Abstract
Chlamydiaceae are obligate intracellular bacterial pathogens characterized by a wide range of vertebrate host, tissue tropism and spectrum of diseases. To get insights into the biological mechanisms involved in these differences, we have put forward a computational and experimental procedure to identify the genome recombination hotspots, as frequent sequence variation allows rapid adaptation to environmental changes. We find a larger potential for recombination in Chlamydophila pneumoniae genomes as compared with Chlamydia trachomatis or Chlamydia muridarum. Such potential is mostly concentrated in a family of seven previously uncharacterized species-specific elements that we named ppp for C.pneumoniae polymorphic protein genes, which have the potential to vary by homologous recombination and slipped-mispair. Experimen tally, we show that these sequences are indeed highly polymorphic among a collection of nine C.pneumoniae strains of very diverse geographical and pathological origins, mainly by slippage of a poly(C) tract. We also show that most elements are transcribed during infection. In silico analyses suggest that Ppps correspond to outer membrane proteins. Given their species specificity, their putative location in the outer membrane and their extreme polymorphism, Ppps are most likely to be important in the pathogenesis of C.pneumoniae and could represent targets for future vaccine development.
INTRODUCTION
Chlamydiaceae are obligate intracellular bacterial pathogens that replicate within membrane-bound vacuoles. A wide range of vertebrate host, tissue tropism and spectrum of diseases characterize them. They are responsible for several major diseases in animals, mainly spontaneous abortion in livestock and systemic disease in birds, and can also infect rodents and cats. In humans, Chlamydiaceae are the leading cause of preventable blindness and sexually transmitted disease; they cause acute respiratory disease and have been associated with cardiovascular disease (1). The biological mechanisms responsible for these differences are unknown. Recently, based on phylogenetic evidence, the Chlamydiaceae family has been split into two genera (Chlamydia and Chlamydo phila) encompassing three (Chlamydia trachomatis, Chlamydia suis, Chlamydia muridarum) and six (Chlamydo phila abortus, Chlamydophila psittaci, Chlamydophila pneumoniae, Chlamydophila pecorum, Chlamydophila felis, Chlamydophila caviae) species, respectively (2). However, virulence traits such as host and tissue tropism are not linked to the phylogenetic groups. As lateral gene transfers seem to be infrequent in Chlamydiae, it has been proposed that the virulence phenotype reflects adaptation of the bacteria to its environment (3).
Frequent sequence variation allows rapid adaptation to environmental changes of pathogenic bacteria (4,5). The sources of such variation are typically constituted by repeats that engage into recombination events, either dependent of RecA (homologous recombination) or independent (illegitimate recombination). Homologous recombination involves exchanges between large segments of DNA molecules of (nearly) identical sequence. This induces the rearrangement, with or without sequence conversion, of DNA sequences both within and between chromosomes (6). Illegitimate recombination between short close or tandem repeats proceeds either by slipped-mispair, at a replication arrest, or single-strand-annealing, at a DNA double-strand break (7). The frequency of illegitimate recombination increases exponentially with the repeat length, from 8 to 20 nt, and decreases with the distance between the two copies, becoming rare for copies >1000 bp apart (8,9).
The completion of five chlamydial genome sequences (three C.pneumoniae, one C.trachomatis, one C.muridarum) has provided new clues on the biology of these organisms (10), among which a better understanding of the protein composition of the chlamydial outer membrane (11). Outer membrane proteins are key elements of host–pathogen interactions as they are engaged in adhesion to, and invasion of, host cells, and host evasion to immune systems. All Chlamydiae encode an abundant protein termed the major outer membrane protein (MOMP, or OmpA) that is surface exposed in C.psittaci and C.trachomatis (12) and is the major determinant for serologic classification of many chlamydial isolates (13). This protein is highly variable within its exposed domains (14) except in C.abortus (15), C.felis (16) and C.pneumoniae (17) where it is extremely conserved.
The C.trachomatis genome encodes a family of nine polymorphic membrane proteins (Pmps), which extends in C.pneumoniae to 21 Pmp paralogs. These proteins are characterized by specific tetrapeptide motifs (GGAI and FXXN) repeated multiple times (18) and resemble members of the autotransporter family (19). All the pmp genes are transcribed, but only a few have been shown to be stably translated into proteins, which are localized in the chlamydial outer membrane (20). It has been shown that these proteins are variable between the C.pneumoniae strains, this polymorphism being linked to the presence of repeats. For example, in CWL029 strain, Pmp6 contains three tandem repeats of 131 amino acids whereas in AR39 and J138 strains it contains only two repeats (20,21). The presence of a simple sequence repeat (SSR) composed of a poly(G) stretch within the coding region of pmp10 leads, by a slipped-strand mechanism altering the number of G residues, to a differential expression of Pmp10 between and within strains (20,22). Given their polymorphisms, surface exposure, antigenicity and phase-variable nature, it has been proposed that Pmps play a role in the virulence and pathogenesis of Chlamydiae (11). Thus, a balance could exist between the polymorphisms of the different outer membrane proteins to ensure a high level of diversity in the surface of the chlamydial cell.
The works cited above had their origins in the analysis of previously characterized genes. When new variable families are to be unravelled, one has to perform an ab initio analysis aimed at identifying all major recombination hotspots (23). By their own variability, these elements may not be correctly annotated, because the sequenced strain contains a frameshift, because these regions are atypical in sequence or simply because their sequencing is more error-prone. These elements are also frequently under the form of one functional gene plus a set of pseudo-genes, with which recombinational exchange is performed (24). Therefore, we started by cataloguing all major recombination hotspots in the five published genomes of Chlamydiae. This involves searching for elements capable of engaging into homologous recombination (large repeats) or illegitimate recombination (close and tandem repeats). Since close non-tandem repeats provided no interesting results, we further analyzed the genomic context of the tandem and large repeats and their potential biological interest. This in silico approach led us to the discovery of a new family of proteins in C.pneumoniae. We then switched to experimental work to verify that these repeated elements were polymorphic in a collection of C.pneumoniae strains involved in different pathologies. We also checked their expression and the positioning of the translational start by RT–PCR.
Additional material can be found at our web site (http://wwwabi.snv.jussieu.fr/people/erocha/pppweb/).
