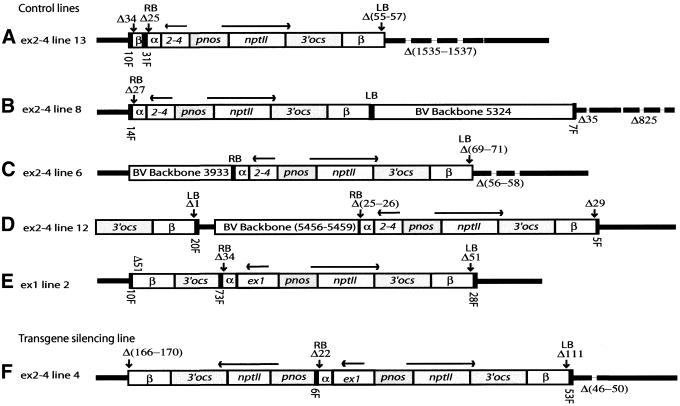
Figure 2.
T-DNA structure and target deletions in selected lines. T-DNAs are shown with intact LB and RB indicated by solid black boxes. Heavy black lines represent flanking genomic DNA, while broken lines represent target deletions. Δ denotes the number of base pairs deleted in targets and T-DNA border sequences. Numbers of base pairs in BVB sequences (open bars) and in filler sequences (F, solid black bars) are given. (A) ex2–4 line 13. Note the truncated inverted T-DNA copy covering 198 bp of the T-DNA β sequence, adjacent to the RB of the intact copy. (B) ex2–4 line 8. Note the 5324 bp BVB sequences continuing directly from the LB. (C) ex2–4 line 6. Note that 3948 bp of BVB sequence has been integrated upstream of the intact right border. (D) ex2–4 line 12. Note the truncated T-DNA copy of at least 1788 bp in direct orientation relative to the intact T-DNA and the 5456–5459 bp (of total 7209) BVB sequence, which has micro-homology at both ends. (E) ex1 line 2. Note the truncated copy of T-DNA (543 bp) comprising the 3′ ocs and β region in inverted orientation to the complete T-DNA copy. Only 1 bp has been deleted at the integration site. (F) ex2–4 line 4. Note the truncated T-DNA copy (2927 bp) comprising the whole nptII gene including the pnos promoter in inverted orientation to the complete T-DNA copy.
At the LB, which is identical in all our constructs, VirD2 is expected to nick the T-DNA strand between the fourth and third nucleotides from the end of the T-DNA, but this end normally suffers further deletion during integration (6). In lines where the T-DNA sequence continued into integrated BVB (see below) the border sequences were intact. In the other lines the number of nucleotides missing from the left end ranged from 1 to 894, with an average of 79 bp and a median value of 44 bp. None of the border deletions were long enough to affect the nptII gene (Fig. 3A and Table 2). All the TS lines as well as the C lines had intact nptII genes.
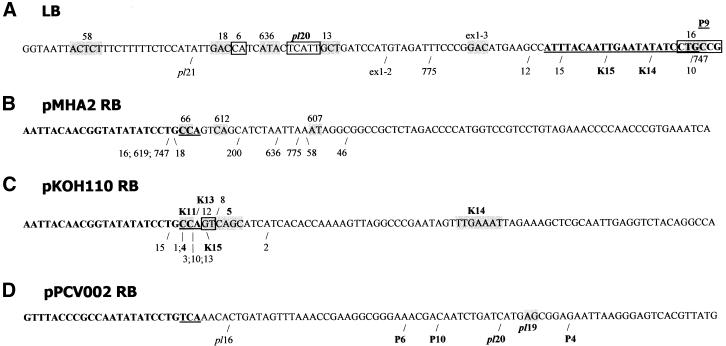
Figure 3.
Right and left border deletions. The left end of all six constructs (A) and the right ends of the vectors pMHA2 (B), pKOH110 (C) and pPCV002 (D) are shown with the border repeats in bold. The nucleotides of the repeats present in the single-stranded T-DNA after VirD2 nicking at the preferred sites are underlined. The right or left end points of the T-DNAs in the different lines are indicated by /. In lines where there is micro-homology between T-DNA and genomic DNA at the target site the homologous nucleotides are boxed. Lines displaying nptII silencing are in bold. (A) All vectors have the same left end. 16, 18, 58, 636, 747 and 775 are pMHA2 lines; 6, 10, 12, 13, 15 are ex2–4 lines; pl20 and pl21 are plant:CpG-rich lines. The ex2–4 line 4 and line P4 have left end deletions of 111 and 178 bp, respectively, and could therefore not be included in the figure. (B) pMHA2 derived lines. (C) pKOH110-derived lines. 1, 2 and 3 are ex1 lines; 4, 5, 8, 10, 13, 15 are ex2–4 lines. (D) pPCV002-derived lines. pl16, pl19 and pl20 are plant:CpG-rich lines.
