Abstract
The 3′ end region of foot-and-mouth disease virus (FMDV) consists of two distinct elements, a 90 nt untranslated region (3′-NCR) and a poly(A) tract. Removal of either the poly(A) tract or both the 3′-NCR and the poly(A) tract abrogated infectivity in susceptible cells in the context of a full-length cDNA clone. We have addressed the question of whether the impairment of RNA infectivity is related to defects at the translation level using a double approach. First, compared to the full-length viral RNA, removal of the 3′ sequences reduced the efficiency of translation in vitro. Secondly, a stimulatory effect of the 3′ end sequences on IRES-dependent translation was found in vivo using bicistronic constructs. RNAs carrying the FMDV 3′ end sequences linked to the second cistron showed a significant stimulation of IRES-dependent translation, whereas cap-dependent translation was not affected. Remarkably, IRES-dependent stimulation exerted by the poly(A) tract or the 3′-NCR seems to be the result of two separate events, as the 3′-NCR alone enhanced IRES activity on its own. Under conditions of FMDV Lb protease-induced translation shut-off, the stimulation of IRES activity reached values above 6-fold in living cells. A northern blot analysis indicated that IRES stimulation was not the consequence of a change in the stability of the bicistronic RNA produced in transfected cells. Analysis of the RNA-binding proteins interacting with a mixture of 3′ end and IRES probes showed an additive pattern. Altogether, our results strongly suggest that individual signals in the viral 3′ end ensure stimulation of FMDV translation.
INTRODUCTION
Internal initiation of translation driven by internal ribosome entry site (IRES) elements is unique in several ways. On the one hand, the RNA structural features differ for particular IRES elements. On the other hand, as a function of the IRES element studied, the functional relevance of eukaryotic initiation factors (eIFs) and other RNA-binding proteins seems to be widely different (1–3).
A stimulatory effect of poly(A) sequences on the internal initiation efficiency driven by IRES elements has been described very recently in cell-free extracts (4–7). The intensity of this enhancing effect was smaller than the synergism found between poly(A) tails and cap-dependent translation initiation (8). This synergism was later explained by a process of RNA circularization mediated by the inter action of poly(A)-binding protein (PABP) with the eukaryotic initiation factor eIF4G (9). Whether a similar mechanism applies to picornavirus IRES-driven translation initiation is not yet known. Both isoforms of eIF4G (eIF4GI and eIF4GII) are proteolytically cleaved by the action of foot-and-mouth disease virus (FMDV) Lb or enterovirus 2A proteases (10,11). As a consequence of this cleavage, the PABP-interacting domain in eIF4G becomes separated from the C-terminal fragment, involved in IRES-dependent translation (12,13). Therefore, PABP–eIF4G-dependent RNA circularization is most likely prevented during FMDV infection. Conversely, it is possible that an RNA–protein bridge could be established between factors interacting with specific sequences in the 3′-non-coding region (NCR) and the IRES. In this respect, there are examples of proteins interacting with viral 3′-NCR that are involved in IRES interaction, providing potential links between 3′ end and IRES sequences (14–17).
FMDV is the causative agent of an acute systemic disease of cloven hoof animals, considered as a major animal health problem world wide (18,19). The FMDV genome consists of a single molecule of messenger-sense RNA of ∼8500 nt containing a single open reading frame (ORF) coding for the viral polyprotein which is subsequently processed into the viral products (20). The 5′-NCR, ∼1200 nt in length, is highly structured and contains several genetic elements necessary to control the replication cycle. This region harbors an IRES element that promotes cap-independent translation initiation of the viral genome (2). Immediately downstream of the stop codon at the end of the polyprotein ORF there is a phylogeneticaly conserved 3′-NCR of ∼90 nt (21) followed by a genome encoded polyadenylate tract (22). At present, the involvement of the FMDV 3′-NCR and/or the poly(A) tract in translation initiation and whether cross-talk exists that communicates between the 3′ end and sequences in the 5′-NCR, including the IRES region, is not known.
The role of the 3′-NCR in aphthovirus infection is poorly understood, though experimental evidence supports its relevance in the FMDV viral cycle. RNA transcripts spanning the 3′ terminal region of the FMDV genome have been reported to inhibit infective particle formation following co-microinjection or co-transfection with viral RNA in BHK-21 cells (23,24). Deletion of the 3′-NCR abolished infectivity and the 3′-NCR of another picornavirus, swine vesicular disease virus, did not restore the infectivity of the chimeric RNA. Furthermore, a major defect in RNA replication was found in the non-infectious RNAs (21). In other picornaviruses it is known that a cis-acting determinant for replication resides in the 3′ end (25–29).
Here we have studied the contribution of the FMDV 3′-NCR and the poly(A) tract to the translation efficiency of full-length viral RNA and bicistronic constructs carrying the FMDV IRES. A stimulatory effect on viral RNA translation was observed in the context of full-length FMDV transcripts in which each of these 3′ end elements was present. This effect was magnified in living cells when the 3′-NCR and/or the poly(A) tract were placed downstream of the second gene in a bicistronic construct, in which the FMDV IRES promotes internal initiation of the second cistron. An IRES-dependent stimulatory effect was strongly enhanced during co-expression of the FMDV Lb protease, mimicking the situation found in infected cells. The pattern of RNA-binding proteins has been analyzed regarding their involvement in IRES-dependent stimulation.