MATERIALS AND METHODS
In silico analyses
Data. The sequence data on the complete genomes of C.pneumoniae strains CWL029 (25), J138 (26) and AR39 (27) and annotation files were downloaded from Entrez genomes (www.ncbi.nlm.nih.gov). For comparative purposes, we also analyzed the complete genomes of C.trachomatis and C.muridarum. Additionally, we performed a BLAST analysis on the nearly finished genome of C.caviae using the TIGR web server (www.tigr.org).
Identification of simple sequencer repeats (SSR). We searched for motifs X of length 1–5 nt (e.g. dinucleotides in CG), with n consecutive copies (e.g. three in CGCGCG), for n high enough so that Xn should not occur by chance in the genome. Considering L, the length of the genome, the probability of not finding Xn anywhere in the genome is:
P = (1 – fXn)L1
Where fX is the relative frequency of the motif X in the genome. Setting a threshold P-value < 0.001, we solved the above equation for all possible motifs X of length 1–5 nt, determining for each motif the threshold length. We then searched for significant SSR elements in all chlamydial genomes using standard pattern matching methods.
Identification of large repeats. We searched the genomes for large repeats capable of engaging into homologous recombination. To compute the threshold length, we used a statistic of extremes that takes into account the composition in nucleotides and the length of the genome (28). For bacteria, the minimal length for which the probability of finding an exact repeat is <0.001, is in the range 21–26 nt (29). In C.pneumoniae the minimal significant length is 23 nt. The search for large strictly identical repeats was done using Reputer (30), which outputs all pairs of repeats larger or equal than the threshold length. Repeats were then clustered to build large non-strict repeats and cross-compared in order to construct families of similarity (29).
Definition and classification of orthologs. Two genes were regarded as homologous if the proteins they code for are similar both in sequence and in size. For this, we made pairwise comparisons of all proteins of all proteome pairs, filtering potential homologs using a threshold in BlastP of E < 10–5 and in maximal difference of protein lengths of 20%. Subsequently, we aligned the sequences, using a variant of the classical dynamic programming algorithm for global alignment, where one counts 0-weight for gaps at both ends of the largest sequence, using the BLOSUM62 matrix (31). Finally, we retained pairs of proteins presenting a similarity >40%. The set of orthologous genes (supposed to have diverged following a speciation event) was defined by adding a further criterion of double best hit, i.e. two genes are defined as orthologs if they are homologous and if they are the best matches of one another in the respective genomes. In the case of the ppp elements, this methodology was slightly changed since all similarity sequence analysis was done using the DNA sequences.
PCR primer design. Given the extensive similarity between the different ppp elements, we had to define a special bioinformatic methodology to choose the regions of larger specificity. This was done by analyzing the number of similar regions for each section of the ppp element, added of 500 bp at each edge. More precisely, we identified for 23 nt windows (1 nt step) the number of similar regions in the genome presenting at most three mismatches with the sequence in the window. With the graphs provided by these analyses we could precisely define the regions presenting specificity for the required sequence.
Experimental work
Bacterial strains. The origins of each C.pneumoniae isolate studied, together with other relevant information, are summarized in Table 4. Chlamydophila pneumoniae strains were grown in mycoplasma-free HEp-2 cells, harvested, pooled and suspended in MEM to an approximate concentration of 106–107 p.f.u. ml–1 and stored at –80°C. Chlamydophila abortus B577, C.psittaci 6BC, C.felis FEPN, C.pecorum LW613 and C.suis S45 strains kindly provided by B. Kaltenboeck (14) were also studied.
Table 4. Characteristics of the C.pneumoniae studied in vitro.
| Straina | Source | Site of isolation | Associated disease | Country of isolation | Date of isolation |
|---|---|---|---|---|---|
| TW-183 | ATCC | Conjunctiva of a child | Trachoma vaccine study | Taiwan | 1965 |
| IOL-207 | C. M. Black | Conjunctiva of a child | Trachoma | Iran | 1967 |
| CWL-029 | B. Kaltenboeck | Throat of adult | Pneumonia | USA | 1987 |
| FML-016 | C. M. Black | Throat of adult | Pneumonia | Norway | 1989 |
| CM-1 | C. Maass | Sputum of adult | Pneumonia | USA | 1990 |
| MUL-1 | C. Maass | Brochoalveolar fluid of adult | Pneumonia | Germany | 1992 |
| MUL-250 | C. Maass | Brochoalveolar fluid of adult | Pneumonia | Germany | 1995 |
| WIEN 1 | C. Maass | Carotid artery of adult | Prolonged reversible ischemic neurologic defect | Austria | 1998 |
| CV-3 | C. Maass | Coronary artery of adult | Coronary sclerosis | Germany | 1996 |
aAll strains are human isolates.
PCR and sequencing. DNA was obtained by lysis of 1 µl of the chlamydial preparation as in Denamur et al. (15) for all the strains except C.pneumoniae CWL-029, IOL-207 and FML-016 from which DNA was purified (<30% of RNA, <2% host cell DNA). PCRs were performed from 5 µl of the DNA lysate solution or 1.3 ng of pure DNA in standard conditions with 1.5 mM MgCl2 and 35 cycles as follows: denaturation, 30 s at 94°C; hybridation, 30 s at 55°C; extension, 1 min at 72°C. The sequences of the primers are given in the additional material. PCR products were directly sequenced without interim cloning or subcloned into the pCR®II vector (InVitrogen, San Diego, CA) following the instructions of the manufacturer. Sequences from the plasmid were performed using the ppp-specific primers. Subcloned ppp PCR products were re-amplified by PCR as above starting from 108 plasmid molecules diluted in 1 ng of Escherichia coli DNA. The obtained PCR products were sequenced directly. Sequence reactions were performed with the Big Dye Terminator method and ABI sequencing (Perkin Elmer, Applied Biosystems) following the manufacturer’s instructions.
RNA analyses. Total RNA was extracted from C.pneumoniae MUL1-infected HEp-2 cells as described in Grimwood et al. (20). The characteristics of the oligonucleotides used for RT–PCR are listed in the Supplementary Material. cDNAs were transcribed from 1 µg of total RNA using random hexamers (40 ng) and Moloney Murine Leukemia Virus reverse transcriptase (SuperScript™II RT, InVitrogen). The DNA was then amplified by PCR in standard conditions as above. The RNA dependence of the amplifications was checked by conducting PCR without the reverse transcription step. RT–PCR products were electrophoresed on a 1.5% agarose gel stained with ethidium bromide and photographed under UV illumination.