The RB deletions also did not distinguish the TS lines from the C lines. Dissimilarities were, however, found between lines derived with different vectors used. In three out of 12 (25%) pMHA2 lines and two out of 15 (13%) of the pKOH110-derived lines, 22 bp of the RB were deleted, leaving the residual three nucleotides 5′-CCA of the border, as expected from the preferred nicking site of VirD2 (Fig. 3B and C, and Table 2). In none of the pPCV002-derived lines did the integrated T-DNA start from this position (Fig. 3D and Table 2). The average number of missing nucleotides counted from the start of the right border was 31 and 28 for pMHA2 and pKOH110-derived lines, respectively. The median was 29 for pMHA2 lines and 25 for pKOH110-derived lines. The number of deleted nucleotides ranged from 22 to 46 in pMHA2. The range was much narrower in the majority of pKOH110-derived lines (21–27 bp; Table 2 and Fig. 3). The pPCV002-derived lines had longer RB deletions: the average, median and range were 59, 62 and 30–75 nt, respectively (Table 2 and Fig. 3).
Micro-homologies without fillers were found both at the left and the right T-DNA/plant DNA junctions
The junctions between T-DNA and plant DNA turned out to be similar in TS and C lines. Fillers between the right border and the genomic plant DNA were observed for 70% (23/33) of the junctions cloned. Fillers were found at the RB junction at a slightly lower percentage in the pMHA2 and pKOH110-derived lines [67% (8/12) and 67% (10/15), respectively] compared with the pPCV002-derived lines (83%, 5/6). Fillers were found in four of the five lines with 22 bp RB deletions. At the LB/plant DNA junctions, fillers were observed for 48% (14/29) of the junctions, but mainly in pKOH110-derived lines (61%, 11/18). LB filler was found in one pMHA2 line only (14%, 1/7). For lines where both left and right border junctions were cloned, fillers were observed at both junctions in 36% (8/22) of the lines (Table 2).
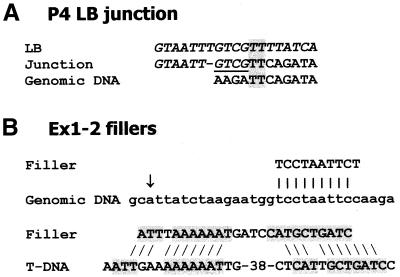
The majority of the fillers were short DNA sequences (1–53 bp; Table 2). In line 609 the RB filler of 113 bp was identical to a sequence from another chromosome, while the 32 bp filler of the plant:CpG-rich line 20 is identical to the genomic sequence 1694 bp from the T-DNA insertion point (Table 2). In ex2–4 line 8, a 14 bp filler at the RB was identical to a sequence found in both chromosomes I and III. Downstream of the BVB on the LB side in this line (see below) a 7 bp filler (TATTAAC) was found of which 6 bp (ATTAAC and TATTAA) were identical to two stretches of genomic DNA 13 and 36 bp downstream for the integration site. In line P4 the 4 bp filler (Fig. 4A, underlined) might rather be a continuation of the LB sequence, since compared with this sequence only a T nucleotide is missing (Fig. 4A). In other cases, the junction sequence can be interpreted either as a filler or a continuation of the genomic sequence where 1 or 2 nt have been lost (not shown). The ex1 line 2 represents a complex case (Fig. 2E and below) where the filler of 73 bp at the RB has an exact match to the sequence 47–120 bp from the left end of the T-DNA. In this line there are another two fillers, one of 10 bp almost identical to a genomic sequence only 17 bp from the site of T-DNA integration (Fig. 4B) and one of 25 bp which patch-wise has high similarity to sequences up to 117 bp from the LB (Fig. 4B). In some cases when the fillers are short (2–4 bp), similarity to T-DNA or genomic DNA close to the junction was observed, e.g. the RB filler of line 200, CTGT, is identical to 4 bp in the T-DNA less than 50 bp from the RB deletion point.
Figure 4.
Examples of junctions and fillers. (A) LB junction of line P4. The T-DNA sequences are given in italic. The putative filler, which alternatively may be regarded as derived from the T-DNA, is underlined. Micro- homologies are boxed. (B) Fillers of ex1 line 2, in one case showing high similarity to genomic DNA close to the T-DNA insertion point (arrow) and in the other case patch-wise high similarity to T-DNA sequences.
It was not always possible to determine exactly where the plant DNA ended and the T-DNA started, due to micro-homology between the T-DNA sequence and the sequences at the integration site (Table 2). Ex2–4 line 5 had micro-homology on both sides. Six lines had micro-homology on one side and filler on the other. Only one junction has been cloned for the remaining of these lines. Micro-homologies were observed both at the RB and the LB; near the right border for eight lines (pMHA2 lines 66, 607 and 612, ex2–4 lines 5 and 12, K lines K11 and K14 and plant:CpG 19) and in 13 cases near the LB (pMHA2 lines 16, 18, 58, 636 and 747, ex1 lines 1 and 3, ex2–4 lines 5, 6, 7 and 13, P line P9 and plant:CpG 20). At such junctions, filler DNA was not observed, with the exception of ex2–4 line 6, which has a 1 bp filler (Fig. 3 and Table 2). When allowing for 1–2 nt gaps in the alignments between the LB, the junction and the genomic target sequence (e.g. line P4, Fig. 4A), it can also be said to have micro-homology and no filler. In two cases only, we found neither filler nor micro-homology (RB pMHA2-747 and ex1 line 3).