MATERIALS AND METHODS
Construction of plasmids
Plasmids pDM and pTAG, described previously (21), are based on a FMDV O1K infectious full-length cDNA clone (30). Subcloning of the 3′ end region present in pTAG yielded the subTAG construct, bearing a unique NotI restriction site downstream of the FMDV insert. Briefly, the 3′-NCR and poly(A) sequences were excised from pTAG by digestion with AvrII and HpaI. The restriction fragment was gel purified, filled-in with T4 DNA polymerase and ligated into Bluescript II SK+ previously linearized with SacI, blunt ended and dephosphorylated.
To prepare bicistronic constructs harboring the FMDV 3′ end sequences, plasmid pBIC (31) was modified to introduce unique AvrII and NotI restriction sites, immediately downstream of the luciferase stop codon. Site-directed mutagenesis was performed using a two-step PCR procedure, essentially as described (32). The external primers ClaI-s (CCGCTGAATTGGAATCGATATTG) and HpaI-as (GCTGCAATAAACAAGTTAACAAC) were used in combination with the mutagenic oligonucleotides AN-as (CGCGGCCGCTACATCCTAGGATTTGGAC) and AN-s (GGATGTAGCGGCCGCCAGCGATGACG) (complementary sequences are underlined and NotI and AvrII restriction sites are in bold). The purified product from the double PCR, digested with HpaI and ClaI, was ligated to pBIC, similarly treated. This sequence modification did not alter its translation properties relative to the unmodified pBIC (data not shown).
The bicistronic construct pBIC-3′NCR-A58 contains the FMDV 3′ end present in pTAG (21). The subTAG plasmid described above was used to amplify by PCR the 3′ end FMDV sequences with the oligonucleotides T7 and 3′NCR-Avr (GAACAAAAGCTGCCTAGGCCC; AvrII site in bold). The PCR product, digested with AvrII, blunt ended and then restricted with NotI, was ligated into the pBIC modified plasmid, similarly treated. Construct pBIC-3′NCR was then generated using oligonucleotides 3′NCR-Avr and as-RVNot (GACCCGCGGCCGCTGGATATCAAGGAAG; NotI and EcoRV sites in bold) with pBIC-3′NCR-A58 as template. The PCR product treated with EcoRV and AvrII was ligated into pBIC, similarly digested. To generate pBIC-A58, including the 58 adenines present in the pDM infectious clone (see Fig. 1), the EcoRV–NotI insert from plasmid subTAG was ligated in pBIC, digested with AvrII, blunt ended and then treated with NotI.
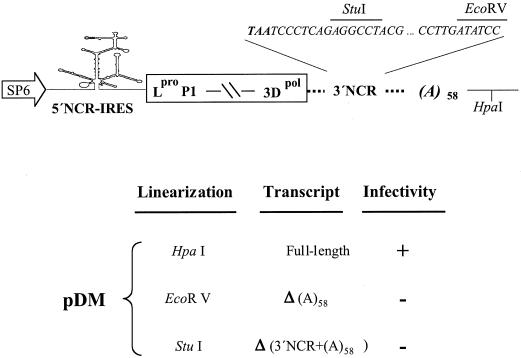
Figure 1.
Effects of 3′ end sequences on FMDV infectivity. Schematic representation of the cDNA present in pDM (21) including relevant restriction sites (top panel). The broken line between P1 and 3Dpol is used for simplicity to represent the coding region. Infectivity of the different transcripts was measured as the ability of an equal amount (4 µg) of the indicated transcripts, transfected into 1–2 × 106 BHK-21 cells, to induce cytopathic effect up to 48 h post-transfection (lower panel). The positive control pDM induced a cytopathic effect 16–20 h post-transfection.
Prior to expression analysis, the nucleotide sequence of the entire length of each region under study was determined using automatic sequencing (ABI PRISM dye terminator cycle sequencing ready reaction kit, Perkin Elmer).
Infectivity assays
Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal bovine serum and 1 mM HEPES, pH 7.4. Semi-confluent BHK-21 or IBRS-2 monolayers were transfected as described (21) using 3–5 µg of transcripts obtained by transcribing HpaI-, EcoRV- or StuI-linearized pDM with SP6 RNA polymerase (see Fig. 1). Transfected cells were incubated at 37°C until an extensive cytopathic effect was observed or longer, up to 48 h post-transfection, for RNAs not inducing a cytopathic effect. In previous assays, neither prolonged incubation of cells transfected with the 3′-NCR-deleted RNA nor three consecutive blind passages of the transfection supernatant in susceptible cells resulted in the formation of pseudorevertants (21).
In vitro translation
Following examination of transcripts generated as above for RNA integrity in agarose gels, 100, 200 and 400 ng of each transcript were translated in 10 µl of rabbit reticulocyte lysate (Promega) in the presence of 10 µCi of [35S]methionine (10 mCi/ml) essentially as described (32). Reaction mixtures were incubated at 30°C for 1 h. Aliquots of the translation products were loaded onto a 12% SDS–polyacrylamide gel. The gels were then dried and exposed to X-ray films. Quantification of data was performed using a phosphorimager.