RESULTS
Comparative analysis of repeats in chlamydial genomes
Repeats are over-represented in the C.pneumoniae strains as compared with C.trachomatis and C.muridarum. We have divided repeats into two categories: simple SSR, which are tandem repeats of small motifs (e.g. CGn); and large repeats (>24 nt). SSR can engage into illegitimate recombination whereas large repeats may engage into homologous recombination. The analysis of the five sequenced genomes indicates that the C.pneumoniae strains contain more repeats of both types than C.trachomatis and C.muridarum (Table 1). This difference is particularly remarkable for the large repeats, and they do not concern duplicated housekeeping genes (such as rDNA). Since these results suggested a larger potential for variation by recombination in C.pneumoniae we set forward a computational and experimental strategy to further unravel potential roles for these repeats.
Table 1. Abundance of repeats in the three strains of C.pneumoniae, C.trachomatis and C.muridarum.
| Genome | SSR | LR | I/D | MR |
|---|---|---|---|---|
| C.pneumoniae CWL029 | 11 (17) | 136 | 0.02 | 1 (7) |
| C.pneumoniae AR39 | 8 (14) | 133 | 0.02 | 1 (7) |
| C.pneumoniae J138 | 10 (16) | 121 | 0.02 | 1 (7) |
| C.trachomatis | 3 (8) | 12 | 0 | 0 |
| C.muridarum | 6 (10) | 63 | 0.34 | 0 |
SSR, simple sequence repeats; LR, large repeats; I/D, ratio inverse/direct repeats; MR, multiple repeats. For SSR, the first numbers indicate the number of SSR able to induce frameshifts, i.e. with motifs variable in length not multiple of 3, whereas the numbers between parentheses correspond to the total number of SSR. LR includes all pairs of long repeats. MR includes the number of multiple repeats and its multiplicity between parentheses.
SSR. We searched the C.pneumoniae strains AR39, CWL029 and J138 for SSR, with motifs ranging from 1 to 5 nt (P < 0.001, see Materials and Methods), and significant tandem repeats of 6 nt (23). This analysis revealed a certain number of these elements, mostly composed of motifs of length 1, but also of lengths 3, 5 and 6 (Table 2). The SSR of trinucleotides and hexanucleotides are inside coding sequences and code for tandem amino acid repeats. Naturally, slippage of such motifs does not induce frameshifts in the coding sequence. The pentanucleotide SSR consist of three tandem elements and are either inside unknown function ORFs (UFOs) or in intergenic regions. The large majority of SSRs consist of tandem nucleotides, and always a series of C or Gs. Contrary to the others, these SSRs are frequently found to be variable among different C.pneumoniae strains, sometimes with variations as high as 5 nt, which strongly suggests hypervariability of these elements. Further analysis showed that two of these elements are within pmp genes, of which the one on pmp_10.2 was already known (20,22). The regions on the edges of the other nucleotide SSR, except the one located in CPn0069, are extremely similar in sequence. This is surprising, since the genome annotations indicated that some elements were on intergenic regions, some in small ORFs and some others in very large ORFs. This is a typical feature of elements capable of engaging homologous recombination for sequence variation (23).
Table 2. Position, characteristics and potentially affected genes by significantly large SSR in the CWL029 strain of C.pneumoniae, as well as the corresponding element in the two other strains.
| Position | SSR | Genes involved | J138 | AR39 |
|---|---|---|---|---|
| 10806 | C14 | Upstream of CPn0008 (UFO) | C14 | C14 |
| 13350 | C14 | Upstream of CPn0010 (UFO) | C13 | C11 |
| 20588 | C14 | pmp_2 | C13 | C14 |
| 58474 | C14 | CPn0043 (UFO) | C14 | C14 |
| 85336 | C14 | CPn0069 (UFO) | C9 | C9a |
| 507200 | G13 | pmp_10.2 | G14 | G13 |
| 1207061 | C13 | CPn1054 (UFO) | C12 | C11 |
| 1209609 | C12 | Upstream of CPn1055 (UFO) | C15 | C16 |
| 607260 | CGT4 + CG | CPn0525 (UFO) | CGT4 + CG | CGT4 + CG |
| 628400 | CAC4 + AC | CPn0542 (ABC transporter) | CAC4 + AC | CAC4 + AC |
| 956212 | GAA5 | yphC (GTPase) | GAA5 | GAA5 |
| 1150530 | CCT4 + CC | ftsH (protease) | CCT4 + CC | CCT4 + CC |
| 258158 | ATGCT3 | ypdP (UFO) | ATGCT3 | ATGCT3 |
| 396387 | TTTCT3 | CPn0352 (UFO) | TTTCT3 | TTTCT3 |
| 407929 | TAATT3 | Upstream of rpsD (sigma factor) | TAATT3 | TAATT3 |
| 1085124 | GCAGCT3 | glgA (glycogen synthase) | GCAGCT3 | GCAGCT3 |
| 492298 | GCAACA3 | pmp_6 | GCAACA3 | GCAACA3 |
The trinucleotide SSRs correspond to repeated codons. Since sometimes these elements end by a partial repeat we indicate this in subscript.
aIn the strain AR39 the gene is annotated as starting further downstream. As a consequence, the stretch of Cs is in the intergenic region.
Large repeats. We were able to identify 13 large two-copy repeats, and one large multiplet of seven copies (Tables 1 and 3). A minimum of 20–25 nt has been found to be required to initiate homologous recombination in Bacillus subtilis and E.coli, for which data is available (32,33). One can thus expect that most of these repeats will indeed be able to engage into homologous recombination. Most doublets consist of small repeats between 23 and 40 nt long, and they include UFO, intergenic regions and pmp genes. The largest repeat consists of the tandemly repeated amino transferase (glmS) and transport tyrP protein coding genes. The large multiplet is distributed in seven different places on the chromosome, with strict identity along regions of >100 nt, and extensive sequence similarity for ∼2500 nt (see below). Furthermore, the early regions of these elements coincide with the regions containing the stretches of Cs, found in the analysis of the SSR elements. That these regions can vary by both illegitimate and homologous recombination, strongly suggests an important role for their sequence variation. Following the observation that these elements have no homologs in GenBank and the demonstration that they suffer sequence variation (see below), we named these seven elements as C.pneumoniae polymorphic proteins (Ppp).
