Abstract
The disease-associated expansion of (CTG)·(CAG) repeats is likely to involve slipped-strand DNAs. There are two types of slipped DNAs (S-DNAs): slipped homoduplex S-DNAs are formed between two strands having the same number of repeats; and heteroduplex slipped intermediates (SI-DNAs) are formed between two strands having different numbers of repeats. We present the first characterization of S-DNAs formed by disease-relevant lengths of (CTG)·(CAG) repeats which contained all predicted components including slipped-out repeats and slip-out junctions, where two arms of the three-way junction were composed of complementary paired repeats. In S-DNAs multiple short slip-outs of CTG or CAG repeats occurred throughout the repeat tract. Strikingly, in SI-DNAs most of the excess repeats slipped-out at preferred locations along the fully base-paired Watson–Crick duplex, forming defined three-way slip-out junctions. Unexpectedly, slipped-out CAG and slipped-out CTG repeats were predominantly in the random-coil and hairpin conformations, respectively. Both the junctions and the slip-outs could be recognized by DNA metabolizing proteins: only the strand with the excess repeats was hypersensitive to cleavage by the junction-specific T7 endonuclease I, while slipped-out CAG was preferentially bound by single-strand binding protein. An excellent correlation was observed for the size of the slip-outs in S-DNAs and SI-DNAs with the size of the tract length changes observed in quiescent and proliferating tissues of affected patients—suggesting that S-DNAs and SI-DNAs are mutagenic intermediates in those tissues, occurring during error-prone DNA metabolism and replication fork errors.
INTRODUCTION
Expansions of (CTG)n·(CAG)n trinucleotide repeat sequences are associated with at least 11 human genetic diseases including myotonic dystrophy type 1 (DM1) and Huntington’s disease (HD) (1). Generally, in the non-affected population the length of the repeat tract ranges from 5 to 24 repeat units. Intermediate- (HD) and pre-mutation alleles (DM1) having 25–34 and 34–90 repeats, respectively, are genetically unstable and can expand to disease-associated lengths of approximately 100 up to thousands of repeats, which exhibit a high degree of genetic instability. The tight association of repeat length with mutation has been termed ‘dynamic mutation’ (2) which describes the increased likelihood for the product of an expansion mutation to undergo a subsequent mutation relative to the precursor (shorter) allele. The increased instability of longer lengths is thought to be due to an increased ability to form mutagenic DNA structures.
Repeat expansion may occur by processes that are dependent, as well as independent, of genome duplication (reviewed in 3). For many of the repeat-associated diseases the repeats can be somatically unstable displaying inter- and intra-tissue differences in length distributions. Length heterogeneities have been observed in proliferating and quiescent tissues (4–12). Expansion of (CTG)·(CAG) tracts can occur at human and primate replication forks (13,14). In non-replicating neural tissues (4,6,8–10) repeat length heterogeneity increased with the age of the patient—suggesting that somatic instability occurred in the absence of genome duplication. Thus, processes associated with and independent of replication forks may contribute to repeat instability.
Unusual DNA structures are thought to be important mutagenic intermediates during repeat instability (3,15–18). Trinucleotide repeats have the propensity to form slipped-strand DNA structures in which the complementary DNA strands display an out-of-register alignment (19). There are two kinds of slipped-strand DNAs: homoduplex slipped DNAs (S-DNAs), (CTG)x·(CAG)y where x = y; and heteroduplex slipped intermediates (SI-DNAs) where x ≠ y (x > y and x < y). Slipped heteroduplex SI-DNAs may be mutagenic intermediates at replication forks, forming between the nascent and template strands (13,14). Differential propensities to form or process different mutagenic intermediates (excess CTG or excess CAG) may explain the tendency to expand or delete at primate replication forks (13,14). In quiescent cells slipped homoduplex S-DNAs may be mutagenic intermediates during error-prone DNA metabolism (repair or recombination). Notably, with increasing mouse age an increased amount of single-stranded DNA has been observed in many non-proliferating tissues (20–22). Aberrant repair of these single-stranded regions may lead to DNA instability of repeated DNAs (20–25). Thus, different processes of tissue-specific repeat instability may involve different DNA structural intermediates.
To elucidate the mechanism of repeat instability and to understand the interaction of proteins with them, it is imperative to understand the structural details of these putative mutagenic intermediates. Different structural features of slipped-structures may critically determine whether and how they are recognized and processed by DNA metabolizing proteins. Structural features might include sequence- and length-specific slip-outs, the tip of the hairpin (where formed), and the junction at which the slipped-out strand extrudes from the Watson–Crick duplex. Based upon analysis of oligonucleotides containing 10–15 repeat units it is known that individual CTG and CAG repeats are capable of forming hairpins (15,16,18,26–28). In these studies the number of repeats were well within the non-diseased and genetically stable range (5 to 33 repeats). Little is known about structures formed by longer, disease-relevant and genetically unstable lengths, which may form structures more complex than hairpins. Thus, hairpins would only be one component of a complex multi-stranded structure formed by repeats embedded within non-repeating sequence. Intermediates of genetic instability involving the gain or loss of some, but not all repeat units would be expected to contain three-way junctions composed of complementary repeats. There is no knowledge about the junctions at which repeats are slipped-out from a duplex of complementary paired repeat strands, i.e. a slip-out of (CTG)n or (CAG)n extruded from two arms of (CTG)n· (CAG)n. The junctions of slipped-out CTG/CAG repeats could theoretically occur at any nucleotide position along the complementary strand. Furthermore, it is possible that one or multiple slip-outs may occur along a given repeat tract. Three-way slip-out junctions may be specifically recognized by repair or recombination proteins—making the slip-out junction analogous to the four-way junction in Holliday recombination intermediates, which are the critical feature recognized and cleaved by specific resolvases (29,30).
We have demonstrated previously that S-DNAs and SI-DNAs are suitable models for mutagenic intermediates of disease-associated repeat instability since the effects of both repeat tract length and tract purity on the propensity of structure formation correlated with their effect on genetic stability in humans (19,31–33). Herein, we characterized at the nucleotide level the major slipped-strand-structures (of S-DNAs and SI-DNAs) formed by (CTG)n·(CAG)n repeats, where n is 30 or 50 repeats, lengths that are typical of normal and premutation/diseased states. Slipped-structures formed by long disease-relevant repeat tracts display unique structural features, including slip-out junctions, multiple as well as individual slip-outs in hairpin and random-coil conformations—many of these features are not detectable in preparations of short oligonucleotides. In addition to structural characterization, we show that individual components of S-DNAs can be recognized by specific proteins.
MATERIALS AND METHODS
Plasmids/DNA
Plasmids contained human DM1 genomic (CTG)n·(CAG)n repeats (n = 30 or 50) and human non-repeating sequences flanking the repeat (sites 417–436 and 451–494 from accession no. S86455) have been described (19,31,33). The repeat tracts are pure. The repeat tract is flanked by unique EcoRI and HindIII restriction sites 59 bp and 54 bp 3′ and 5′ of the CTG tract, respectively. Plasmids were prepared as described (31).
DNA treatments
Except where noted, restriction digestions were performed as specified by the manufacturer (New England Biolabs). Plasmids were linearized with HindIII then radiolabeled on the 5′ or 3′ ends with T4 kinase (USB) and [γ-32P]ATP (NEN) or AMV reverse transcriptase (USB) and [α-32P]dATP, respectively. Samples were reduplexed or heteroduplexed (see below); the repeat-containing fragment was then liberated by a secondary restriction digestion with EcoRI. For some experiments plasmids were linearized with EcoRI and followed by HindIII. Reaction products were resolved on a 4% polyacrylamide gel and the bands corresponding to the main HindIII–EcoRI linear, S-DNA, and SI-DNA forms were excised and purified by electroelution as described previously (19). Linear forms were gel-purified from samples that had never been denatured, the S-DNA forms were purified from individual DNAs that had been reduplexed, and only the SI-DNAs were purified from heteroduplexed DNA samples. For details see the Supplementary Material.