The presence of BVB sequences is not correlated with silencing in single-copy lines
The fragments amplified by IPCR, PCR or genome walking were investigated for the presence of BVB sequences. At the left end all three vectors have identical 713 bp. pKOH110 and pMHA2 have ∼3 kb in common at this end and in addition ∼5 kb common BVB. Continuous integration of BVB sequences was encountered in 10% (3/29) of the LBs, in one TS (K11) and two C lines (pMHA2 200 and ex2–4 line 8). Two of these lines harbour a pKOH110-derived construct (Table 2). The length of the BVB sequences ranges from 1239 to 5324 bp (Fig. 2B and Table 2). In one C line (ex2–4 line 6) the right border continued directly into 3933 bp of BVB sequence (Fig. 2C and Table 2).
The C line ex2–4 line 12 was particularly complex, with 5459 bp BVB outside the RB of an intact T-DNA copy (Fig. 2D). However, this BVB sequence is not a continuation from the RB, but rather the BVB sequence adjacent to the LB. A truncated T-DNA fragment encompassing the β and 3′-ocs sequences was found 124 bp from the end of the BVB. This truncated copy lies in direct orientation relative to the intact T-DNA copy.
Truncated T-DNA sequences were found in addition to intact copies in both TS and C lines
Sequencing of right border flanking regions revealed that two ex2–4 lines (lines 4 and 13), as well as ex1 line 2, contained truncated T-DNA sequences, which had not been detected by Southern analysis (Fig. 2A, E and F, and Table 2). Only line 4 is a TS line. In all cases the truncated T-DNAs lack the human sequences, but have more or less intact left regions and are positioned in opposite orientation with respect to the complete T-DNA copy. In line 4, 2927 bp of T-DNA was found next to a 6 bp filler at the RB of the complete copy. The truncated T-DNA comprises the part of the T-DNA that includes the nptII gene with the pnos promoter and ocs terminator. There are 166–170 bp missing from the LB of the truncated copy, which has 5 bp of micro-homology to the genomic target sequence (Fig. 2F). In ex2–4 line 13, the whole right border is deleted in the complete copy and a filler of 31 bp separates the truncated and the complete T-DNAs (Fig. 2A and Table 2). The truncated copy covers 543 bp left of the ocs terminator. The truncated copy is separated from the plant DNA by a filler of 10 bp. In the ex1 line 2, there is a filler of 73 bp between the complete and the truncated T-DNAs, which is 1620 bp and covers the ocs terminator (Fig. 2D). Between the LB of the truncated T-DNA and the plant DNA 10 bp of filler was found.
Line P6 (not shown) and ex2–4 line 7 (Table 2) turned out to have more complex integration patterns, with right and left fragments that are not parts of a continuous T-DNA. Truncated T-DNA copies upstream of the right border were not observed for any pMHA2 lines (Table 2).
Methylation of the pnos promoter is not correlated with transgene silencing
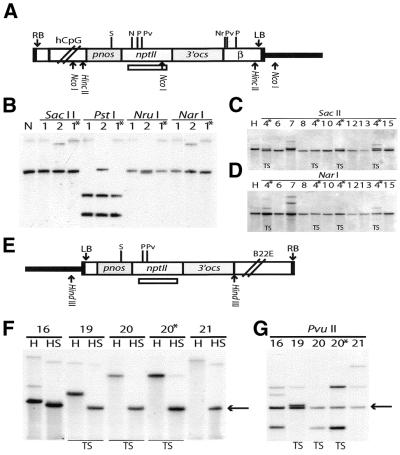
We have earlier reported that the SacII/SstII site in the pnos promoter of the nptII gene was unmethylated in the lines K11, P4 and P6, while it was partially or fully methylated in multicopy lines showing silencing (24). We have extended the methylation analysis to the five novel K and P lines presented here and the CpG and Pl:CpG lines. None of the novel K and P lines showed methylation of this site (data not shown). However, in the ex1 lines 1 and 2, the SacII/SstII site was fully methylated in the T3 generation (Fig. 5A and B), although the nptII gene was active. In addition, the NarI and NruI sites in the nptII and β regions, respectively, were completely methylated in ex1 line 1, while the PstI site in the nptII gene was unmethylated (Fig. 5B). In ex1–2, the NarI site was methylated, while partial methylation was observed for the NruI and PstI sites (Fig. 5B). In the ex2–4 lines, the PstI sites were unmethylated in all lines (not shown). In contrast, the NarI and NruI sites were methylated in all lines (Fig. 5D and data not shown). Differences in methylation status between individual lines were observed for the SacII/SstII site, which was partially methylated in line 4 and fully methylated in line 7 (Fig. 5C). In addition, partial methylation was seen at a PvuII site in line 7 (data not shown). While nptII silencing has been observed in line 4 (Table 1), line 7 showed no silencing for four generations. In the plant:CpG-rich constructs, three sites were assayed in the T2 and T3 generations. The SacII/SstII site was unmethylated in all lines (Fig. 5F). In contrast, PstI and PvuII detected partial methylation for two lines (16 and 21) in T2 and an increasing number of lines displaying partial methylation in T3 (Fig. 5G and data not shown). Only two of these lines (19 and 20) have shown nptII silencing (Table 1).
Figure 5.