Table 3. Position of the large two-copy repeats of C.pneumoniae CWL029, including the elements, where they are (gene or intergenic), the length of the repeat, and the existence of a repeat in the other two sequenced strains.
| Position 1 | Position 2 | Type | Length | Element 1 | Element 2 | Other strains |
|---|---|---|---|---|---|---|
| 26238 | 29415 | D | 23 | pmp_4.2 | pmp_5.2 | None |
| 234959 | 236693 | D | 27 | oppA_1 | oppA_2 | J138 AR39 |
| 259232 | 259385 | D | 26 | Intergenic | tgt | J138 AR39 |
| 290023 | 292838 | D | 40 | CPn0255 | Intergenic | J138 AR39 |
| 415142 | 416513 | D | 31 | CPn0369 | CPn0370 | J138 AR39 |
| 495909 | 498766 | D | 23 | pmp_7 | pmp_8 | J138 AR39 |
| 501979 | 514804 | D | 24 | pmp_9 | pmp_13 | J138 AR39 |
| 522778 | 525176 | D | 28 | CPn0457 | CPn0458 | J138 AR39 |
| 528528 | 530945 | D | 29 | CPn0461 | CPn0462 | J138 AR39 |
| 1111630 | 1113279 | D | 1650 | glmS tyrP_1 | yccA tyrP_2 | AR39 |
| 207095 | 208884 | I | 35 | CPn0165 | CPn0169 | J138 AR39 |
| 493543 | 506266 | I | 23 | pmp_6 | Intergenic | J138 AR39 |
| 954974 | 955029 | I | 32 | CPn0843 | CPn0843 | J138 AR39 |
D stands for direct repeats, and I for inverted repeats. The ppp elements are not displayed in this table because they are seven copy repeats (see Fig. 1).
Characterization of the ppp family
Identification of the elements. The elements were originally identified due to the cytosine SSRs and the large multiplet of repeats, as described above. We then made BLAST comparisons on the genome followed by the clustering of the regions of similarity, which were further examined with the help of dot-plots. This allowed the identification of six ppp elements, of ∼2500 nt, and one other element of 1600 nt (with a large deletion at the interior), in each strain of C.pneumoniae (Fig. 1). Since the start of these elements is associated with the large C-SSR, and this element shows significant sequence variation, the definition of the exact edges of these elements is not trivial. We have defined ppp as a sequence homologous to the largest gene annotated in these regions. This corresponds to the 2436 nt long CPn1054 gene of the CWL029 strain (CPj1054 in J138). This UFO has a putative start, just 30 nt upstream of the stretch of Cs (Fig. 2). The analysis of all the elements in the three strains shows a strong conservation at this putative start, with the exception of the stretch of C, which suffers extensive sequence variation. Thus, we could easily identify the equivalent putative starts in the other elements of the family in each of the three C.pneumoniae strains. The identification of the end of these elements was more difficult, due to the smaller conservation of this region. Initially, we restricted our attention to the strongly conserved region in all elements, which ends ∼50 nt before the stop. We then used a multiple alignment to extend this region, given the strong similarity between the orthologs and the relatively weaker similarity between different elements. As a matter of definition, we considered these elements to stop at the stop codon in the CPn1054 gene, although sometimes this does not correspond to a stop codon (in phase with the start codon), because of the SSR downstream of the start codon and the two other variable regions inside the gene (see below). The G + C content of the ppp elements is 42%, which is not significantly different from the genome (40.6%).
Figure 1.
The family of ppp elements, its position in the three strains, the number of Cs in the SSR and the genes annotated for their region. The vertical bar in the box for ppp7 indicates a deletion at the interior of the element (see text). ppp elements are represented in numerical order from 1 (left) to 7 (right).
Figure 2.
Multiple alignment of the start region for the ppp elements. When the ppps elements are identical in the three strains only one copy is shown. Otherwise, the three variants of the polymorphic elements of this region are indicated.
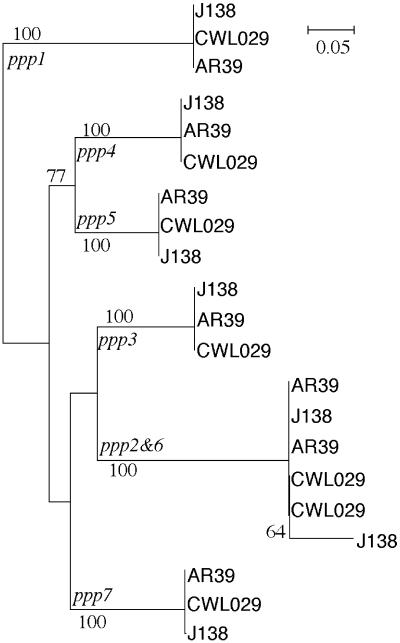
Similarity between the orthologous elements. The similarity between the orthologous elements in the three sequenced strains is very strong. With one single exception, this similarity is >99%, as expected given the high sequence similarity between the chromosomes (26,27). This similarity extends beyond the edges of the elements (data not shown). The maximum likelihood tree for the ppp elements shows that, with a single exception, the orthologs group together, typically with bootstrap values of 100% (out of 1000 replicates) (Fig. 3). The exception to this trend is constituted by ppp6, which presents very similar elements between the strains CWL029 and AR39 (99.8% similarity), but a more divergent element in strain J138 (95%). The former elements cluster together very well with the ppp2 elements, with similarities >99%. The J138 ppp6 shows a larger divergence only in the first 500 nt of the J138 ppp6 element (81% of similarity). In the remaining J138 ppp6 element, the similarity with the other ppp6 and ppp2 elements is the expected one (98%).
Figure 3.
Maximal likelihood tree of the ppp elements, obtained from 1000 bootstrap replicates, using the HKY model (42). The tree was computed with phylowin (43), after manual inspection of the multiple alignment of the complete elements. Gaps were excluded. The families are indicated at the origin of the branches that separate them from the remaining elements. All bootstrap values better than 50% are shown. We obtained a tree with a similar topology using maximum parsimony (data not shown).