Structure formation
Homoduplex slipped-structures (S-DNAs) of 30 or 50 repeat-containing DNAs uniquely 32P-labeled on the (CTG) or (CAG) strand were formed by alkaline denaturation/ renaturation as described in detail (19,31). Heteroduplex SI-DNAs were prepared as follows: DNAs uniquely 32P-labeled on the (CTG)50 or (CAG)50 strand were mixed with an equimolar amount of unlabeled (CTG)30·(CAG)30 and heteroduplexed by denaturation/renaturation. Similarly, DNAs uniquely 32P-labeled on the (CTG)30 or (CAG)30 strand were mixed with an equimolar amount of unlabeled (CTG)50·(CAG)50 and heteroduplexed by denaturation/ renaturation. For details see the Supplementary Material.
BbvI, mung bean nuclease and T7 endonuclease I treatments
Gel-purified repeat-containing DNAs that were uniquely radiolabeled were probed with BbvI or mung bean nuclease (MBN) or T7 endonuclease I (T7endoI). For each enzyme reaction (20 µl) we selected reaction conditions, which for the linear duplex form (0.04–0.65 ng/µl) resulted in an even distribution of ladder bands within the trinucleotide repeat tract—these conditions approximated single-hit kinetics. For each treatment equal concentrations of the S-DNA and SI-DNA structures were treated under identical conditions. Digestions with BbvI (New England Biolabs) were performed in 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 10 mM Tris–HCl (pH 7.9), with 0.009–0.05 U/reaction, and reactions were incubated at 37°C for 40 min. Digestions with MBN (New England Biolabs) were performed in 50 mM NaCl, 10 mM Tris–HCl (pH 7.5), with 0.25–1 U/reaction and reactions were incubated at 37°C for 40 min. Digestions with T7endoI (New England Biolabs) were performed in 50 mM potassium acetate, 20 mM Tris–acetate (pH 7.9), 10 mM magnesium acetate, 1 mM dithiothreitol, with 0.05–2 U/reaction and reactions were incubated at 37°C for 15 min. All reactions were stopped by phenol–chloroform extraction and ethanol precipitation, then resuspended in formamide loading buffer and analyzed on 5% sequencing gels. Autoradiographs from multiple experiments were scanned for densitometric analyses. The relative sensitivities of each nucleotide within a given strand to either MBN or T7endoI were determined by densitometric analysis of individual gel bands within a lane. Densitometric readings of multiple gels were converted to a bar length shown in Figure 4. Relative sensitivities between strands to each enzyme were determined by comparing the densitometric analyses of individual gel bands relative to the undigested material. In this fashion, the relative bar lengths between strands were normalized. Such comparisons were possible since reaction conditions (DNA and enzyme concentrations and reaction times) were identical. As a result of the hyper-reactive sequence-specificity of BbvI, the relative sensitivities of each nucleotide within a given strand were determined by densitometric analysis of reaction products of high and low BbvI concentrations.
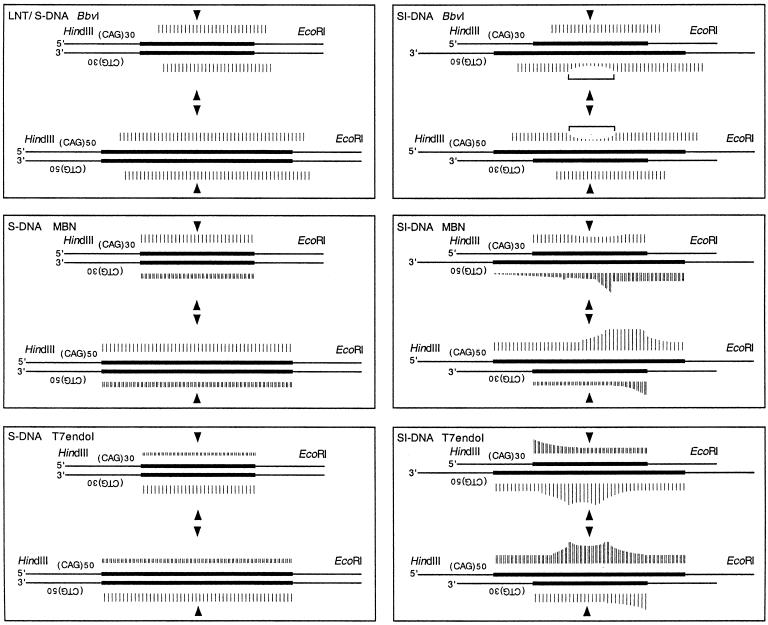
Figure 4.
Summary of enzymatic probing of S-DNAs. (A) Gel-purified DNAs were structurally probed with BbvI, MBN or T7endoI, as shown in Figures 11–3. Each of the complementary strands (CTG and CAG) containing either 30 or 50 repeats were individually analyzed in each of the gel-purified structures; linear form (L), slipped homoduplex S-DNAs (S) and slipped intermediate heteroduplexes SI-DNAs (SI). The (CT/AG)30 and (CT/AG)50 strands are shown with the non-repeating flanks as thin lines and the repeat tracts as bold lines. The center of the repeat tracts is indicated by arrowheads. For the SI-DNAs, each of the complementary strands is aligned from the center of the repeat tracts. Each scissile site is indicated by a vertical bar; the relative intensity of cleavage is represented by the height of the bar (Materials and Methods).
Electron microscopic analysis
Gel-purified DNA samples were analyzed by electron microscopy (EM) as described (32). Binding reactions with bacterial single-strand binding protein (SSB) were carried out in a 50 µl reaction mixture containing 8 mM NaCl, 20 mM HEPES (pH 7.5) and 300 ng SSB for 10 min at room temperature. Complexes were fixed with 0.6% glutaraldehyde (v/v) for 10 min at room temperature, followed by filtration through a 2-ml column of Bio-Gel A5m (Bio-Rad) to remove excess glutaraldehyde and free proteins. Factions containing DNA–protein complexes were prepared for EM. Briefly, the indicated gel-purified DNAs or SSB–DNA complexes were mixed in a buffer containing 2 mM spermidine, adsorbed to glow-charged carbon-coated grids, washed with a water/graded ethanol series and rotary shadow cast with tungsten. Samples were examined using a Philips 420 electron microscope. Micrographs are shown in reverse contrast. A Cohu CCD camera attached to a Macintosh computer programmed with NIH IMAGE and Adobe Photoshop was used to form montages of the images and to measure the contour length of DNA molecules.
RESULTS
Formation of slipped-strand DNAs
We have demonstrated previously that the linear, S-DNAs and SI-DNAs electrophoretically migrate as distinct species (19,31–33). Since S-DNAs resolved into multiple and distinct electrophoretic species, suggested the formation of a wide range of different slipped isomers (33). In contrast, since each of the sister SI-DNAs (excess of CTG and excess of CAG repeats) resolved into predominantly one distinct electrophoretic product, suggested the formation of a limited set of preferred structures (19). Furthermore, the differential migration of sister SI-DNAs indicated the existence of structural differences between the two. Such differences may involve the geometries of the three-way slip-out junctions; and/or the conformation of the slipped-out CAG compared with the slipped-out CTG (random-coils versus hairpins and hairpin loops); and/or the location of the slip-out extrusion may vary between sister SI-DNAs. Each of these possibilities was experimentally addressed below by structurally characterizing individual strands in isolated structures.
Slipped-strand DNAs were prepared from DM1 patient-derived DNA clones containing 30 or 50 (CTG)·(CAG) repeats (for details see the Supplementary Material). Tracts of 30 are within the normal genetically stable range, while tracts of 50 repeats are within the genetically unstable expanded range. The repeat tracts are flanked by unique EcoRI and HindIII restriction sites 59 bp and 54 bp 3′ and 5′ of the CTG tract, respectively. Clones with 30 and 50 (CTG)·(CAG) repeats were linearized with HindIII digestion and 32P-radiolabeled on either the 5′ or 3′ ends. These radiolabeled DNAs were used to prepare linear forms, homoduplex S-DNAs and sister SI-DNA heteroduplexes. Homoduplex S-DNAs and heteroduplex SI-DNAs were prepared from individual plasmids of a given length or mixtures of plasmids with different lengths, respectively. The repeat-containing fragment was released by a secondary restriction digestion with EcoRI. Depending upon whether the n = 30 or the n = 50 plasmid was radiolabeled as well as which strand was radiolabeled (5′ or 3′) permitted the production of 12 samples, representing six different DNA structures (two linear forms, two S-DNAs and two sister SI-DNAs), each uniquely 32P-labeled on either the CTG or CAG strand. Each of the linear duplexes, homoduplex S-DNAs [(CTG)30·(CAG)30 and (CTG)50·(CAG)50] and sister heteroduplex SI-DNAs [(CTG)30·(CAG)50 and (CTG)50·(CAG)30] were individually resolved on polyacrylamide gels and gel-purified (for an example see figure 2 in ref. 19). The isolated structures displayed remarkable biophysical stability with minimal inter-conversion to other structural isoforms (31,32), permitting their structural characterization, described below.