In vivo IRES-dependent translation activity
Bicistronic plasmids carrying the FMDV IRES between the chloramphenicol acetyltransferase (CAT) and luciferase genes, and the FMDV 3′ end sequences downstream of the second cistron were assayed in BHK-21 cells. Transfection of 80–90% confluent monolayers was carried out using cationic liposomes 1 h after infection with the vaccinia virus VTF7-3, expressing T7 RNA polymerase (33). Extracts from 2 × 105 cells were prepared 20 h after transfection in 100 µl of 50 mM Tris–HCl, pH 7.8, 120 mM NaCl, 0.5% NP-40. Luciferase and CAT activities were measured as described (34). Plasmid pLb, encoding the FMDV Lb protease, was co-transfected in assays designed to shut down cap-dependent translation (31). Assays were performed at least three times.
Northern blot analysis
BHK-21 cells (∼1 × 106) transfected with bicistronic plasmids, co-transfected or not with pLb, were prepared at 12 h following transfection, as described above for IRES-dependent translation activity assays. Infection with VTF7-3 was omitted to prepare a DNA control and mock-transfected cells. The resulting lysates were then centrifuged at 12 000 r.p.m. for 5 min and total cytoplasmic RNA was extracted with TRI Reagent (Roche) and resuspended in 20 µl of TE. Aliquots (6 and 12 µl) of each RNA were loaded in a formaldehyde–agarose gel and blotted onto a nylon Zeta probe membrane (Bio-Rad). The probe was prepared by in vitro transcription using SP6 RNA polymerase (Promega) in the presence of 50 µCi of [α-32P]UTP (400 Ci/mmol; Amersham) and 500 ng of a pGEM-IRES plasmid containing the full-length IRES of FMDV C-S8 (35), linearized with EcoRI. The probe was subsequently purified using a microSpin G25 column (Amersham). The resulting probe is an RNA fragment of 470 nt complementary to the FMDV IRES. Hybridization was performed in 50% formamide, 0.12 M Na2HPO4, pH 7.2, 0.25 M NaCl, 7% SDS, containing 0.5 × 106 c.p.m./ml, and incubated overnight at 42°C. The membrane was then washed three times for 10 min each at room temperature with 2× SSC, 0.1% SDS, 0.5× SSC, 0.1% SDS and 0.1× SSC, 0.1% SDS and exposed to X-ray film.
RNA–protein interaction assays
S10 extracts from BHK-21 cells were prepared basically as described (36). The probe corresponding to the FMDV 3′ end was prepared from construct subTAG linearized with NotI, and the FMDV IRES probe was generated from a pGEM clone containing the full-length IRES region (35). RNA transcripts were labeled to a specific activity of ∼107 c.p.m./pmol using [α-32P]CTP (400 Ci/mmol) and incubated with RQ1 DNase (1 U/µg RNA; Promega). Unincorporated [32P]CTP was eliminated by exclusion chromatography on Sephadex G 50–80 (Sigma) columns equilibrated with 10 mM Tris–HCl pH 8, 1 mM EDTA. RNAs were then phenol/chloroform extracted and ethanol precipitated. RNA integrity was determined by 6% acrylamide–6 M urea denaturing gel electrophoresis. UV-crosslinking assays were performed as described (13) using 40 µg of native proteins present in S10 extracts of BHK-21 cells and the specific 32P-labeled RNA (0.03 pmol). The RNA–protein crosslinked lysate was digested with an excess of RNase A during 30 min at 37°C, followed by addition of SDS loading buffer, heating for 2 min at 95°C and electrophoresis in SDS–polyacrylamide gels. Then, dried gels were used to visualize the 32P-labeled proteins by autoradiography.
RESULTS
The poly(A) tract is essential for FMDV RNA infectivity
The 3′ end region of FMDV consists of a 90 nt untranslated region (3′-NCR) and a poly(A) tract, genetically encoded. We had previously shown that the 3′-NCR is essential for viral infectivity (21). Thus, we decided to analyze the role of the poly(A) tract in the infectious cycle, alone or in combination with the 3′-NCR. To this end, we made use of an FMDV O1K infectious clone, pDM, in which three unique restriction sites flank the 3′-NCR and poly(A) tract: StuI and EcoRV flank the 3′-NCR; HpaI is downstream of the poly(A) (Fig. 1). Using these restriction sites, we synthesized in vitro RNAs which selectively removed the poly(A) tract alone or both the 3′-NCR plus the poly(A) tract. The biological relevance of each viral element in the context of the full-length RNA was assayed upon transfection of BHK-21 or IBRS-2 monolayers and monitoring of the cytopathic effect. As expected, the full-length transcript derived from linearized pDM was infectious (Fig. 1). For full-length RNA-transfected cells, monolayer detachment was clearly observed at 16–20 h post-transfection. No symptoms of infectivity could be observed in monolayers transfected with transcripts lacking the poly(A) tract or both the 3′-NCR and the poly(A) tract up to 48 h post-transfection (Fig. 1). Thus, both the 3′-NCR (21) and the poly(A) tract are separate essential elements for FMDV infectivity.