Methylation analysis of sites in the pnos–nptII region in CpG and Pl:CpG lines. DNA from transgenic plants generated with the constructs ex1 and ex2–4 (A) and plant:CpG-rich (E) was digested with a methylation- sensitive enzyme in combination with a delimiting methylation-insensitive enzyme. Digested DNA was subjected to Southern hybridisation with a nptII coding region probe [indicated in (A) and (E)]. (A) The nptII region common for both constructs is depicted (refer to Fig. 1). Sites for non-methylation-sensitive limiting enzymes, NcoI for ex1 and HincII for ex2–4, are shown. (B) Methylation analysis of ex1 lines 1 and 2 after digestion with NcoI (N) or this enzyme in combination with SacII/SstII, PstI, NarI or NruI. The latter sites are methylated when the hybridising fragment has the same size as the delimiting fragment (N). The presence of this fragment and in addition smaller fragments indicate partial methylation. (C) Methylation analysis of ex2–4 lines using the delimiting enzyme HincII (H) alone or in combination with SacII. A slightly shorter hybridising band is generated when the SacII site is unmethylated. (D) Methylation analysis of ex2–4 lines using the delimiting enzyme HincII (H) alone or in combination with NarI. Note that the hybridising fragment is identical in size to the delimiting fragment for all lines. (E and F) Methylation analysis in plant:CpG-rich lines with HindIII (H) as the delimiting enzyme. The SacII (S) site was unmethylated in all lines as indicated by a hybridising fragment of 1660 bp depicted by an arrow. (G) Methylation analysis in the PvuII site in plant:CpG-rich lines using HindIII as the delimiting enzyme. Note the partial methylation in all lines as indicated by fragments that are larger than the fragment of the delimiting enzyme, marked by an arrow. Thick lines, plant DNA; N, NarI; Nr, NruI; P, PstI; Pv, PvuII; S, SacII/SstII. Asterisks, siblings of same line; TS, transgene silencing lines.
DISCUSSION
We have cloned the regions flanking the T-DNAs in a collection of transformed lines comprising transgene silencing (TS) and control lines (C) not displaying silencing, to investigate whether we could find distinguishing differences. Small deletions at the integration site, as well as at the RB and LB of the T-DNAs, were observed (Table 2), in agreement with earlier findings (1). The right end of the T-DNA coincided with the preferred nicking site of the VirD2 protein for one-third of the lines. The TS lines were in these respects indistinguishable from the C lines.
We did, however, note a difference between the constructs (Fig. 3). For the lines transformed with pKOH110-derived T-DNAs, the RB deletions are, with one exception (line K14), clustered at the RB, while in the pMHA2 lines and even more so in the pPCV002-derived lines a larger base pair range is seen in the deletions. Less efficient nicking by the VirD2 protein at the border sequences may occur if the borders are not the natural context (18). Only the first 35 bp are common at the right border of the pKOH110 and pMHA2 vectors. The shorter natural sequence context of the inner right border of pMHA2 might influence the length of the RB deletion. pMHA2 and pKOH110 have borders from an octopine Agrobacterium strain, while the pPCV002 vector has a nopaline right border. Differences in right border deletions may therefore be due to the differences between the VirD2 proteins of the nopaline- and octopine-based Agrobacterium strains used.
Micro-homologies at the right border may mediate T-DNA insertions
Although T-DNA/plant DNA junctions have been sequenced in a number of studies, most studies have characterised a limited number of integration events (9,36,37) or multicopy or complex T-DNA integrations (7,8,16,17). The cloning of the T-DNA/plant DNA junctions in our collection of single copy lines has made it possible to evaluate the current models for T-DNA integration. The ‘LB first’ model of Tinland (6,38) implies that the 3′ end of the single-stranded T-DNA invades plant DNA. The T-DNA is anchored near the 3′ end to plant DNA through micro-homology, i.e. a short (2–5 bp) contiguous stretch of homologous nucleotides. The nucleotide of the T-DNA strand downstream of the micro-homology is degraded. Only the displaced strand of the plant DNA is nicked at the synapsis. According to this model perfect micro-homology without any filler DNA should be found at the left T-DNA/plant DNA junction. In our material, 12 lines have junctions compatible with this model.
To account for the presence of fillers, which in our material were found at ∼50% of the left junctions, the model must be modified. As shown in Figure 4A and B, some junctions can either be regarded as fillers or as non-perfect micro-homologies. These may have arisen by the repair of heteroduplexes generated from short stretches of non- contiguous micro-homologies between T-DNA and plant target DNA (9). Others of our fillers are identical to short stretches of genomic or T-DNA sequences close to the junctions (Fig. 4C) and, in one case (line 609), identical to a sequence of another chromosome. Filler DNA can be captured in cis from the same molecule, in trans from another molecule or from genomic DNA, and in cis capture seems most efficient (39). Formation of fillers is thought to be a common result of repair of double-strand breaks (DSB) (10). In fact T-DNA has been found to integrate into DSB generated experimentally in somatic plant cells (40). In our material, fillers at both junctions were observed in one-third of the lines, and may indicate insertion of T-DNA in DSB present in plant DNA due to a variety of DNA-breaking factors.