Figure 2.
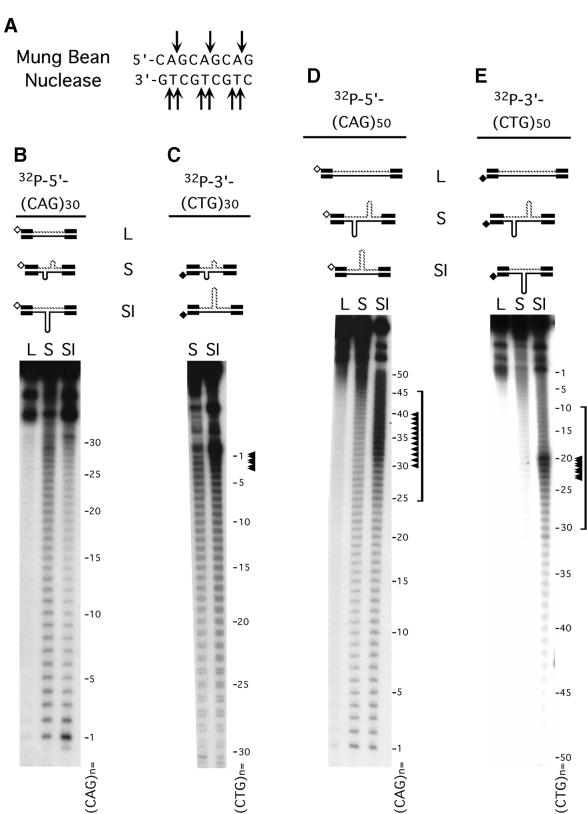
S-DNAs contain regions of single-stranded DNA. (A) MBN specifically recognizes single-strand DNA; arrows indicate the observed sites of scission. (B–E) Gel-purified HindIII–EcoRI linear form (L), S-DNA (S) and SI-DNAs (SI) structures uniquely radiolabeled at the HindIII site on either the CTG or the CAG strands were digested with MBN (Materials and Methods) and products resolved on denaturing acrylamide gels. Shown are schematics of the purified DNAs and the location of the unique radiolabels, indicated by hollow or filled diamonds. Each of the complementary strands (CTG and CAG) containing either 30 or 50 repeats were individually analyzed in each of the purified structures. Sequencing ladders (not shown) indicated nucleotide positions and indicated repeat numbers are read 5′→3′ such that the first CTG repeat unit corresponds to the last CAG repeat unit. No digestion products were detected for the linear forms. Only the longer 50 repeat strands in the SI-DNAs displayed specific regions of hypersensitivity (indicated by arrowheads and brackets). While the hypersensitive sites are localized, there are more on the (CAG)50 strand than on the (CTG)50 strand. The analyses of each strand in each of the structural conformations are summarized in Figure 4.
Slipped-strand DNAs have regions resistant to a double-strand-specific enzyme: BbvI
In an attempt to detect non-B features each of the structures was probed with the restriction enzyme BbvI which can digest duplex, but not single-stranded, DNA (34). BbvI recognizes the site:
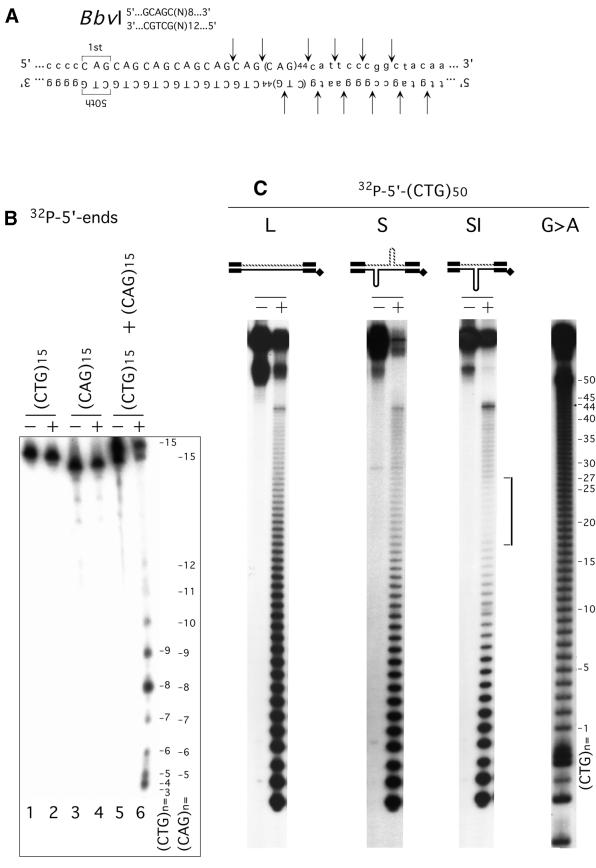
which is multiply repeated within the (CTG)·(CAG) repeat tract and is not present in the flanking sequences (Fig. 1A). We assessed both the sequence- and structural-specificity of BbvI. Neither (CTG)15 nor (CAG)15 oligonucleotides, which form hairpins, were susceptible to scission by excess BbvI (Fig. 1B, lanes 2 and 4). Only when the two oligonucleotides were mixed—producing a Watson–Crick duplex—was digestion observed (Fig. 1B, lane 6). Thus, BbvI digestion required complementary duplex DNAs, while individual CTG or CAG repeats in the single-stranded or hairpin conformations were resistant. These results are in agreement with previous characterization of BbvI (34) and permitted its use as a probe for non-B-DNA structures within (CTG)·(CAG) repeats.
Figure 1.
S-DNAs contain regions of non-B-DNA. (A) The recognition sequence of the restriction enzyme BbvI is multiply repeated within the CTG/CAG repeat tract. Flanking non-repeating sequences are shown in lower case and the repeats are shown in upper case. Arrows indicate the expected and confirmed sites of scission. (B) 32P-Labeled (CTG)15 or (CAG)15 oligonucleotides were digested with BbvI individually or as a hybridized mixture. Products were resolved on a sequencing gel. Alone, neither were susceptible to BbvI digestion, while the duplexed (CTG)15·(CAG)15 was. Starting DNAs (–) and BbvI digestion products (+). Repeat location is indicated for both the 32P-labeled (CTG)15 or (CAG)15 oligonucleotides; relative product intensity varies due to the end effect. (C) Gel-purified EcoRI–HindIII linear form (L), S-DNA (S) and SI-DNAs (SI) structures uniquely radiolabeled at the EcoRI site on either the CTG or the CAG strands were digested with BbvI (Materials and Methods) and products resolved on denaturing acrylamide gels. Shown are schematics of the purified DNAs and the location of the unique radiolabels, indicated by filled diamonds. Each of the complementary strands (CTG and CAG) containing either 30 or 50 repeats were individually analyzed in each of the six possible structures. Maxam & Gilbert sequencing ladders (G>A) indicated nucleotide positions and indicated repeat number, which are read 5′→3′ such that the first CTG repeat unit corresponds to the last CAG repeat unit. Only the (CTG)50 and (CAG)50 strands in the SI-DNA displayed specific regions of protection (indicated by bracket). Shown is the analysis of the CTG strand 2P-labeled on the 5′ end (EcoRI). Analysis of the CAG strand yielded similar results. The analyses of each strand in each of the structural conformations is summarized in Figure 4.