Effect of the 3′-NCR and poly(A) tract on the in vitro translation efficiency of FMDV RNA
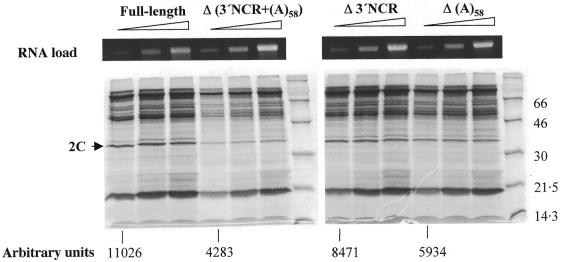
In common with all picornavirus genomes, the FMDV viral RNA also acts as mRNA. The lack of infectivity observed for the viral transcript in which the poly(A) tract had been deleted prompted us to study if a translation defect was involved. Hence, equal amounts of RNAs lacking the poly(A) tract, the 3′-NCR or both were used to program in vitro translation reactions. Figure 2 shows a representative example of the translation products obtained with RNA concentrations between 10 and 40 µg/ml. Measurement of the intensity of the 2C viral protein at the lower RNA concentration showed a 2.5-fold reduction in the translation efficiency when both elements were removed. Interestingly, a decrease of about 1.3- to 1.9-fold was observed in the reactions loaded with RNAs that lacked the 3′-NCR or the poly(A) tract relative to the full-length infectious RNA derived from pDM. Altogether these results suggest that the simultaneous presence of these regions have a relevant function in translation.
Figure 2.
Translation efficiency of FMDV transcripts harboring deleted 3′ end sequences. Synthetic FMDV RNAs of ∼8.5 kb, bearing the indicated deletions, were used to program in vitro translation reactions. Δ3′NCR transcripts were derived from the previously described pΔ74 construct (21). Numbers below each RNA indicate the intensity of protein 2C in arbitrary units. Mobility of molecular weight markers is shown on the right of each gel autoradiograph. The upper panel shows the ethidium bromide staining of an agarose gel loaded with aliquots of the RNAs (100, 200 and 400 ng each).
Enhancement of FMDV IRES-dependent translation in bicistronic constructs depends on the presence of the 3′-NCR and poly(A) sequences
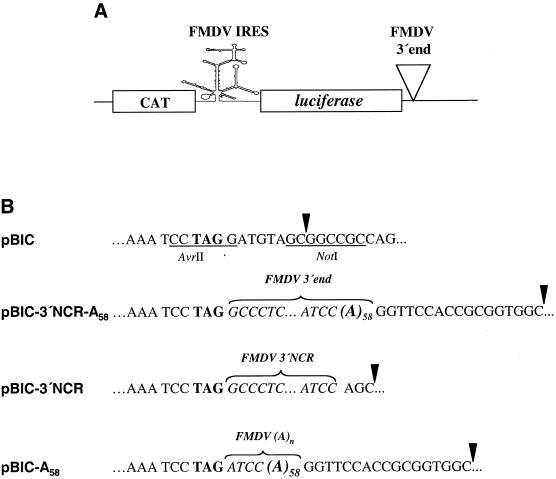
To further analyze the involvement of the FMDV 3′ end sequences in translation, the effect of these RNA regions on IRES-dependent translation was studied in living cells, in the context of bicistronic constructs (Fig. 3A). The FMDV 3′-NCR, poly(A) tract or both sequences were attached downstream of the luciferase stop codon (Fig. 3B) such that linearization of pBIC, pBIC-A58, pBIC-3′NCR and pBIC-3′NCR-A58 with NotI allowed us to study the effect of the two elements of FMDV 3′ end sequences.
Figure 3.
Bicistronic constructs harboring FMDV 3′ end sequences. (A) Diagram of the bicistronic RNA, CAT–FMDV IRES–luciferase. (B) Differences in the 3′ end sequences in the bicistronic constructs used in transfection assays. Sequences in pBIC generating unique AvrII and NotI restriction sites are underlined. The luciferase stop codon TAG is in bold. The 58 nt poly(A) present in some constructs is denoted as (A)58. A brace embracing residues in italic denotes the FMDV 3′ end sequences included in each bicistronic construct, pBIC-3′NCR-A58, pBIC-3′NCR and pBIC-A58. An arrowhead denotes the last residue of transcripts obtained after NotI linearization of the respective constructs.
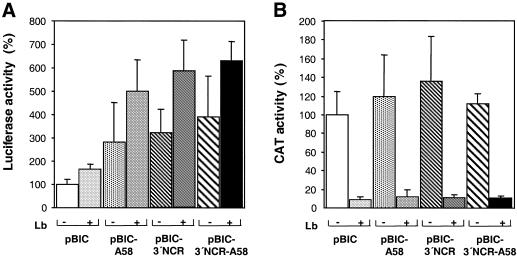
The efficiency of FMDV IRES-dependent translation in living cells, measured by the levels of luciferase translation, was stimulated by the presence of the poly(A) tract in pBIC-A58 compared to the control pBIC lacking any 3′ end viral region (Fig. 4A). This result is in agreement with recent reports on the effect of the poly(A) tract on the IRES of encephalomyocarditis virus (EMCV), hepatitis A virus (HAV) and poliovirus in cell-free assays (4,6). Interestingly, the FMDV 3′-NCR also stimulated in vivo IRES activity when present alone (pBIC-3′NCR) or in combination with the poly(A) tract (pBIC-3′NCR-A58), with stimulation values above those obtained for the poly(A) tract alone.