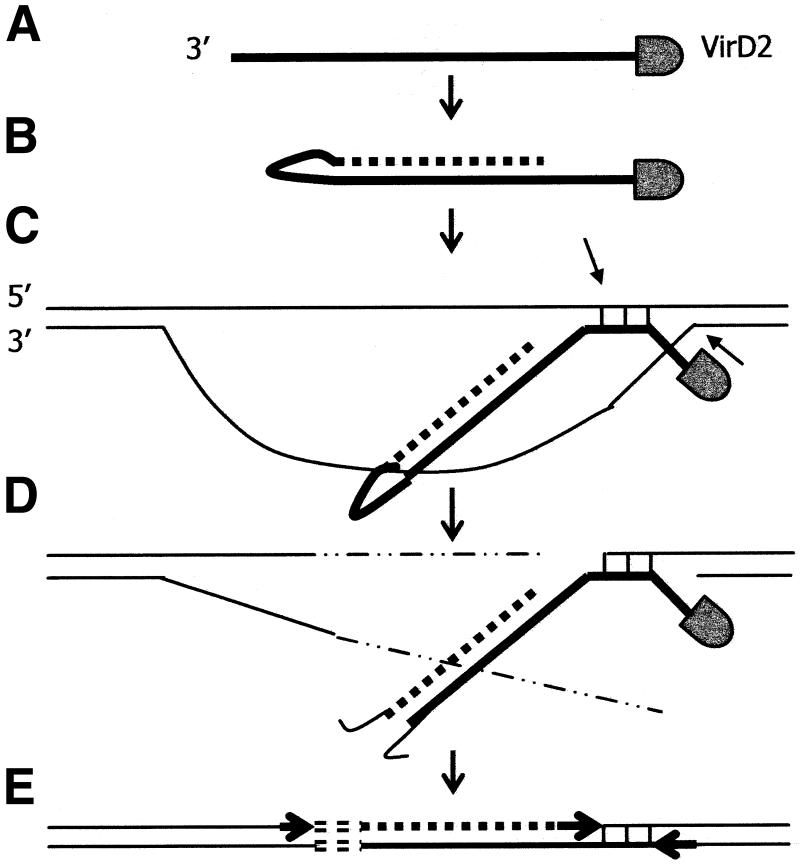
Another interesting finding in our material is that perfect micro-homologies without fillers are common not only near the LB, but also near the RB. At the right end, this homology was in two cases found at the processing site of VirD2 (lines 66 and K11). These two lines fit the Tinland model, where VirD2 attached to the 5′ end nicking site screen for micro-homologies on the complementary target strand and exchanges the phosphotyrosine bond at the 5′ end for a phosphodiester bond with the target strand (38). However, in six of our cases, the micro-homology is more distant from the RB, especially in lines 607, K14 and plant:CpG 19 (Fig. 3). Mayerhofer et al. (37) found two lines with micro-homologies at both the right and left ends without fillers. Their model, suggesting annealing of both T-DNA ends to genomic DNA followed by deletion of the genomic DNA between the regions of base pairing, is applicable to our ex2–4 line 5. However, we also have an example (K14) in which micro-homology without filler is found at the RB, while a filler is found at the left junction. This may indicate that T-DNA can associate with plant DNA by a ‘RB first’ mechanism, either in DSB or by invasion of intact DNA (Fig. 6A). If so, no free 3′-OH plant DNA end can prime the synthesis of the complementary T-DNA strand. It has been indicated that primases could synthesise primers fitting the left end, allowing complementary strand synthesis (8). Alternatively, the left end of the T-DNA can loop back on itself to prime the synthesis of double-stranded T-DNA (Fig. 6B). This could account for the shortening of the T-DNA at the left end. Opening of the single-stranded loop at this end, followed by repair and ligation, would explain the finding of a filler at the left T-DNA/plant DNA junction (Fig. 6C–E).
Figure 6.
T-DNA integration model based on micro-homologies between T-DNA and genomic DNA near the T-DNA RB (see also text). (A) Single-stranded T-DNA enters the nucleus covered with VirE2 protein (not shown) and with VirD2 attached to the 5′ end. (B) The 3′ end loops back and anneals to itself, thus providing a 3′-OH end to prime synthesis of a complementary T-DNA strand (broken line), thereby removing VirE2 (not shown). (C) T-DNA, still single stranded near the 5′ end, invades genomic DNA and finds a short stretch of micro-homology (vertical lines). Nicks will occur in the genomic DNA as indicated by the arrows. (D) The 3′ end loop in double-stranded T-DNA is degraded resulting in a left end deletion. Genomic DNA is degraded (thin broken lines), resulting in a target deletion. (E) VirD2 and the 5′ end of T-DNA upstream of the micro-homology is removed. Single-stranded gaps are repaired (arrows) and the DSB between the T-DNA left end and the genomic DNA is repaired, thereby generating filler DNA (short open bars).
Integration of the BVB is not sufficient to induce transgene silencing
Although Southern hybridisation had indicated that our lines harboured single, non-rearranged T-DNAs, the investigation revealed the presence of undiscovered truncated T-DNA copies in three lines, five cases of BVB integration and two lines with complex integration patterns.
The presence of complex arrangements of T-DNA and integrated BVB have been proposed to be factors leading to silencing of transgenes in T-DNAs by homology-dependent transcriptional or conformational silencing (41,42). In most publications such proposals have been based on a low number of lines and these lines have rarely been compared to lines not displaying silencing. A configuration where the RB leads directly into the binary vector sequences has for instance been reported for two unstably expressed transgene loci in tobacco (42). Our ex2–4 line 6 has a similar arrangement, with almost 4 kb of BVB at the RB. However, this line was tested for five successive generations and does not display silencing (26). Neither did ex2–4 line 12, with an even more complex structure, show silencing over five generations.