Using BbvI, both the linear, slipped homoduplex (S-DNA) and slipped intermediate (SI-DNA) forms were probed for unusual DNA structures (Fig. 1C and summarized in Fig. 4). Reactions were performed under conditions approximating single-hit kinetics, which enabled the mapping at the nucleotide level of any sites resistant to cleavage by BbvI. Each of the complementary strands containing either 30 or 50 repeats was individually analyzed in each of the various structural forms. In the linear form both the (CTG)50 and (CAG)50 strands were sensitive throughout the repeat tract to BbvI restriction digestion as revealed by the ladder bands (Fig. 1C, see lanes labeled ‘L’, and Fig. 4), which indicated B-form duplex DNA. Similar results were observed for the linear form (CTG)30 and (CAG)30 strands (see Fig. 4). In the n = 30 and n = 50 S-DNAs both the CTG and CAG strands were equally sensitive throughout the repeat tract to BbvI digestion resulting in a ladder of bands (Fig. 1C, see lanes labeled ‘S’, and Fig. 4). However, multiple experiments indicated that the sensitivity of S-DNA to BbvI digestion was slightly greater than that of the linear forms suggesting that within S-DNA molecules non-duplex regions of enhanced reactivity were randomly located throughout the duplex repeat tract (Fig. 1C, compare relative amount digested and undigested at the top of the gel between lanes labeled ‘L’ and lanes labeled ‘S’). The enhanced scission of S-DNAs may be similar to the enhanced reactivity of Z-DNA to MboI relative to the B-form (35). The enhanced reactivity of the S-DNAs to BbvI digestion supports the presence of non-B-DNA structures along the repeat tract.
Distinct and specific regions of resistance to digestion by BbvI were evident only on the longer, slipped-out strand (CTG)50 or (CAG)50 in each of the SI-DNA heteroduplexes (Fig. 1C, see bracket; summarized in Fig. 4). The regions of maximal protection from BbvI digestion occurred at repeat 25 on either the (CTG)50 or (CAG)50 strands in their respective SI-DNA heteroduplexes. In both cases the protected region spanned approximately 10 repeats, with slightly greater protection 3′ of the central 25th CAG or CTG repeat. It is likely that the protected regions are caused by a non-B-DNA configuration, possibly a three-way DNA junction and/or single-stranded or hairpin conformations. In contrast to the n = 50 repeat strands, neither of the n = 30 repeat strands in the SI-DNA forms displayed any differences in the intensities of BbvI scissile sites (Fig. 4). The shorter strands in both SI-DNA heteroduplexes were digested by BbvI throughout their length, suggesting that the localized protection from digestion of the longer repeat was due to the inability of BbvI to recognize or digest the imperfectly base-paired slipped-out strand and not due to inaccessibility to restriction sites at the junction. Thus, both of the Watson–Crick branches of a putative three-way DNA junction have B-DNA characteristics and only the slipped-out strand is protected. The protected regions were localized and spanned approximately 10 repeats, this suggested that many of the excess repeats were contained in a single slip-out that extruded from a specific location along the Watson–Crick duplex. We conclude that since BbvI is unable to digest the individual CTG or CAG strands (Fig. 1B, lanes 2 and 4), the region resistant to BbvI digestion in both sister SI-DNAs belies a localized non-B-DNA structure within the repeat tracts. The sites of BbvI cleavage on each of the strands in each of the structural forms are summarized in Figure 4.
The location of the slipped-out repeats cannot readily be mapped by the boundaries of the BbvI protection since some class-II restriction enzymes, of which BbvI is a member, can restrict sites flanking or spanning either single-stranded regions (36,37), three-way DNA junctions (38), four-way DNA junctions (37) or Z-DNA (35). The ability of BbvI to cleave across DNA junctions is unknown. Experiments described below using MBN, T7endoI and EM were more useful in mapping the location of slip-out extrusion.
Slipped-strand DNAs have single-stranded character: mung bean nuclease
Slipped-out repeats may be in a hairpin or an unwound random-coil conformation, both of which would contain single-stranded regions. We used the single-strand specificity of MBN to determine if there were any regions of single-stranded character in the various structural forms of the trinucleotide repeat DNAs (Fig. 2 and summarized in Fig. 4). Neither the CTG nor CAG strands in the linear form displayed any sites sensitive to MBN (Fig. 2B–E, see lanes labeled ‘L’), indicating a B-DNA character (31). However, in the S-DNA forms both the CAG and CTG strands were sensitive to MBN, with each repeat unit throughout the tract displaying a similar susceptibility to scission (Fig. 2B–E, see lanes labeled ‘S’). The CAG strand was cut at the ApG linkages, while the CTG strand was cut at both the CpT and TpG linkages (Fig. 2A and data not shown). Comparing the relative amount of digested and undigested material between multiple experiments revealed that in the S-DNA form, the CAG strand was more susceptible to MBN cleavage than the CTG strand [Fig. 2, compare lanes labeled ‘S’ between B and C and between D and E], in agreement with crude analyses on unpurified samples (31). The even distribution of MBN-sensitive sites in the homoduplex S-DNAs revealed the existence of single-stranded structures, and that such structures were randomly located throughout the duplex repeat tract. This conclusion is further supported by the BbvI reactivity of each repeat unit in S-DNAs (see above and Fig. 1).
Distinct and specific regions of hypersensitivity to MBN were evident on the longer, slipped-out strand (CTG)50 or (CAG)50 in each of the SI-DNA heteroduplexes (Fig. 2 and summarized in Fig. 4). For the (CAG)50 tract the region of maximal MBN sensitivity spanned approximately 20 repeats (between the 25th and 45th repeats) with peak sensitivity occurring between the 30th and 40th repeats (Fig. 2D, see bracket and arrowheads; repeat unit numbers are read 5′→3′). For the (CTG)50 tract the region of MBN sensitivity spanned approximately 20 repeats (between the 10th and 30th repeats) with peak sensitivity occurring predominantly on the 20th repeat (Fig. 2E, see bracket and arrowheads; repeat unit numbers are read 5′→3′). The peak scission sites on the (CTG)50 were more acute than those on the (CAG)50 strand in their respective SI-DNA heteroduplexes. The (CAG)50 strand presented a window of numerous tandem hypersensitive sites (Fig. 2D, see bracket). Since cleavage of synthetic CTG and CAG hairpins by single-strand-specific nucleases occurred at the hairpin tips, and not at T-T or A-A mismatches along the hairpin stems (17,18), it is possible that the MBN hypersensitive sites observed herein represent the tips of hairpins formed by slipped-out repeats, with the multiple peaks of scission representing the preferred locations for the tips of the hairpins. An alternative explanation, particularly for the (CAG)50 strand, is that the MBN hypersensitive sites represent a single-stranded random-coil conformation. This notion is further supported by electron microscopic analyses below.
Both the (CTG)30 and (CAG)30 repeat strands in their respective SI-DNA forms displayed a relatively even distribution of MBN scissile sites (Fig. 2B and C). Slightly reduced scission occurred between the 10th and 25th repeat units of the (CAG)30 strand (Fig. 2B, compare lane labeled ‘SI’ with lane labeled ‘S’; repeat unit numbers for both strands are read 5′→3′). Increased scission occurred at the first to third repeat units of the (CTG)30 strand (Fig. 2C, see arrowheads). The sites of MBN cleavage on each of the strands in each of the structural forms are summarized in Figure 4.
Slipped-strand DNAs have distinct three-way DNA junctions: T7 endonuclease I
It has been hypothesized that slipped-out repeats, especially those capable of forming intra-strand hairpin structures, may form stable three-way DNA junctions (26,31,39,40). However, the existence of such junctions has not been reported. To identify and map at the nucleotide level the location of slip-out extrusions, we used the junction-specific phage T7endoI to probe each of the strands in each of the structural forms (linear, S-DNA and SI-DNA). T7endoI is highly structure-specific, preferring three-way and four-way DNA junctions, typically cutting multiple arms of the junctions. While T7endoI can digest single-stranded DNA (41) on DNA junctions, it cleaves exclusively at the base of the junctions and does not recognize the single-stranded tips of hairpins (42–44).