Figure 4.
Effects of FMDV 3′ end sequences on cap- and IRES-dependent translation efficiency in living cells. (A) IRES-dependent translation was estimated as the luciferase activity, measured in extracts from BHK-21 cells transfected with NotI-linearized plasmids, relative to the value obtained in pBIC, that was set at 100%. Plus or minus symbols indicate whether or not cells were co-transfected with pLb plasmid. (B) Cap-dependent translation was estimated by CAT activity, measured in the same extract as luciferase activity. Error bars correspond to the SEM, from three to six experiments.
To confirm that the stimulation of expression of the second cistron exerted by the viral 3′ end sequences was IRES dependent, we co-expressed the FMDV Lb protease together with the bicistronic RNAs of interest. Stimulation of IRES activity in the Lb-transfected monolayers was observed in pBIC-A58, pBIC-3′NCR and pBIC-3′NCR-A58 (5-, 6- and 6.3-fold increase, respectively) (Fig. 4A). As expected, cap-dependent translation was shut down to values below 10% of those found in the cells transfected only with the bicistronic constructs, which, on the other hand, remained fairly constant between assays (Fig. 4B; note the different scales in Fig. 4A and B). These results allowed us to conclude that two elements present at the 3′ end of the FMDV RNA, the 3′-NCR and the poly(A) tract, separately stimulate translation. Therefore, IRES stimulation was strongly enhanced following eIF4G cleavage induced by the FMDV Lb protease, as is the case during viral infection. These results are in full agreement with previous data that have demonstrated resistance of the FMDV IRES-dependent translation to eIF4G cleavage (13,31).
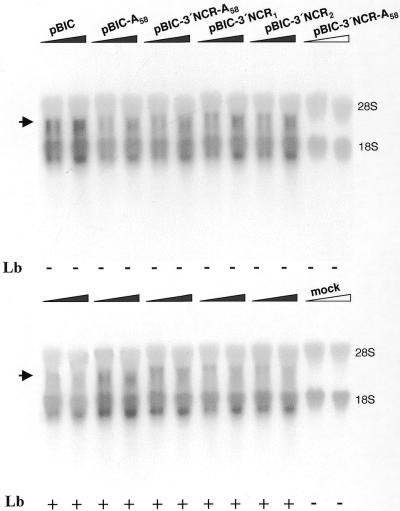
In order to address whether the stimulation exerted by the 3′ end FMDV sequences was mediated by an improved stability of the transcripts, a northern blot analysis of the RNAs extracted from transfected cells was carried out. A transcript of the expected size (3, 3.1 or 3.2 kb) was detected in samples extracted from cells transfected with each construct, in the presence or absence of pLb. No signal at this position was observed for the DNA control sample, in which no T7 RNA polymerase was expressed, or in the mock-transfected cells (Fig. 5). Samples from cells transfected with constructs carrying 3′ end sequences showed a similar amount of probe retained in the 18S rRNA to the controls. A small variation in RNA intensity was observed, likely due to changes in the efficiency of transfection. Remarkably, the levels of RNA present in cells 12 h after transfection did not correlate with the stimulatory effects observed on IRES-dependent translation for each construct (Fig. 4A), strongly suggesting a functional role for the 3′ end sequences, unrelated to differences in RNA stability.
Figure 5.
Northern blot analysis of RNAs extracted from BHK-21 cells at 12 h after transfection with bicistronic constructs. A filled triangle, as opposed to an empty triangle, depicts RNA extracted from cells expressing T7 RNA polymerase. For each construct two lanes were loaded, containing in the right lane a double amount of RNA compared to the left lane. pBIC-3′NCR1 or pBIC-3′NCR2 corresponds to two independent transfections carried out with two plasmid samples of identical sequence. Below each lane is indicated whether or not co-transfection with pLb was performed. The blot was probed with a [α-32P]UTP-labeled RNA fragment complementary to nucleotides 1–462 of the FMDV IRES. The positions of 28S and 18S rRNA have been marked as migration and loading controls. The mobility of the RNAs derived from the bicistronic plasmids is indicated with an arrow.
IRES RNA–protein interaction in the presence of 3′ end FMDV sequences
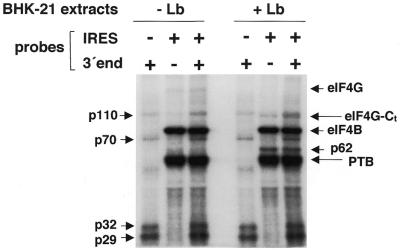
We have previously shown that the FMDV IRES activity depends upon eIF4G interaction with specific sequences in domain 4 (13). Therefore, we hypothesized that the stimulatory effect of the 3′ end sequences could be a consequence of an increase in the binding efficiency of proteins known to interact with the FMDV IRES. To test this possibility, we carried out a UV-crosslink assay using as probes the FMDV IRES alone or in conjunction with the 3′ end sequences (Fig. 6).
Figure 6.