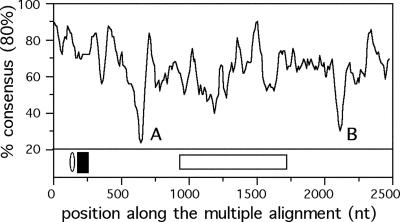
Similarity between the different elements. The analysis of the ppp elements, by means of a maximum likelihood phylogenetic tree (Fig. 3), shows a robust separation of the different elements, with exception for the ppp2 and ppp6 elements described above. This similarity is not homogenous along the ppp elements. Indeed, some regions are very similar and a consensus sequence can easily be established, whereas others regions are highly variable (Fig. 4). The start region is the best conserved, with exception for the stretch of Cs, which is highly variable. One less conserved region concerns an insertion/deletion in some of the elements, and the other concerns a region rich in small stretches of A and T (Fig. 4). The seven elements are similarly organized in the three complete genomes. They are located in three regions of the chromosome, one containing ppp1 and ppp2, another containing ppp3, ppp4 and ppp5, and a third location containing ppp6 and ppp7 (Fig. 1). The three regions are <100 kb apart in the 1.2 Mb genome. All the seven elements are coded in the leading replicating strand, which is significant considering that half of the genes are coded in each strand of C.pneumoniae (P < 0.001, binomial test).
Figure 4.
Analysis of concordance of the columns of the multiple alignment of the ppp family. The curves represent the percentage of columns for which a base is present in at least 80% of the sequences. These values are calculated in sliding windows of 50 and 10 bp steps. The minima indicated by A and B represent regions with an insert (A) and rich in A-T sequences (B). In the lower panel, the ellipse indicates the position of the signal peptide and the black box indicates the predicted transmembrane domain. The white box indicates the large deletion in ppp7.
The ppp elements contain the largest potential for recombination in the chromosome. Since the ppp elements seem to be subject to variation, we have tried to evaluate what is their share of the recombination potential of the chromosome. They contain the majority of the large SSR elements capable of inducing frameshifts by slipped-mispair (Table 2). To determine their share in terms of the potential for homologous recombination we computed the cumulated length of the strict repeats with at least 23 nt present in the complete chromosome. This resulted in 17 194 nt for CWL029. We then computed the cumulated length of the repeats inside ppp elements. This resulted in 9776 nt, i.e. 56% of the total length of large repeats, which is extremely significant considering that they only occupy 1.3% of the genome (P < 0.001, χ2 test). This percentage increases to 63% when the edges of the ppp elements are also included in the analysis. As a matter of comparison, we did the same analysis on the complete set of Pmp proteins, currently regarded as major variant proteins in C.pneumoniae (11). Although the corresponding genes occupy 5% of the genome, the repeats found in these elements only constitute 3.9% of the total. Thus, the recombination potential of ppp seems to be much larger than the one of pmp, either in terms of homologous or of illegitimate recombination. Whether such potential results in effective variation was then experimentally checked in several C.pneumoniae strains.
ppp sequences are highly polymorphic due to the poly(C) tract in C.pneumoniae isolates
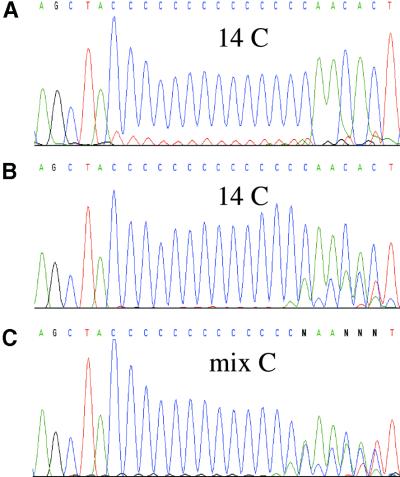
We first amplified by PCR the regions of the ppps containing the five significant SSRs, i.e. ppp1, ppp2, ppp4, ppp6 and ppp7 in order to analyze for variation in the number of cytosine residues. PCR was performed from genomic DNA of a collection of nine C.pneumoniae strains isolated all over the world during the past 40 years and involved in distinct pathologies (Table 4). According to the high level of homology between the different elements, the PCR primers were designed using the analysis cited in the Materials and Methods. Sequencing of the PCR products demonstrated a variable number of C residues in the poly(C) tract in all the studied strains except for ppp4 in TW-183 and IOL-207 strains, which was not amplifiable by PCR (Fig. 5C and data not shown). To eliminate artefacts due to the slippage of the Taq polymerase in vitro, during the PCR reaction, which can mimic the polymorphism observed in the sequences of the PCR products, we performed the following controls: we cloned the PCR products of ppp2, ppp4 and ppp6 in a plasmid and (i) sequenced the plasmid directly or (ii) did a ppp PCR on 108 copies of the plasmid, this number corresponding roughly to the number of ppp targets in the genomic DNA preparation used for the initial PCR, and then sequenced this PCR product. As shown in Figure 5A, the sequence after the poly(C) tract is monomorphic when the sequencing reaction is done directly on the plasmid, corresponding to a finite number of C. Although less clear than the sequence done directly on the plasmid, the sequence of the PCR product done from the plasmid still shows a fixed number of C (Fig. 5B), opposite to the sequences of the PCR product obtained from the genomic DNA which have a variable number of C followed by a polymorphic sequence (Fig. 5C). Thus, the observed polymorphism in the sequences after the poly(C) tract in the ppp1, ppp2, ppp4 and ppp7 is due to the presence in the genomic DNA of DNA molecules with variable numbers of C and not from a PCR artefact. We also studied the polymorphism of a tract of six cytosine residues and of an impure poly(C) tract (CCCCCCTCCCC) in ppp5 and ppp3, respectively (Fig. 2), in the nine C.pneumoniae strains. These two regions are located at the same position as the significant SSRs in the remaining ppp genes, i.e. eight (nine for ppp5) codons after the more upstream ATG. None of these regions were polymorphic (Fig. 6 and data not shown).
Figure 5.