In the linear forms neither the CTG nor CAG strands displayed any sites sensitive to T7endoI (not shown), supporting a B-DNA duplex. However, in the S-DNAs both CAG and CTG strands were sensitive to T7endoI cutting, with each repeat unit displaying a similar susceptibility to scission (Fig. 3B and C, and summarized in Fig. 4). T7endoI cut the CAG strand at the ApG and the GpC linkages, while the CTG strand was cut at the CpT linkages (Fig. 3A). Comparing the relative amount of digested and undigested material between multiple experiments revealed that in the S-DNA form, the CTG strand was more susceptible to T7endoI cleavage than the CAG strand (Fig. 3B and C, compare lanes labeled ‘S’). This latter point, in conjunction with the increased MBN sensitivity of the CAG strand in S-DNAs and SI-DNAs (see above and Fig. 2), suggested that the slip-out junctions formed by the CTG strand may be biophysically more stable than those formed by the CAG strand.
Figure 3.
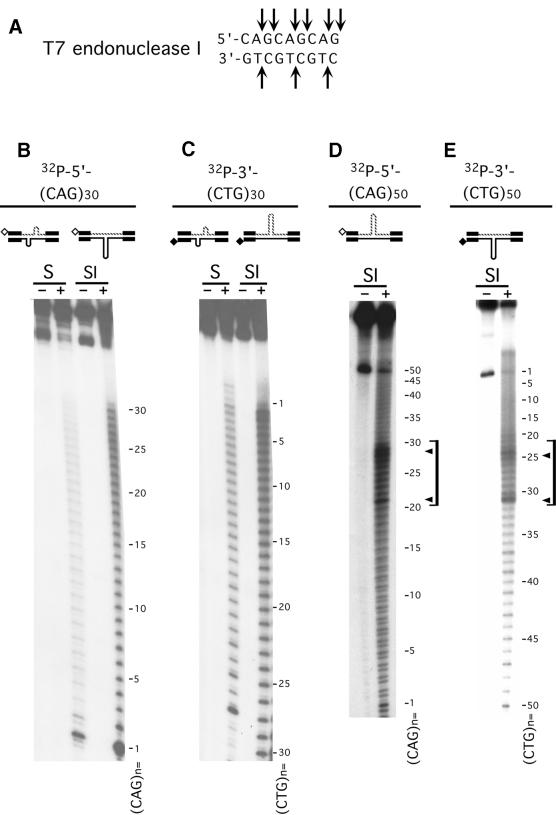
S-DNAs contain DNA junctions. (A) The bacteriophage T7endoI specifically recognizes three-way and four-way DNA junctions; arrows indicate the sites of scission. (B–E) Gel-purified HindIII–EcoRI linear form (L), S-DNA (S) and SI-DNA (SI) structures uniquely radiolabeled at the HindIII site on either the CTG or the CAG strands were digested with T7endoI (Materials and Methods) and products resolved on denaturing acrylamide gels. Shown are schematics of the purified DNAs and the location of the unique radiolabels, indicated by hollow or filled diamonds. Each of the complementary strands (CTG and CAG) containing either 30 or 50 repeats were individually analyzed in each of the purified structures. Starting DNAs (–) and T7endoI digestion products (+). Sequencing ladders (not shown) indicated nucleotide positions and indicated repeat numbers are read 5′→3′ such that the first CTG repeat unit corresponds to the last CAG repeat unit. No digestion products were detected for the linear forms (not shown). Only the longer 50 repeat strand in the SI-DNAs displayed specific regions of hypersensitivity (indicated by brackets) demarcated by peak sites (indicated by arrowheads). The analyses of each strand in each of the structural conformations are summarized in Figure 4.
Distinct and specific regions of hypersensitivity to T7endoI cutting were evident on only the longer (CAG)50 and (CTG)50 strands in their respective SI-DNA heteroduplexes (Fig. 3D and E, see arrowheads and brackets). These hypersensitive sites were 2–5-fold more reactive than the scissile sites evident for each repeat unit along the tracts. The regions of hypersensitivity to T7endoI digestion were demarcated by peak sites at the 20th and 30th CAG repeats and the 24th and 31st CTG repeats of the n = 50 strands (Fig. 3D and E, see brackets and arrowheads; repeat unit numbers for both strands are read 5′→3′). The T7endoI hypersensitive sites support the presence of preferred locations for the extrusion of slipped-out repeats from the Watson–Crick duplex repeat tract. Each repeat unit of the (CAG)30 and (CTG)30 strands in their respective SI-DNA heteroduplexes could be cleaved (Fig. 3B and C). Each showed mildly increased scission at the first to the 10th repeats. The sites of T7endoI cleavage on each of the strands in each of the structural forms are summarized in Figure 4.
The ability of T7endoI to cleave throughout the repeat tracts in both the S-DNA homoduplexes and SI-DNA heteroduplexes suggested that junctions of slipped-out repeats could exist at multiple locations along the repeat tracts. T7endoI can bind and cut short branches as well as single base mismatches (43,45). The T7endoI data (Fig. 3), the MBN data (Fig. 2) and that of other experiments (33) show that in S-DNAs there are numerous slip-outs, each containing a few repeats. However, for SI-DNAs, the hypersensitive sites of T7endoI scission revealed that there are preferred locations for the extrusion of the excess repeats. Since only two peaks of hypersensitivity were observed (both on the n = 50 strand), indicating that only two of the three arms of the three-way slip-out junction were preferentially susceptible to T7endoI cleavage. We conclude that the excess repeats of SI-DNAs slip-out at defined and localized three-way junctions. These results reveal that slip-out junctions formed by CTG/CAG repeats can be recognized and processed by a junction-specific resolvase.
Electron microscopic analysis of SI-DNAs: slipped-out CAG is preferentially bound by SSB
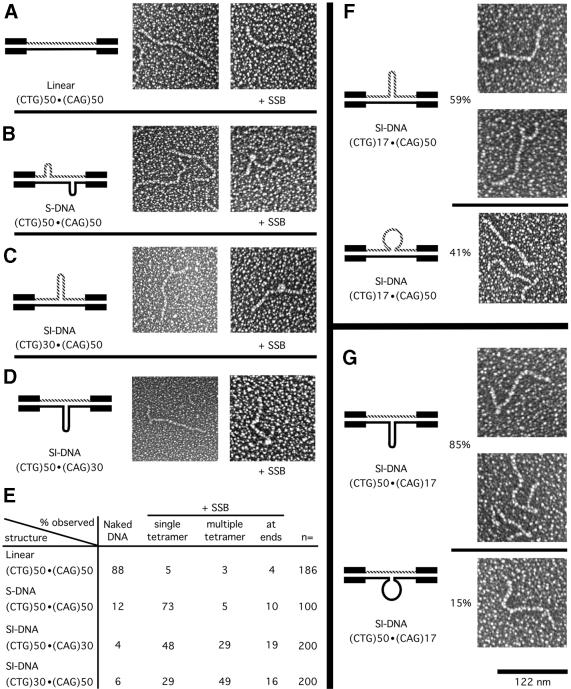
To gain a better structural understanding of the S-DNAs we analyzed each of the gel-purified DNAs by EM. The individual n = 30 or n = 50 repeat-containing DNAs in the linear fully duplexed form appeared as long molecules with smooth contours identical to random sequence DNA (Fig. 5A). In agreement with previous analyses (33), many of the homoduplex S-DNA molecules contained one or more kinks, which are typical of DNA junctions of short slipped-out repeats (Fig. 5B). In contrast, many SI-DNA molecules having an excess of CAG or an excess of CTG repeats appeared to contain a single branched structure (Fig. 5C and D). Based upon the MBN sensitivity of both S-DNAs and SI-DNAs we investigated their ability to bind the bacterial SSB. Using our conditions SSB bound as tetrameric complexes that can recognize 30–35 nt of ssDNA (46). Neither of the linear forms bound SSB to a significant degree (Fig. 5A and E). Both of the homoduplex S-DNAs were bound by SSB, with the majority binding as single-SSB tetramers (Fig. 5B and E). Both sister SI-DNAs were also bound by SSB. However, the majority of the (CTG)30·(CAG)50 SI-DNA bound multiple-SSB tetramers in tandem, while the majority of the (CTG)50·(CAG)30 SI-DNA bound single-SSB tetramers (Fig. 5C and D, summarized in E). A 30 bp hairpin, formed by the excess 20 repeats, would not readily bind more than a single-SSB tetramer. Thus, the binding of multiple, tandemly located SSB tetramers indicated that the slipped-out CAG repeats might be in a random-coil conformation rather than a hairpin. This is consistent with the preferential MBN digestion of the CAG strand in the (CTG)30·(CAG)50 SI-DNA (see Fig. 2). Since single- and multiple-SSB tetramers were detected for each SI-DNA, the slipped-out repeats may be in a dynamic equilibrium between the random-coil and hairpin conformations. Based upon the relative distribution of multiple- and single-SSB tetramers (Fig. 5E) the SI-DNAs having slipped-out CAG and slipped-out CTG repeats may exist predominantly in the random-coil and hairpin conformations, respectively. We conclude that there were indeed structural differences between the sister SI-DNAs and that a biologically important protein could detect these differences.