Of the three lines where LB BVB was found, only one (K11) displayed silencing. We have shown that the human CpG island of the ex1 and ex2–4 lines is protected from methylation when introduced into the plant genome (26). Our data also indicate that the human CpG island, but not any CpG-rich sequence, e.g. the barley B22E gene plant:CpG-rich construct, protect neighbouring transgenes from silencing (26). One should, however, note that the pMHA2 line 200, which is devoid of human CpG sequences, harbours more than 1200 bp of BVB without any silencing effect. In this line, as in other lines, the T-DNA was integrated in a region with a GC content of ∼39%, while the GC content of the BVB is 57.6% (T.J.Meza, M.Skårn, I.S.Mercy, D.A.Nymoen, M.C.Eike and R.B.Aalen, in preparation). Thus, even with strongly divergent GC content between target and foreign DNA, the presence of BVB is not sufficient to induce silencing mechanisms.
ex2–4 line 6, containing nearly 4 kb of BVB extending from the right border, has no backbone sequences at the LB junction. It therefore seems unlikely that the whole binary vector had been transferred during transformation (12,18). Pseudoborders upstream of the RB have been suggested occasionally to function as targets for VirD2 nicking and the transfer start point (13,43). No pseudoborder sequence was detected at the end of the integrated BVB. In ex2–4 line 6, it is therefore more likely that a left to right T-DNA transfer has occurred. If VirD2 nicks at the LB, associates with the 5′ end of the non-T-DNA strand (13,44,45) and then skips the RB, it is conceivable that BVB in continuation with RB is transferred to the plant cell.
In ex2–4 line 12, the BVB has micro-homology to the intact T-DNA at one end and to the genomic DNA at the other end. This indicates a recombination event between T-DNA, BVB and plant DNA during integration.
Integration of BVB extending from the LB is thought to result from skipping of the LB during the production of the T-DNA strand in Agrobacterium (14,18). In our material, BVB was more frequent at the LB side (3/29, 10%) than on the RB side (1/30, 3%). This is in agreement with De Buck et al. (18), but in contrast to Kononov et al. (14). The overall incidence of BVB integration found in our material is lower than reported by these groups [20–50% (18) and 75% (14)]. This might result from the fact that we, in contrast to others (13,14,18), have selected single-copy lines.
Partial inverted T-DNA repeats are not sufficient to induce transgene silencing
Truncated T-DNAs, organised in inverted orientation compared with the intact T-DNA copy, were encountered in three cases in the CpG lines. The truncated T-DNAs are most likely remnants of ordinary T-DNA integrations where the right part of the T-DNA to a lesser or greater extent has been degraded during integration. Inverted repeats have been considered a trigger of transgene silencing and to be prone to de novo methylation (46,47). We have previously screened 110 transgenic lines for nptII silencing and found transgene silencing in nearly half of them. Of 12 lines segregating T-DNA as a single Mendelian locus, nine harboured more than one copy and four of these had two T-DNAs organised as inverted repeats (24). However, in the three CpG lines shown to contain partial inverted repeats, only one (ex2–4 line 4) displayed silencing (Fig. 2) (26), suggesting that the presence of inverted T-DNA sequences in itself is not sufficient to trigger transgene silencing. A notable difference between line 4 and the two other lines is that the truncated T-DNA of line 4 encompasses the whole nptII gene with its pnos promoter. There is no possibility of the production of an antisense transcript from endogenous promoters of annotated genes flanking the T-DNA in any of the lines comprising partially inverted repeats (T.J.Meza, M.Skårn, I.S.Mercy, D.A.Nymoen, M.C.Eike and R.B.Aalen, in preparation). This may indicate that silencing is only induced from inverted repeats when there are two identical promoters in the repeat. Stam et al. (47) have shown that silencing of transgenes in inverted repeats with two complete T-DNA copies in most cases is due to transcriptional silencing and that the silenced nptII genes were heavily methylated. The SacII/SstII site in the pnos promoter, which has been considered diagnostic for silencing (48,49), is partially methylated in line 4. However, we have not found any correlation between the presence of inverted repeats and methylation, nor between silencing and methylation (Fig. 5). The plant:CpG-rich lines are particularly illustrative; while only two out of four lines show silencing, none are methylated in the SacII/SstII site and all are partially methylated at sites in the nptII coding region. In the CpG lines, displaying methylation of the SacII/SstII site, but no silencing, the CpG island possibly keeps the transgenic construct in an open chromatin structure despite methylation (26).
In investigating TS lines, in most cases one will find multiple copies, complex integrations and/or the presence of BVB. We have focused on single-copy lines displaying silencing and included in our study a large control group. By taking this approach we are confident to conclude that the BVB, short inverse repeats and methylation are factors that in themselves are not the cause of transgene silencing of single-copy transgenes. We infer that silencing in our lines is due to position effects, e.g. specific features in the surrounding plant DNA, possibly in interaction with the integrated T-DNA. A thorough analysis of the plant DNA flanking these insertions is underway.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Z. Salehian, S.H. Engebretsen and R. Falleth for technical assistance. This work was supported by grant no. 129525/420 from the Norwegian Research Council.
REFERENCES
- 1.Gheysen G., Angenon,G. and Van Montagu,M. (1998) Agrobacterium-mediated plant transformation: a scientifically intriguing story with significant applications. In Lindsey,K. (ed.), Transgenic Plant Research. Harwood Academic Publishers, Amsterdam, The Netherlands, pp. 1–33.