Electrophoregrams of ppp2 sequencing reactions of the CWL-029 C.pneumoniae strain performed (A) directly from the plasmid with the cloned PCR product, (B) from the PCR product obtained from the plasmid as template and (C) from the PCR product obtained from the genomic DNA as template.
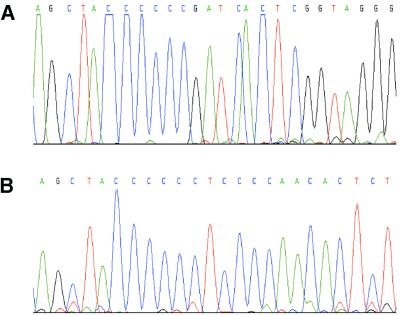
Figure 6.
Electrophoregrams of (A) ppp5 and (B) ppp3 sequencing reaction products of the CWL-029 C.pneumoniae strain. Note the absence of polymorphism in the poly(C) tracts.
As observed for ppp4, no PCR product was obtained for ppp5 in TW-183 and IOL-207 strains suggesting that both genes are deleted in these two strains. Given the genome organization of these elements (Fig. 1), the absence of ppp4 and ppp5 may be the result of intra-chromosomal recombination between ppp3 and ppp5, whose outcome may be the deletion of one of the elements (ppp5) and of the intervening sequence (ppp4).
The ppp elements are C.pneumoniae specific. BlastP and BlastN searches on the complete genomes and on the complete GenBank/EMBL/DDBJ database provided no significant hit at an E < 10–10, outside Ppp sequences of C.pneumoniae. Blast searches on the TIGR web site against the fully sequenced, but still non-annotated, genome of C.caviae, also failed to provide homologs. Since available sequence data do not fully cover all known Chlamydiaceae, we performed PCR experiments using the ppp-specific primers of all the seven ppp genes on C.abortus B577, C.psittaci 6BC, C.felis FEPN, C.pecorum LW613 and C.suis S45 strains. No PCR product was obtained in any of the experiments (data not shown). Thus, Ppp elements seem specific to C.pneumoniae.
Characterization of the protein
We then characterized Ppp elements, based on the protein sequence of the Ppp6 (CPn1054) putative peptide. Ppp6 contains 811 residues, has an average molecular mass of 93.4 kDa and a predicted isoelectric point of 6.1. A multiple alignment of the putative peptides, as well as the sequences of the elements, can be found in the additional material at our web site. We have searched for Prosite motifs on the peptide without success. Since Pmp proteins have been found to contain motifs typical of autotransporters (19), we searched for these motifs in Ppp. The two most important motifs are GGAI and FXXN, but neither of them was found in Ppp.
Ppp contain an excess of residues, such as cysteine, characteristic of outer membrane proteins of C.pneumoniae (e.g. found in Pmp) (34). The hydrophobicity profile, and the use of PSORT (35) allowed the identification of a signal peptide to be cleaved at residue 51. Also, a transmembrane domain is predicted in residues 68–84. The prediction of the membrane topology of the peptide indicates that the N-terminal side should be inside, and the C-terminus outside. Similar results were obtained by using Top-pred (36). Thus, bioinformatic analyses consistently suggest that the Ppp peptide is a membrane protein with one transmembrane segment, separating a small cytoplasmic N-terminus from the majority of the peptide (C-terminus), which is surface exposed.
RNA analysis of the ppp
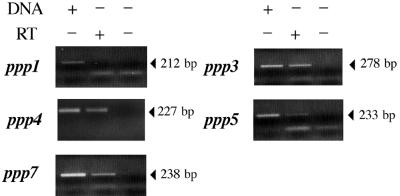
We analyzed the ppp RNA to determine the transcription start sites of the genes. To do this, we designed, for RT–PCR, upstream primers with the 5′ region of the primer containing the ATG located eight codons upstream the poly(C) tracts (see Supplementary Material). The 3′ primers were those used for the PCR performed from the genomic DNA. Using this strategy, ppp2 and ppp6 could not be studied, as the sequences of both the 5′ and 3′ primers were absolutely identical in both genes. The PCR from the genomic DNA gave the expected size. No bands were obtained without the RT step confirming the absence of contaminant DNA. ppp3, ppp4, ppp5 and ppp7 are transcribed during the infection whereas no signal was obtained from ppp1 using either the ppp1 RT primer or the ppp1 RT bis primer which is located 21 codons downstream of the poly(C) tract and starts with a GTG codon (Table 1) (Fig. 7). These results are in agreement with the in silico predictions that the initiation codon of ppp7 is located eight codons upstream of the poly(C) tract, and not downstream of this tract as suggested by the first annotations of the sequences.
Figure 7.
Analysis of ppp transcription of the MUL-1 C.pneumoniae strain. Each ppp transcript was specifically amplified by RT–PCR using primers listed in the Supplementary Material. Chlamydophila pneumoniae genomic DNA was added in some of the reactions, as indicated, to serve as a positive control. RNA obtained from C.pneumoniae-infected Hep-2 cells was used as the template with (+) or without (–) RT. PCR products were electrophoresed on a 1.5% agarose gel. Lower bands correspond to the primers.
DISCUSSION
Bacteria have specific highly mutable loci named ‘contingency loci’ (4), which have been selected to rapidly generate phenotypic diversity, thus allowing faster adaptation. This is particularly useful in pathogenic bacteria, which have to cope with different environments and the polymorphic nature of their hosts’ immune responses. Since these interactions typically take place at the outer membrane, contingency loci are usually associated with the expression of proteins present therein, such as adhesins, transport systems and lipoproteins (37,38). Most of the molecular mechanisms generating diversity within these loci involve recombination events in repeated sequences either mediated by RecA (homologous recombination) or not (illegitimate recombination). These different types of recombination target different types of repeats. Thus, if one wants to make an inventory of recombination hotspots in a genome, all types of repeats must be taken into account (23). Further, since these elements are frequently under the form of pseudo-genes, a thorough analysis should be done ab initio, by searching all repeats, regardless of genome annotations. Only when the complete record of repeats is available, do annotations become essential for the understanding of the genomic context of repeats.