Figure 5.
Electron microscopic analyses of S-DNAs +/– SSB. (A–D) Electron microscopic analyses of gel-purified (CTG)50·(CAG)50 DNAs; linear form (L), slipped homoduplex S-DNAs (S) and the (CTG)50·(CAG)30 and (CTG)30·(CAG)50 slipped intermediate heteroduplexes (SI). The repeat tracts are contained on an NdeI–HindIII fragment which has 265 and 63 bp 5′ and 3′, respectively, of the (CTG)n tract. The left-most panels show schematic representations of the DNA structures, the middle panels show micrographs of the gel-purified naked DNA samples, and the right-most panels show micrographs of the same DNAs in the presence of single-strand binding protein (+ SSB). See text for details. Micrographs are shown in reverse contrast. Size bar in the lower right is 122 nm. (E) Summary of single-strand binding analyses; the qualitative and quantitative analyses of SSB binding were determined and tabulated. (F and G) Electron microscopic analyses of gel-purified sister SI-DNAs (CTG)50·(CAG)17 and (CTG)17·(CAG)50. The repeats are contained on the same NdeI–HindIII fragment as used for (A–D). The magnification for all panels is the same. The left-most panels show schematic representations of the DNA structures. The upper images of (F) and (G) show branched molecules with defined three-way slip-out junction with the slip-out in the hairpin conformation. The lower panels show ‘branchless’ molecules observed in the same gel-purified SI-DNA sample. Both a ‘branchless’ and a ‘branched’ molecule are seen in the lower micrograph of (F). The ‘branchless’ molecules had lengths similar to that of the n = 17 linear form (not shown), which electrophoretically migrated considerably faster than the SI-DNAs. The relative proportion of branched and branchless species in a given sample is indicated.
To further examine the structures formed by slipped-out repeats, we analyzed SI-DNAs having a greater difference in repeat numbers to increase EM resolution (Fig. 5F and G). Gel-purified SI-DNAs formed between n = 17 and n = 50 repeats could result in single-stranded loops of 33 repeats (99 nt) or hairpins of 48 bp (33 repeats, one repeat in loop). Most molecules in the purified (CTG)50·(CAG)17 SI-DNA and many in the (CTG)17·(CAG)50 SI-DNA preparations contained hairpins, with notable differences (see below). All hairpins mapped within the repeat tract and a peak of distribution was observed that was similar to the cleavage pattern generated by MBN (Fig. 4). Length measurements of the hairpins on the sister SI-DNAs revealed that the slipped-out CTG was similar to the slipped-out CAG repeats: ∼12 ± 3 nm with a maximum size of 15 nm (corresponding to 50 bp), which is roughly equivalent to a hairpin of 33 repeats—the difference between the two strands. All these observations suggest that most SI-DNAs contain a single slip-out of the excess repeats.
Structures with an excess of CAG repeats were distinct from those with an excess of CAG repeats. Notably, 85% of the purified (CTG)50·(CAG)17 SI-DNA molecules contained detectable branches, while only 59% of the purified (CTG)17·(CAG)50 SI-DNA molecules contained detectable branches (Fig. 5F and G, compare upper and lower portions). The EM method used cannot detect stretches of single-stranded DNA, and only the duplex portion of molecules containing a long single-stranded loop would be visible. Since the lengths of the ‘branchless’ molecules in the SI-DNA preparations were approximately that of the n = 17 linear form it is likely that these gel-purified heteroduplex molecules contained slipped-out repeats in the single-stranded conformation (Fig. 5F and G, lower portions). The relative distribution of branched and non-branched molecules for the slipped-out CAG and slipped-out CTG repeats correlated well with the relative distribution of SI-DNAs containing single- and multiple-SSB tetramers (Fig. 5F and G, compare relative percentages with those in E). This further supported the notion that slipped-out CAG and CTG repeats may be in a conformational equilibrium existing predominantly in the random-coil and hairpin conformations, respectively.
DISCUSSION
We have characterized at the nucleotide level slipped-strand structures formed by disease-relevant lengths of CTG/CAG repeats flanked by non-repeating sequences. The structures studied contained all predicted components including slipped-out repeats and three-way DNA junctions. This represents the first characterization of these components together as part of one structure. This analysis has revealed surprising characteristics of slipped-strand DNAs.
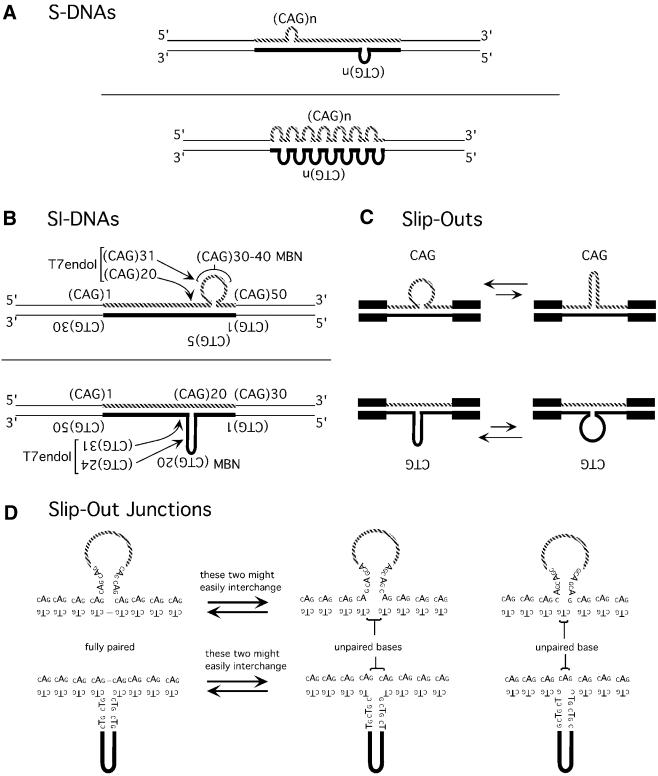
Slip-outs extruded at preferred locations in SI-DNAs but not S-DNAs. Theoretically a slip-out of repeats can extrude at virtually any point along the repeat tract. This suspicion was confirmed for slipped homoduplex S-DNAs, which have predominantly short slip-outs of 1 to 10 repeats, varying in number and occurring anywhere along the duplex repeat tract (Fig. 6A). In contrast to homoduplex S-DNAs, the SI-DNA heteroduplexes formed a limited set of isomers; each of which contained a unique slipped-out region that extruded at specific preferred locations from the Watson–Crick duplex (Fig. 6B). Points of extrusion were off-center such that the SI-DNA with an excess of CAG repeats extruded the 25th to the 45th repeats, while the SI-DNA with an excess of CTG repeats extruded the 10th to the 30th repeats. The preferred locations of slip-out extrusion are a curious observation. We have shown previously that the pattern and amount of S-DNA formed by a given length of repeats can vary depending upon the sequence of the non-repeating DNAs flanking the repeat (compare patterns for SCA1 and DM1 DNAs in refs 32,19, respectively). However, unlike S-DNAs, SI-DNAs formed by SCA1 clones formed distinct electrophoretic species much like those formed by DM1 clones—thereby arguing against a role for flanking sequences in the number and size of slip-outs in SI-DNAs (unpublished results; 19). It is unclear as to why all of the excess repeats extruded at distinct locations in the SI-DNAs and was not distributed in multiple short slip-outs along the complementary strand. It is possible that during strand annealing the exchange levels of inter- and intra-strand pairings are increased for two complementary strands having different numbers of repeats than for two strands having the same number of repeats.