Analysis of the interaction of the FMDV 3′ end and FMDV IRES sequences with cellular proteins. The radiolabeled transcripts (0.03 pmol) were used in UV-crosslinking assays with native proteins present in S10 extracts from BHK-21 cells transfected or not with pLb plasmid. When indicated, probes corresponding to the FMDV IRES and 3′ end were co-incubated. Following RNase A treatment, proteins were fractionated by 10% SDS–PAGE. The apparent molecular masses of the polypeptides crosslinked to the 3′ end probe are indicated on the left and those crosslinked to the IRES probe on the right side of the autoradiograph. Identification of p220 as eIF4G, p80 as eIF4B, p110 as a component of eIF3, and p57 as PTB is described in López de Quinto (11) and references therein.
The probes including the 3′ end interacted specifically with four polypeptides of 110, 70, 32 and 29 kDa. The mobility of p70 closely resembles that of PABP (37), suggesting that the 58 adenine residues encoded in the FMDV infectious transcript have the potential to interact with PABP. Two proteins of unknown identity, p29 and p32, bound with high affinity to the 3′ end probe. The use of an unrelated probe did not allow the detection of any of these proteins.
When the IRES and 3′ end probes were co-incubated and used to interact with the protein extract, the IRES-binding proteins p220 (eIF4G), p116/p110 (eIF3), p80 (eIF4B) and p57 (PTB) were readily detected. This pattern was superimposed on that seen with the 3′ end probe alone, including p70, p32 and p29 (Fig. 6). The simultaneous presence of the IRES and 3′ end probes did not induce a significant change in the pattern of the IRES-binding polypeptides. Only the intensity of p32 and p29 was slightly stimulated.
The use of extracts prepared from cells that had been transfected with pLb showed a lack of interaction of p220 (eIF4G), concomitant with the appearance of p110, corresponding to the C-terminal fragment of eIF4G (13). This polypeptide was detected as a doublet with p110, already interacting with the 3′ end probe alone (Fig. 6). Additionally, a polypeptide of ∼62 kDa appeared in the Lb-processed extracts. Once again, the simultaneous presence of 3′ end and IRES sequences did not induce a significant change in the IRES-interacting protein pattern. Preincubation of the 3′-NCR with the IRES probe under permissive conditions for RNA–RNA interaction (35) prior to addition of protein extracts led to a small stimulation in p29 and p32 RNA binding (data not shown). We conclude that the stimulation of IRES-dependent translation observed in Lb-transfected cells is mediated by a mechanism not requiring an enhanced interaction between eIF4G and/or PABP with FMDV RNA. At present, a functional RNA–protein bridge involving p32 or p29 cannot be ruled out.
DISCUSSION
We have examined the role of the 3′ end region on FMDV translation, a picornavirus for which no information regarding translation effects of the 3′-NCR was available. Our results show that RNAs lacking the poly(A) tract or both the 3′-NCR and poly(A) tract were not infectious when transfected into susceptible cells, as no cytopathic effect could be observed and we were unable to recover infectious virus after prolonged incubation times after transfection. Thus, presence of both elements appears to be a strict requirement for infectivity.
The impairment of viral growth was associated with a lower translation capacity, as removal of the 3′-NCR, the poly(A) tract or both reduced the efficiency of FMDV RNA translation in the context of full-length FMDV transcripts. This effect was magnified in living cells when the 3′ end sequences were placed in a chimeric bicistronic construct, in which the FMDV IRES promotes internal translation initiation of the second cistron. Analysis of the levels of the bicistronic RNAs present in transfected cells indicated that stimulation of relative IRES activity was not the consequence of a higher stability of the corresponding transcripts. Thus, the specific IRES stimulatory effect exerted by the 3′-NCR relative to that exerted by the poly(A) tract alone indicates that different signals ensure efficient translation initiation of the viral RNA. Moreover, the fact that maximum stimulation of IRES-dependent translation required the presence of the 3′-NCR strongly suggests a biological role in mediating a functional bridge with the IRES.
To our knowledge, this is the first evidence of the role of the 3′-NCR sequences on FMDV IRES stimulation. In contrast to the poly(A) tract stimulation of cap-dependent translation, the IRES-dependent stimulatory effect of the 3′-NCR is not only resistant, but enhanced during co-expression of the FMDV Lb protease. Under this situation, eIF4G is cleaved (13) and the FMDV IRES is fully active (31,38). Processing of the eIF4G initiation factor by Lb leads to separation of the N-terminal fragment that contains the PABP-binding site from the C-terminal fragment required for IRES activity (2). Therefore, stimulation of translation via PABP–eIF4G interaction is expected to be impaired in FMDV-infected cells or in cells expressing the Lb protease. In agreement with this, eIF4G cleavage induced by human rhinovirus 2A protease abrogates poly(A) tract stimulation of the poliovirus or EMCV IRES (7).