We have focused on the search of large repeats and tandem repetitive SSRs, since the analysis of close non-tandem repeats provided no interesting results (data not shown). Our analysis suggests that C.pneumoniae has the highest potential for recombination among fully sequenced Chlamydiaceae and that the majority of recombination hotspots of the C.pneumoniae genome are concentrated on a new family of polymorphic proteins, the Ppp elements. Contrary to Pmps, which are known to be more abundant in C.pneumoniae than in C.trachomatis and C.abortus strains (11), Ppp elements seem to be C.pneumoniae-specific. The concentration of such a large fraction of large repeats and SSRs in these elements shares resemblances with the variation of immunodominant proteins in mycoplasmas (23). One is then inclined to speculate a very important role for these elements in the evolution of C.pneumoniae pathogenicity.
Our analysis indicates that most elements are transcribed, thus variation can be achieved either by differentially silencing genes or by differential protein dosage. These elements may also change by homologous recombination, either by conversion or by deletion/duplication, which can easily occur between elements close in the chromosome. Our experimental analysis suggests that some elements (ppp4 and ppp5) are deleted in the strains TW-183 and IOL-207, which, interestingly, are the only strains from our collection, which have been isolated from child conjunctiva (Table 4). Whether this is a significant link will have to be clarified in further studies. Since duplication may also arise from recombination between tandem Ppp elements, one may also suppose the existence of more than seven elements in some other strains. Further, the analyses of the alignments and of the phylogenetic tree suggest frequent intra-chromosomal recombination between the ppp2 and ppp6 elements, since the stability of the genome and the organization of the elements does not seem to indicate a recent duplication (the other possible explanation for the unresolved branches of the tree). Recombination between ppp2 and ppp6 elements has probably resulted in gene conversion, making ppp6 elements similar to ppp2 in AR39 and CWL029. Mutation or partial recombination with other ppp seems to have resulted in larger divergence in the early regions of J138 ppp6. Such larger divergence is likely to preclude frequent recombination between the early regions of ppp2 and ppp6 in strain J138.
The ppp elements are placed in the chromosome <100 kb apart. All elements are in the leading replicating strand, which is a favorable location for highly expressed genes (39). Naturally, if one supposes that the origin of these elements is proceeded by horizontal transfer followed by tandem duplication, such configuration becomes less unexpected since tandem duplication would place the genes in the same strand. The clustering together of ppp4 and ppp5 in the phylogenetic trees reinforces this scenario, but the other elements do not cluster accordingly. Naturally, the conversion events that seem to be taking place between ppp6 and ppp2 may have also occurred between the other elements, which would have changed the topology of the tree, hiding the traces of the original duplications.
The existence of a signal peptide, one subsequent transmembrane domain and the composition of the protein, suggest that Ppps are outer membrane proteins. These proteins are relatively abundant in the bacteria as proteome analysis of the C.pneumoniae elementary body identified one of them, i.e. Ppp6 (CPN1054) (40). OmpA and Pmp variations do not correlate with host cell niche or epidemiologic success (21,41). Thus, it has been suggested that they should not be the major ligand responsible for directing infection of various human cell types (21). Whether Ppp elements may perform this function remains to be tested, since further work will be required to determine exactly the function and cellular location of this family and its role in the virulence and diversity of the different strains. Although the function of Ppps is unknown, it can be assumed that, given their C.pneumoniae specificity, their putative location and their extreme polymorphism, these proteins are important in the pathogenesis of C.pneumoniae. They could also be used to type the bacteria and eventually as a candidate for vaccine development. However, before this becomes possible, further experimental work will be required, especially the generation of specific antibodies to the different Ppps. Also, the understanding of the cellular role of Ppps might become clearer through the identification of cell machinery elements that interact with Ppps. We have started to work on some of these issues.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Bernhard Kaltenboeck, Carolyn Black and Matthias Maass for providing strains, to Alain Blanchard, Bernhard Kaltenboeck and Florence Hommais for fruitful comments on the manuscript and to Jacques Elion for constant encouragement. This work was supported by grants from INSERM and ActivBiotics, Inc.
REFERENCES
- 1.Stephens R.S. (ed.) (1999) Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. ASM Press, Washington, DC.
- 2.Everett K.D., Bush,R.M. and Andersen,A.A. (1999) Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol., 49, 415–440. [DOI] [PubMed] [Google Scholar]
- 3.Bush R. and Everett,K. (2001) Molecular evolution of the Chlamydiaceae. Int. J. Syst. Evol. Microbiol., 51, 203–220. [DOI] [PubMed] [Google Scholar]
- 4.Moxon E.R., Rainey,P.B., Nowak,M.A. and Lenski,R.E. (1994) Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol., 4, 24–33. [DOI] [PubMed] [Google Scholar]
- 5.van Belkum A., Scherer,S., van Alphen,L. and Verbrugh,H. (1998) Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev., 62, 275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd R.G. and Low,K.B. (1996) Homologous recombination. In Curtiss,R., Ingraham,J.L., Lin,E.C.C., Brooks Low,K., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 2236–2255.
- 7.Michel B. (1999) Illegitimate recombination in bacteria. In Charlebois,R.L. (ed.), Organization of the Prokaryotic Genome. ASM Press, Washington, DC, pp. 129–150.