Figure 6.
Structural features of slipped-strand DNAs. Schematic models for S-DNAs, SI-DNAs, slipped-out repeats, and slip-out junctions based upon the BbvI, MBN, T7endoI probing (Figs 1–3, summarized in Fig. 4), SSB binding and electron microscopic analyses (Fig. 5). (A) Homoduplex S-DNAs with multiple small slip-outs (3–10 repeats) formed throughout the repeat tract. (B) Heteroduplex, SI-DNAs. The (CTG)30·(CAG)50 SI-DNA extruded the 25th to the 45th CAG repeats, while the sister SI-DNA (CTG)50·(CAG)30 extruded the 10th to the 30th CTG repeats. The preferred sites of MBN and T7endoI cleavage are indicated by repeat unit numbers (read 5′→3′ such that the first CTG repeat unit corresponds to the last CAG repeat unit). (C) Schematic representation of two possible conformations of the slipped-out repeats. The size of the arrows are representative of the observed relative proportions of each. (D) Slip-out junction conformations. Shown are three possible base-pairing schemes at the three-way slip-out junctions. Depending upon the nucleotide at the interchange of Watson–Crick and slip-out arms, the junctions can be fully base-paired, or can contain one or two mismatched bases on the strand opposite the slipped out repeats. The fully paired and the two unpaired base junction isoforms might easily interchange.
Slip-outs can form intra-strand hairpins and unpaired loops. In SI-DNAs the slipped-out CAG and CTG repeats existed as unpaired loops and hairpins, respectively (Fig. 6C). Scissile site selection by MBN is sensitive to the sequence of the single stranded DNA (47) as well as hairpin tip structure (48) suggesting that MBN cleavage of CTG and CAG repeats (Fig. 2A) may reflect the unwound and/or hairpin tip conformations. The observation that slipped-out CAG repeats assumed a random-coil conformation was unexpected as all other studies, which used short oligonucleotides, have reported hairpin structures for both CAG and CTG repeats (15–18). It may be that in the context of its complementary strand at the slip-out junction, the long tract of excess CAG repeats behaves differently than it would as an isolated oligonucleotide. A slip-out that is unhindered by intra-strand base pairings may ‘creep’ (49) or ‘inchworm’ (50) along the Watson–Crick duplex—such slip-out migration would lead to a net translational slippage of the two strands. The unpaired nature of slipped-out CAG repeat may permit higher frequencies of errors with this strand than slipped-out CTG.
We present the first evidence for slipped-strand DNA junctions where each arm of the three-way junction is composed of CTG or CAG repeats. Junctions were visualized by EM and mapped by T7endoI scission (Fig. 6B). Assuming that the MBN hypersensitive sites defined the center of the slipped-out repeats, the T7endoI hypersensitive sites on each SI-DNA were located at approximately four to five repeats 5′ of the CTG hairpin tip, approximately four to five repeats 5′ of the center of the slipped-out CAG repeats and within the Watson–Crick duplex at approximately one to five repeats 5′ of the slip-out junction (Fig. 6B). The preferred locations of T7endoI cleavage (Fig. 3A) may reflect either the junction conformations or sequence preferences of the enzyme (41,44,45). What is the base pairing at the three-way slip-out junctions? For each sister SI-DNA there are three possible schemes (Fig. 6D) each defined by the first 5′ base of the slipped-out strand being C or T/A or G. The junctions formed by these would contain two, one or no unpaired bases on the strand immediately opposite the slipped-out repeats. Unpaired bases at heterologous three-way DNA junctions can increase the biophysical stability of the junctions (51) and may facilitate coaxial helix–helix stacking (52). However, the even scission pattern of MBN and BbvI on the non-slipped n = 30 strand at the slip-out extrusion point argues against any unpaired bases. While we are unable to exclude the presence of unpaired bases at the three-way slip-out junctions our results favor a slip-out junction containing no unpaired bases.
Implications
Current results suggest that S-DNAs and SI-DNAs may participate in different types of trinucleotide repeat instability observed within patients. We have shown previously that the propensity to form slipped-strand DNAs is directly affected by repeat tract length and purity—both factors directly affect the genetic instability of the repeats in humans (19,31–33). Here we report observations that support the participation of SI-DNAs and S-DNAs in replication- and repair-based mutations, respectively.
The preferred location for the extrusion of excess repeats in SI-DNAs may contribute to repeat instability at primate replication forks (13,14). Both the sequence (determined by replication direction) and the location of the repeat tract within the single-stranded Okazaki Initiation Zone may determine whether a mutagenic intermediate will form. The preferred location for the extrusion of excess slipped-out repeats in SI-DNA observed herein may explain the location-sensitivity of repeat length instability at primate replication (13).
The lengths of the slip-outs formed on S-DNAs and SI-DNAs correlate with the magnitudes of repeat length alterations reported in certain tissues of diseased patients. Slip-outs in S-DNAs ranged from one to 10 repeats, while individual slip-outs in SI-DNAs can be as great as the total difference in repeat numbers between the two strands. Large meiotic and somatic (CTG)·(CAG) expansions (increases of 20–500 repeats) (11,12) may occur through large jumps involving DNA mutagenic intermediates such as SI-DNA which can contain large individual slip-outs. SI-DNAs may be formed by replication slippage (19), during unequal crossover recombination between sister chromatids (53) or homologous recombination (54). In contrast a relatively short range (1 to 23 repeats) of intra-tissue repeat lengths has been detected in non-proliferating neural tissues such as the brains of HD, DRPLA, SCA3 patients (4,6,8–10). These short length changes may be incurred during processing of the short slip-outs of homoduplex S-DNAs formed within the large amounts of single-stranded DNA observed in aging neural tissues (20–22). Notably, use of the sensitive small pool PCR has revealed rare large expansions in the brains of transgenic mice and human HD brains (P.F.Shelbourne, personal communication; 55)—which may be the result of multiple rounds of damage and repair. However, the majority of repeat length changes in post-mitotic tissues are relatively short. The correlation of the size of the slip-outs in SI-DNA and S-DNAs with the size of tract length changes in proliferating and non-proliferating tissues, respectively, might suggest that SI-DNAs and S-DNAs are mutagenic intermediates in proliferative and non-proliferative processes of instability, respectively.
Protein recognition of either the slipped-outs, the hairpin tips or slip-out junctions may critically determine whether slipped-structures are repaired and/or recombined. Such processing, or lack thereof, may contribute to repeat maintenance or instability. Differential processing of mutagenic intermediates with either an excess of CTG or CAG repeats may explain the tendency to expand or delete repeats at primate replication forks (14). Herein we have shown that SSB binds preferentially to slipped-out CAG repeats over slipped-out CTG repeats. SSB is ubiquitous (46) and involved in DNA replication, repair and recombination. In bacteria SSB stabilizes CTG/CAG repeats in a replication direction-dependent manner (56). Similarly, the human mismatch repair protein, hMSH2, also binds preferentially to SI-DNAs with an excess of CAG repeats (19). MSH2 and MSH3 are required for CTG/CAG instability in transgenic mice (57–59). Preferential binding of CAG over CTG by replication or repair proteins may reflect differential processing of one strand over another and may explain different instabilities observed for different replication directions (17,18).