In recent reports, EMCV, HAV, poliovirus and hepatitis C virus (HCV) IRES-dependent translation had been shown to be stimulated by poly(A) sequences in cell-free extracts (4,6,7). In these cases, a protein–protein bridge has been proposed to mediate this stimulation by their mutual interaction with the poly(A) tail and IRES sequences, bringing together the required signals present in both RNA ends. Our findings indicate that interaction between eIF4G and PABP is not the only explanation for RNA end communication during IRES stimulation by the FMDV 3′ end. The enhancing effects on translation associated with the presence of the poly(A) tract or with the 90 residues present in the 3′-NCR strongly support a redundant role in translation initiation. UV-crosslinking experiments have shown that a 70 kDa protein, likely PABP, binds to the 3′ end region. In addition, three polypeptides of 110, 32 and 29 kDa interacted with this region. Co-incubation of IRES and 3′ end sequences yielded a combined pattern without detectable changes in intensity of the 220, 110, 80, 72 and 57 kDa polypeptides bound to the probes. This result opened the possibility of additional factors acting as a bridge between the RNA-binding proteins interacting with the IRES and the 3′ end sequences. Preincubation of IRES and 3′ end RNAs under permissive conditions for RNA–RNA interactions (35) prior to addition of the S10 extract led to a slight stimulation of the intensity of p32 and p29. This result suggests that 5′–3′ crosstalk may likely be mediated by protein bridges involving RNA–RNA contacts. However, as the assay carried out with separate IRES and 3′ end probes involved intermolecular interactions, it remains to be studied whether placing both RNA ends in cis will lead to a stronger modification in the pattern of IRES–protein interaction.
In addition to the enhancing effect of 3′ end sequences, stimulation of FMDV IRES-dependent translation in Lb-processed extracts could be associated with different causes. On the one hand, the C-terminal fragment of eIF4G may interact with higher affinity with the FMDV IRES than intact eIF4G. On the other hand, the absence of competitor mRNAs due to cap-dependent translation shut down could lead to an enrichment of the pool of available RNA-binding proteins, including eIFs, when the majority of the mRNAs are not engaged in the translation machinery.
Our data have underscored a new role for FMDV 3′-NCR sequences in the stimulation of IRES-dependent translation. The X region of the 3′-NCR of HCV has also been reported to enhance HCV IRES translation (6,14,15). However, a down-regulatory effect was observed when the entire 3′-untranslated region was present in full-length cDNA clones (39). Likewise, suppression of translation has been reported in chimeric RNAs that contain the 3′ end of West Nile virus RNA (40), possibly related to the interaction of translation factors with specific sequences within the 3′-untranslated region (41,42). In another example, the interaction of hnRNP could potentially be involved in communicating between the 3′ and 5′ ends of mouse hepatitis virus RNA (43).
The interaction of the 5′ and 3′ RNA ends during translation and replication in picornavirus is currently being investigated. In poliovirus, inter-regulation of both processes seems to be mediated by the formation of specific RNA–protein complexes in the cloverleaf structure present at the 5′ end (44,45). We have previously reported a strict requirement for the 3′-NCR for the initial rounds of FMDV replication (21). The effect on the efficiency of translation initiation described here may contribute to the lack of dispensability of the 3′-NCR. However, involvement of other signals present in the 5′ end needs further investigation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank E. Cano for his excellent technical assistance in the preparation of plasmids and C. Gutiérrez for helpful suggestions. M. Sáiz is the holder of a grant from the Ministerio de Ciencia y Tecnología (Programa Ramón y Cajal). This work was supported by grants from DGES (PM 98.0122) and CICYT (BIO 99-0833-C02-01) and an Institutional grant from Fundación Ramón Areces.
REFERENCES
- 1.Belsham G.J. and Jackson,R.J. (2000) Translation initiation on picornavirus RNA. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 869–900.
- 2.Martínez-Salas E., Ramos,R., Lafuente,E. and López de Quinto,S. (2001) Functional interactions in internal translation initiation directed by viral and cellular IRES. J. Gen. Virol., 82, 973–984. [DOI] [PubMed] [Google Scholar]
- 3.Hellen C.U. and Sarnow,P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev., 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 4.Bergamini G., Preiss,T. and Hentze,M.W. (2000) Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA, 6, 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michel Y.M., Poncet,D., Piron,M., Kean,K.M. and Borman,A.M. (2000) Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem., 275, 32268–32276. [DOI] [PubMed] [Google Scholar]
- 6.Michel Y.M., Borman,A.M., Paulous,S. and Kean,K.M. (2001) Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol., 21, 4097–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svitkin Y.V., Imataka,H., Khaleghpour,K., Kahvejian,A., Liebig,H.D. and Sonenberg,N. (2001) Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA, 7, 1743–1752. [PMC free article] [PubMed] [Google Scholar]
- 8.Gallie D.R. (1991) The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev., 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- 9.Tarun S.Z. Jr and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 10.Gradi A., Svitkin,Y.V., Imataka,H. and Sonenberg,N. (1998) Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl Acad. Sci. USA, 95, 11089–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López de Quinto S., Lafuente,E. and Martínez-Salas,E. (2001) IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA, 7, 1213–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomakin I.B., Hellen,C.U.T. and Pestova,T.V. (2000) Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol., 20, 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López de Quinto S. and Martínez-Salas,E. (2000) Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA, 6, 1380–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito T., Tahara,S.M. and Lai,M.M. (1998) The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol., 72, 8789–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito T. and Lai,M.M. (1999) An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology, 254, 288–296. [DOI] [PubMed] [Google Scholar]