- 2.Zupan J., Muth,T.R., Draper,O. and Zambryski,P. (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J., 23, 11–28. [DOI] [PubMed] [Google Scholar]
- 3.Joos H., Timmermann,B., Van Montagu,M. and Schell,J. (1983) Genetic analysis of transfer and stabilization of Agrobacterium DNA in plant cells. EMBO J., 2, 2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jen G. and Chilton,M.-D. (1986) The right border region of pTiT37 T-DNA is intrinsically more active than the left border region in promoting T-DNA transformation. Proc. Natl Acad. Sci. USA, 83, 3895–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caplan A., Van Montagu,M. and Schell,J. (1985) Genetic analysis of integation mediated by single T-DNA borders. J. Bacteriol., 161, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tinland B. (1996) The integration of T-DNA into plant genomes. Trends Plant Sci., 1, 178–184. [Google Scholar]
- 7.De Buck S., Jacobs,A., Van Montagu,M. and Depicker,A. (1999) The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J., 20, 295–304. [DOI] [PubMed] [Google Scholar]
- 8.De Neve M., De Buck,S., Jacobs,A., Van Montagu,M. and Depicker,A. (1997) T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J., 11, 15–29. [DOI] [PubMed] [Google Scholar]
- 9.Gheysen G., Villarroel,R. and Van Montagu,M. (1991) Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev., 5, 287–297. [DOI] [PubMed] [Google Scholar]
- 10.Gorbunova V.V. and Levy,A.A. (1999) How plants make ends meet: DNA double-strand break repair. Trends Plant Sci., 4, 263–269. [DOI] [PubMed] [Google Scholar]
- 11.Denis M., Renard,M. and Krebbers,E. (1995) Isolation of homozygous transgenic Brassica napus lines carrying a seed-specific chimeric 2S albumin gene and determination of linkage relationships. Mol. Breeding, 1, 143–153. [Google Scholar]
- 12.Wenck A., Czako,M., Kanevski,I. and Marton,L. (1997) Frequent collinear long transfer of DNA inclusive of the whole binary vector during Agrobacterium-mediated transformation. Plant Mol. Biol., 34, 913–922. [DOI] [PubMed] [Google Scholar]
- 13.Ramanathan V. and Veluthambi,K. (1995) Transfer of non-T-DNA portions of the Agrobacterium tumefaciens Ti plasmid pTiA6 from the left terminus of TL-DNA. Plant Mol. Biol., 28, 1149–1154. [DOI] [PubMed] [Google Scholar]
- 14.Kononov M.E., Bassuner,B. and Gelvin,S.B. (1997) Integration of T-DNA binary vector ‘backbone’ sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J., 11, 945–957. [DOI] [PubMed] [Google Scholar]
- 15.Martineau B., Voelker,T.A. and Sanders,R.A. (1994) On defining T-DNA. Plant Cell, 6, 1032–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krizkova L. and Hrouda,M. (1998) Direct repeats of T-DNA integrated in tobacco chromosome: characterization of junction regions. Plant J., 16, 673–680. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S. and Fladung,M. (2000) Transgene repeats in aspen: molecular characterisation suggests simultaneous integration of independent T-DNAs into receptive hotspots in the host genome. Mol. Gen. Genet., 264, 20–28. [DOI] [PubMed] [Google Scholar]
- 18.De Buck S., De Wilde,C., Van Montagu,M. and Depicker,A. (2000) T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol. Breeding, 6, 459–468. [Google Scholar]
- 19.Kooter J.M., Matzke,M.A. and Meyer,P. (1999) Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci., 4, 340–347. [DOI] [PubMed] [Google Scholar]
- 20.Stam M., Mol,J.N.M. and Kooter,J.M. (1997) The silence of genes in transgenic plants. Ann. Bot., 79, 3–12. [Google Scholar]
- 21.Kumpatla S.P., Chandrasekharan,M.B., Iyer,L.M., Li,G.F. and Hall,T.C. (1998) Genome intruder scanning and modulation systems and transgene silencing. Trends Plant Sci., 3, 97–104. [Google Scholar]
- 22.Matzke M.A., Matzke,A.J.M. and Eggleston,W.B. (1996) Paramutation and transgene silencing: a common response to invasive DNA? Trends Plant Sci., 1, 382–388. [Google Scholar]
- 23.Day C.D., Lee,E., Kobayashi,J., Holappa,L.D., Albert,H. and Ow,D.W. (2000) Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev., 14, 2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meza T.J., Kamfjord,D., Håkelien,A.M., Evans,I., Godager,L.H., Mandal,A., Jakobsen,K.S. and Aalen,R.B. (2001) The frequency of silencing in Arabidopsis thaliana varies highly between progeny of siblings and can be influenced by environmental factors. Transgenic Res., 10, 53–67. [DOI] [PubMed] [Google Scholar]
- 25.Meyer P., Linn,F., Heidmann,I., Meyer,H., Niedenhof,I. and Saedler,H. (1992) Endogenous and environmental factors influence 35S promoter methylation of a maize A1 gene construct in transgenic petunia and its colour phenotype. Mol. Gen. Genet., 231, 345–352. [DOI] [PubMed] [Google Scholar]