- 8.Peeters B.P., de Boer,J.H., Bron,S. and Venema,G. (1988) Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol. Gen. Genet., 212, 450–458. [DOI] [PubMed] [Google Scholar]
- 9.Lovett S.T., Gluckman,T.J., Simon,P.J., Sutera,V.A. and Drapkin,P.T. (1994) Recombination between repeats in E.coli by a recA-independent, proximity-sensitive mechanism. Mol. Gen. Genet., 245, 294–300. [DOI] [PubMed] [Google Scholar]
- 10.Rockey D.D., Lenart,J. and Stephens,R.S. (2000) Genome sequencing and our understanding of Chlamydiae. Infect. Immun., 68, 5473–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephens R.S. and Lammel,C.J. (2001) Chlamydia outer membrane protein discovery using genomics. Curr. Opin. Microbiol., 4, 16–20. [DOI] [PubMed] [Google Scholar]
- 12.Cadwell H.D., Krombort,J. and Schaechter,J. (1981) Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun., 31, 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens R.S., Wagar,E.A. and Schoolnik,G. (1988) High-resolution mapping of serovar-specific and common antigenic determinants of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med., 167, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaltenboeck B., Kousalas,K.G. and Storz,J. (1993) Structures of and allelic diversity and relationships among the major outer membrane protein (ompA) genes of the four Chlamydial species. J. Bacteriol., 175, 487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denamur E., Sayada,C., Souriau,A., Orfila,J., Rodolakis,A. and Elion,J. (1991) Restriction pattern of the major outer-membrane protein gene provides evidence for a homogeneous invasive group among ruminant isolates of Chlamydia psittaci. J. Gen. Microbiol., 137, 2525–2530. [DOI] [PubMed] [Google Scholar]
- 16.Sayada C., Andersen,A., Rodriguez,P., Eb,F., Milon,A., Elion,J. and Denamur,E. (1994) Homogeneity of the major outer membrane protein gene of feline Chlamydia psittaci. Res. Vet. Sci., 56, 116–118. [DOI] [PubMed] [Google Scholar]
- 17.Gaydos C.A., Quinn,T.C., Bobo,L.A. and Eiden,J.J. (1992) Similarity of Chlamydia pneumoniae strains in the variable domain IV region of the major outer membrane protein gene. Infect. Immun., 60, 5319–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimwood J. and Stephens,R.S. (1999) Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics, 4, 187–201. [DOI] [PubMed] [Google Scholar]
- 19.Henderson I.R. and Lam,A.C. (2001) Polymorphic proteins of Chlamydia spp.—autotransporters beyond the Proteobacteria. Trends Microbiol., 9, 573–578. [DOI] [PubMed] [Google Scholar]
- 20.Grimwood J., Olinger,L. and Stephens,R.S. (2001) Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect. Immun., 69, 2383–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirai M., Hirakawa,H., Ouchi,K., Tabuchi,M., Kishi,F., Kimoto,M., Takeuchi,H., Nishida,J., Shibata,K., Fujinaga,R. et al. (2000) Comparison of outer membrane protein genes omp and pmp in the whole genome sequences of Chlamydia pneumoniae isolates from Japan and the United States. J. Infect. Dis., 181 (Suppl. 3), S524–S527. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen A.S., Christiansen,G. and Birkelund,S. (2001) Differential expression of Pmp10 in cell culture infected with Chlamydia pneumoniae CWL029. FEMS Microbiol. Lett., 203, 153–159. [DOI] [PubMed] [Google Scholar]
- 23.Rocha E.P.C. and Blanchard,A. (2002) Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res., 30, 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenri T., Taniguchi,R., Sasaki,Y., Okazaki,N., Narita,M., Izumikawa,K., Umetsu,M. and Sasaki,T. (1999) Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun., 67, 4557–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens R.S., Kalman,S., Lammel,C., Fan,J., Marathe,R., Aravind,L., Mitchell,W., Olinger,L., Tatusov,R.L., Zhao,Q. et al. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science, 282, 754–759. [DOI] [PubMed] [Google Scholar]
- 26.Shirai M., Hirakawa,H., Kimoto,M., Tabuchi,M., Kishi,F., Ouchi,K., Shiba,T., Ishii,K., Hattori,M., Kuhara,S. and Nakazawa,T. (2000) Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res., 28, 2311–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Read T.D., Brunham,R.C., Shen,C., Gill,S.R., Heidelberg,J.F., White,O., Hickey,E.K., Peterson,J., Utterback,T., Berry,K. et al. (2000) Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res., 28, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlin S. and Ost,F. (1985) Maximal segmental match length among random sequences from a finite alphabet. In Cam,L.M.L. and Olshen,R.A. (eds), Proceedings of the Berkeley Conference in Honor of Jerzy Neyman and Jack Kiefer. Wadsworth, Inc., Vol. I, pp. 225–243.
- 29.Rocha E.P.C., Danchin,A. and Viari,A. (1999) Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol. Biol. Evol., 16, 1219–1230. [DOI] [PubMed] [Google Scholar]
- 30.Kurtz S. and Schleiermacher,C. (1999) REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics, 15, 426–427. [DOI] [PubMed] [Google Scholar]
- 31.Erickson B.W. and Sellers,P.H. (1983) Recognition of patterns in genetic sequences. In Sankoff,D. and Kruskal,J.B. (eds), Time Warps, String Edits, and Macromolecules: the Theory and Practice of Sequence Comparison. Addison-Wesley, Reading, MA, pp. 55–91.
- 32.Shen P. and Huang,H.V. (1986) Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics, 112, 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohan F.M. (1994) Genetic exchange and evolutionary divergence in prokaryotes. Trends Ecol. Evol., 9, 175–180. [DOI] [PubMed] [Google Scholar]
- 34.Melgosa M.P., Kuo,C.C. and Campbell,L.A. (1993) Outer membrane complex proteins of Chlamydia pneumoniae. FEMS Microbiol. Lett., 112, 199–204. [DOI] [PubMed] [Google Scholar]
- 35.Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- 36.Claros M.G. and von Heijne,G. (1994) TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci., 10, 685–686. [DOI] [PubMed] [Google Scholar]
- 37.Chambaud I., Wroblewski,H. and Blanchard,A. (1999) Interactions between mycoplasma lipoproteins and the host immune system. Trends Microbiol., 7, 493–499. [DOI] [PubMed] [Google Scholar]
- 38.Saunders N.J., Jeffries,A.C., Peden,J.F., Hood,D.W., Tettelin,H., Rappuolli,R. and Moxon,E.R. (2000) Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol., 37, 207–215. [DOI] [PubMed] [Google Scholar]
- 39.McLean M.J., Wolfe,K.H. and Devine,K.M. (1998) Base composition skews, replication orientation and gene orientation in 12 prokaryote genomes. J. Mol. Evol., 47, 691–696. [DOI] [PubMed] [Google Scholar]
- 40.Vandahl B.B., Birkelund,S., Demol,H., Hoorelbeke,B., Christiansen,G., Vandekerckhove,J. and Gevaert,K. (2001) Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis, 22, 1204–1223. [DOI] [PubMed] [Google Scholar]
- 41.Stothard D.R., Boguslawski,G. and Jones,R.B. (1998) Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun., 66, 3618–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasegawa M., Kishino,H. and Yano,T. (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol., 22, 160–174. [DOI] [PubMed] [Google Scholar]
- 43.Galtier N., Gouy,M. and Gautier,C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 12, 543–548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.