- 16.Gosert R., Chan,K.H., Rijnbrand,R., Yi,M., Sangar,D.V. and Lemon,S.M. (2000) Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol., 20, 1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood J., Frederickson,R.M., Fields,S. and Patel,A.H. (2001) Hepatitis C virus 3′ X region interacts with human ribosomal proteins. J. Virol., 75, 1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitching R.P. (1999) Foot-and-mouth disease: current world situation. Vaccine, 26, 1772–1774. [DOI] [PubMed] [Google Scholar]
- 19.Sobrino F., Sáiz,M., Jiménez-Clavero,M.A., Núñez,J.I., Rosas,M.F., Baranowski,E. and Ley,V. (2001) Foot-and-mouth disease virus: a long known virus, but a current threat. Vet. Res., 32, 1–30. [DOI] [PubMed] [Google Scholar]
- 20.Ryan M.D., Belsham,G.J. and King,A.M. (1989) Specificity of enzyme-substrate interactions in foot-and-mouth disease virus polyprotein processing. Virology, 173, 35–45. [DOI] [PubMed] [Google Scholar]
- 21.Sáiz M., Gómez,S., Martínez-Salas,E. and Sobrino,F. (2001) Deletion or substitution of the aphthovirus 3′ NCR abrogates infectivity and virus replication. J. Gen. Virol., 82, 93–101. [DOI] [PubMed] [Google Scholar]
- 22.Chatterjee N.K., Bachrach,H.L. and Polatnick,J. (1976) Foot-and-mouth disease virus RNA. Presence of 3′-terminal polyriboadenylic acid and absence of amino acid binding ability. Virology, 69, 369–377. [DOI] [PubMed] [Google Scholar]
- 23.Gutiérrez A., Martínez-Salas,E., Pintado,B. and Sobrino,F. (1994) Specific inhibition of aphthovirus infection by RNAs transcribed from both 5′ and 3′ noncoding regions. J. Virol., 68, 7426–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigeriego P., Rosas,M.F., Zamora,E., Martínez-Salas,E. and Sobrino,F. (1999) Heterotypic inhibition of foot-and-mouth disease virus infection by combinations of RNA transcripts corresponding to the 5′ and 3′ regions. Antiviral Res., 44, 133–141. [DOI] [PubMed] [Google Scholar]
- 25.Rohll J.B., Moon,D.H., Evans,D.J. and Almond,J.W. (1995) The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol., 69, 7835–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todd S., Nguyen,J.H. and Semler,B.L. (1995) RNA-protein interactions directed by the 3′ end of human rhinovirus genomic RNA. J. Virol., 69, 3605–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todd S. and Semler,B.L. (1996) Structure-infectivity analysis of the human rhinovirus genomic RNA 3′ non-coding region. Nucleic Acids Res., 24, 2133–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellits K.H., Meredith,J.M., Rohll,J.B., Evans,D.J. and Almond,J.W. (1998) Binding of a cellular factor to the 3′ untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J. Gen. Virol., 79, 1715–1723. [DOI] [PubMed] [Google Scholar]
- 29.Teterina N.L., Egger,D., Bienz,K., Brown,D.M., Semler,B.L. and Ehrenfeld,E. (2001) Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol., 75, 3841–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zibert A., Maass,G., Strebel,K., Falk,M.M. and Beck,E. (1990) Infectious foot-and-mouth disease virus derived from a cloned full-length cDNA. J. Virol., 64, 2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez-Salas E., Sáiz,J.C., Dávila,M., Belsham,G. and Domingo,E. (1993) A single nucleotide sustitution in the internal ribosome entry site of foot-and-mouth disease virus leads to enhanced cap-independent translation in vivo. J. Virol., 67, 3748–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López de Quinto S. and Martínez-Salas,E. (1999) Involvement of the aphthovirus RNA sequence located between both functional AUGs in start codon selection. Virology, 255, 324–336. [DOI] [PubMed] [Google Scholar]
- 33.Fuerst T.R., Niles,E.G., Studier,F.W. and Moss,B. (1986) Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 83, 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López de Quinto S. and Martínez-Salas,E. (1997) Conserved structural motifs located in distal loops of the aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol., 71, 4171–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramos R. and Martínez-Salas,E. (1999) Long-range interactions in aphthovirus internal ribosome entry site elements. RNA, 5, 1374–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamarnik A.V. and Andino,R. (1997) Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA, 3, 882–892. [PMC free article] [PubMed] [Google Scholar]
- 37.Adam S.A., Nakagawa,T., Swanson,M.S., Woodruff,T.K. and Dreyfuss,G. (1986) mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol. Cell. Biol., 6, 2932–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belsham G.J. and Brangwyn,J.K. (1990) A region of the 5′ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J. Virol., 64, 5389–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami K., Abe,M., Kageyama,T., Kamoshita,N. and Nomoto,A. (2001) Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch. Virol., 146, 729–741. [DOI] [PubMed] [Google Scholar]
- 40.Li W. and Brinton,M.A. (2001) The 3′ stem loop of the West Nile virus genomic RNA can suppress translation of chimeric mRNAs. Virology, 287, 49–61. [DOI] [PubMed] [Google Scholar]
- 41.Blackwell J.L. and Brinton,M.A. (1995) BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol., 69, 5650–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blackwell J.L. and Brinton,M.A. (1997) Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol., 71, 6433–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang P. and Lai,M.M. (2001) Heterogeneous nuclear ribonucleoprotein a1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol., 75, 5009–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gamarnik A.V. and Andino,R. (1998) Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev., 12, 2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herold J. and Andino,R. (2001) Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell, 7, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]