- 26.Meza T.J., Enerly,E., Børud,B., Larsen,F., Mandal,A., Aalen,R.B. and Jakobsen,K.S. (2002) A human CpG island randomly inserted into a plant genome is protected from methylation. Transgenic Res., 11, 133–142. [DOI] [PubMed] [Google Scholar]
- 27.Mandal A., Lång,V., Orczyk,W. and Palva,E.T. (1993) Improved efficiency for the T-DNA-mediated transformation and plasmid rescue in Arabidopsis thaliana. Theor. Appl. Genet., 86, 621–628. [DOI] [PubMed] [Google Scholar]
- 28.Koncz C. and Schell,J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet., 204, 383–396. [Google Scholar]
- 29.Steinum T.M., Berner,H.S., Stacy,R.A.P., Salehian,Z. and Aalen,R.B. (1998) Differential regulation of the barley (Hordeum vulgare) transcripts B22E and B12D in mature aleurone layers. Physiol. Plant, 102, 337–345. [Google Scholar]
- 30.Klemsdal S.S., Hughes,W., Lonneborg,A., Aalen,R.B. and Olsen,O.-A. (1991) Primary structure of a novel barley gene differentially expressed in immature aleurone layers. Mol. Gen. Genet., 228, 9–16. [DOI] [PubMed] [Google Scholar]
- 31.Mandal A., Sandgren,M., Holmstrom,K.-O., Gallois,P. and Palva,E.T. (1995) Identification of Arabidopsis thaliana sequences responsive to low temperature and abscisic acid by T-DNA tagging and in-vivo gene fusion. Plant Mol. Biol. Rep., 13, 243–254. [Google Scholar]
- 32.Deblaere R., Bytebier,B., De Greve,H., Deboeck,F., Schell,J., Van Montagu,M. and Leemans,J. (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res., 13, 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellaporta S.L., Wood,J. and Hicks,J.B. (1983) A plant DNA minipreparation: version II. Plant Mol. Biol. Rep., 1, 19–21. [Google Scholar]
- 34.Baumbusch L.O., Sundal,I.K., Hughes,W.D., Galau,G.A. and Jakobsen,K.S. (2001) Efiicient protocols for CAPS-based mapping in Arabidopsis. Plant Mol. Biol. Rep., 19, 137–149. [Google Scholar]
- 35.Devic M., Albert,S., Delseny,M. and Roscoe,T.J. (1997) Efficient PCR walking on plant genomic DNA. Plant Physiol. Biochem., 35, 331–339. [Google Scholar]
- 36.Matsumoto S., Ito,Y., Hosoi,T., Takahashi,Y. and Machida,Y. (1990) Integration of Agrobacterium T-DNA into a tobacco chromosome: possible involvement of DNA homology between T-DNA and plant DNA. Mol. Gen. Genet., 224, 309–316. [DOI] [PubMed] [Google Scholar]
- 37.Mayerhofer R., Koncz-Kalman,Z., Nawrath,C., Bakkeren,G., Crameri,A., Angelis,K., Redei,G.P., Schell,J., Hohn,B. and Koncz,C. (1991) T-DNA integration: a mode of illegitimate recombination in plants. EMBO J., 10, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tinland B. and Hohn,B. (1995) Recombination between prokaryotic and eukaryotic DNA: integration of Agrobacterium tumefaciens T-DNA into the plant genome. Genet. Eng., 17, 209–229. [PubMed] [Google Scholar]
- 39.Gorbunova V. and Levy,A.A. (1997) Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res., 25, 4650–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salomon S. and Puchta,H. (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J., 17, 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fladung M. (1999) Gene stability in transgenic aspen (Populus). I. Flanking DNA sequences and T-DNA structure. Mol. Gen. Genet., 260, 574–581. [DOI] [PubMed] [Google Scholar]
- 42.Iglesias V.A., Moscone,E.A., Papp,I., Neuhuber,F., Michalowski,S., Phelan,T., Spiker,S., Matzke,M. and Matzke,A.J. (1997) Molecular and cytogenetic analyses of stably and unstably expressed transgene loci in tobacco. Plant Cell, 9, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miranda A., Janssen,G., Hodges,L., Peralta,E.G. and Ream,W. (1992) Agrobacterium tumefaciens transfers extremely long T-DNAs by a unidirectional mechanism. J. Bacteriol., 174, 2288–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Graaff E., den Dulk-Ras,A. and Hooykaas,P.J. (1996) Deviating T-DNA transfer from Agrobacterium tumefaciens to plants. Plant Mol. Biol., 31, 677–681. [DOI] [PubMed] [Google Scholar]
- 45.Durrenberger F., Crameri,A., Hohn,B. and Koukolikova-Nicola,Z. (1989) Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc. Natl Acad. Sci. USA, 86, 9154–9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selker E.U. (1999) Gene silencing: repeats that count. Cell, 97, 157–160. [DOI] [PubMed] [Google Scholar]
- 47.Stam M., Viterbo,A., Mol,J.N. and Kooter,J.M. (1998) Position-dependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: implications for posttranscriptional silencing of homologous host genes in plants. Mol. Cell. Biol., 18, 6165–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matzke M.A., Primig,M., Trnovsky,J. and Matzke,A. (1989) Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J., 8, 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kilby N.J., Leyser,H.M.O. and Furner,I.J. (1992) Promotor methylation and progressive transgene inactivation in Arabidopsis. Plant Mol. Biol., 20, 103–112. [DOI] [PubMed] [Google Scholar]