Slip-out junctions may be structural features recognized by DNA repair or recombination resolvases—making slip-out junctions analogous to four-way junctions in Holliday recombination intermediates (29,30). Mutagenic intermediates involving the gain or loss of some, but not all repeat units would be expected to contain three-way junctions composed of complementary repeats (Fig. 6D). We have shown that the junctions formed by slip-outs of (CTG)n or (CAG)n extruded from two arms of (CTG)n·(CAG)n are recognized and resolved by T7endoI, a member of the MutH/Vsr repair endonuclease family. Hypersensitive scission occurred only on the strand containing an excess of repeats, which differs from that expected of heterologous three-way junctions, where each junction strand is cut without preference (42–44). Preferential scission of only the slipped-out strand suggests that the unique features of the CTG/CAG junctions afforded their differential processing. In yeast, where the repeats tend to delete, it has been shown that heteroduplex DNAs with intra-strand hairpins composed of (CTG)10 or (CAG)10 escaped repair (60). We note that the repeat structures analyzed in yeast were true heteroduplexes such that they contained only CTG or CAG repeats in the complete absence of CAG or CTG repeats in the complementary strand (60). Slip-out junctions of (CTG)n or (CAG)n extruded from two arms of (CTG)n·(CAG)n may be processed by human cells in a manner that leads to expansions. Mammalian (61) but not yeast cells (60) are capable of repairing palindromic heteroduplexes. It is unknown if slipped-out CTG or CAG repeats escape repair in mammalian cells as they do in yeast (60). It is of interest to know if human cells are capable of metabolizing slipped-strand trinucleotide repeat structures and what role, if any, the slip-out and CTG/CAG three-way junctions may play.
Understanding if and how slipped mutagenic intermediates are repaired or recombined will shed light on the mechanism(s) of disease-associated repeat instability. Detailed biophysical characterization of the individual features (slip-outs, loop-outs, hairpin stems, hairpin tips and slip-out junctions) now permits further studies aimed at understanding which proteins may act upon slipped-strand DNAs, the commonly hypothesized mutagenic intermediates.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Paul Sadowski, Richard Collins and Gagan Panigrahi for their comments on the manuscript. We thank Peggy Shelbourne for communication of her results prior to publication. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and the Muscular Dystrophy Association USA to C.E.P. and from the NIH (CA85826) and the March of Dimes to Y.-H.W. C.E.P. is a CIHR Scholar, a PREA Scholar, and a Canadian Genetic Disease Network Scholar. Y.-H.W. is a Basil O’Connor March of Dimes Fellow. J.D.C. is supported by CIHR doctoral research award, M.T. by Samuel Lunenfeld Summer Research Student Training Program and S.E.M. by The Hospital for Sick Children Graduate Scholarship.
REFERENCES
- 1.Cummings C.J. and Zoghbi,H.Y. (2000) Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet., 9, 909–916. [DOI] [PubMed] [Google Scholar]
- 2.Richards R.I. and Sutherland,G.R. (1992) Dynamic mutations: a new class of mutations causing human disease. Cell, 70, 709–712. [DOI] [PubMed] [Google Scholar]
- 3.Sinden R.R. (2001) Neurodegenerative diseases. Origins of instability. Nature, 411, 757–758. [DOI] [PubMed] [Google Scholar]
- 4.Chong S.S., McCall,A.E., Cota,J., Subramony,S.H., Orr,H.T., Hughes,M.R. and Zoghbi,H.Y. (1995) Gametic and somatic tissue-specific heterogeneity of the expanded SCA1 CAG repeat in spinocerebellar ataxia type 1. Nature Genet., 10, 344–350. [DOI] [PubMed] [Google Scholar]
- 5.Martorell L., Monckton,D.G., Gamez,J., Johnson,K.J., Gich,I., de Munain,A.L. and Baiget,M. (1998) Progression of somatic CTG repeat length heterogeneity in the blood cells of myotonic dystrophy patients. Hum. Mol. Genet., 7, 307–312. [DOI] [PubMed] [Google Scholar]
- 6.Telenius H., Kremer,B., Goldberg,Y.P., Theilmann,J., Andrew,S.E., Zeisler,J., Adam,S., Greenberg,C., Ives,E.J., Clarke,L.A. et al. (1994) Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nature Genet., 6, 409–414. [DOI] [PubMed] [Google Scholar]
- 7.Thornton C.A., Johnson,K. and Moxley,R.T.,III (1994) Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann. Neurol., 35, 104–107. [DOI] [PubMed] [Google Scholar]
- 8.Hashida H., Goto,J., Kurisaki,H., Mizusawa,H. and Kanazawa,I. (1997) Brain regional differences in the expansion of a CAG repeat in the spinocerebellar ataxias: dentatorubral-pallidoluysian atrophy, Machado-Joseph disease, and spinocerebellar ataxia type 1. Ann. Neurol., 41, 505–511. [DOI] [PubMed] [Google Scholar]
- 9.Takano H., Onodera,O., Takahashi,H., Igarashi,S., Yamada,M., Oyake,M., Ikeuchi,T., Koide,R., Tanaka,H., Iwabuchi,K. and Tsuji,S. (1996) Somatic mosaicism of expanded CAG repeats in brains of patients with dentatorubral-pallidoluysian atrophy: cellular population-dependent dynamics of mitotic instability. Am. J. Hum. Genet., 58, 1212–1222. [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka F., Sobue,G., Doyu,M., Ito,Y., Yamamoto,M., Shimada,N., Yamamoto,K., Riku,S., Hshizume,Y. and Mitsuma,T. (1996) Differential pattern in tissue-specific somatic mosaicism of expanded CAG trinucleotide repeats in dentatorubral-pallidoluysian atrophy, Machado-Joseph disease, and X-linked recessive spinal and bulbar muscular atrophy. J. Neurol. Sci., 135, 43–50. [DOI] [PubMed] [Google Scholar]
- 11.Martorell L., Johnson,K., Boucher,C.A. and Baiget,M. (1997) Somatic instability of the myotonic dystrophy (CTG)n repeat during human fetal development. Hum. Mol. Genet., 6, 877–880. [DOI] [PubMed] [Google Scholar]
- 12.Wohrle D., Kennerknecht,I., Wolf,M., Enders,H., Schwemmle,S. and Steinbach,P. (1995) Heterogeneity of DM kinase repeat expansion in different fetal tissues and further expansion during cell proliferation in vitro: evidence for a casual involvement of methyl-directed DNA mismatch repair in triplet repeat stability. Hum. Mol. Genet., 4, 1147–1153. [DOI] [PubMed] [Google Scholar]
- 13.Cleary J.D., Nichol,K., Wang,Y. and Pearson,C.E. (2002) Evidence of cis-acting factors in replication mediated trinucleotide repeat instability in primate cells. Nature Genet., 31, 37–46. [DOI] [PubMed] [Google Scholar]
- 14.Panigrahi G.B., Cleary,J.D. and Pearson,C.E. (2002) In vitro (CTG).(CAG) expansions and deletions by human cell extracts. J. Biol. Chem., 277, 13926–13934. [DOI] [PubMed] [Google Scholar]
- 15.Bowater R.P. and Wells,R.D. (2001) The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol., 66, 159–202. [DOI] [PubMed] [Google Scholar]
- 16.Wells R.D. (1996) Molecular basis of genetic instability of triplet repeats. J. Biol. Chem., 271, 2875–2878. [DOI] [PubMed] [Google Scholar]
- 17.Pearson C.E. and Sinden,R.R. (1998) Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol., 8, 321–330. [DOI] [PubMed] [Google Scholar]
- 18.McMurray C.T. (1999) DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl Acad. Sci. USA, 96, 1823–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson C.E., Ewel,A., Acharya,S., Fishel,R.A. and Sinden,R.R. (1997) Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet., 6, 1117–1123. [DOI] [PubMed] [Google Scholar]
- 20.Chetsanga C.J., Boyd,V., Peterson,L. and Rushlow,K. (1975) Single-stranded regions in DNA of old mice. Nature, 253, 130–131. [DOI] [PubMed] [Google Scholar]
- 21.Nakanishi K., Shima,A., Fukuda,M. and Fujita,S. (1979) Age associated increase of single-stranded regions in the DNA of mouse brain and liver cells. Mech. Ageing Dev., 10, 273–281. [DOI] [PubMed] [Google Scholar]
- 22.Price G.B., Modak,S.P. and Makinodan,T. (1971) Age-associated changes in the DNA of mouse tissue. Science, 171, 917–920. [DOI] [PubMed] [Google Scholar]
- 23.Johnson R. and Strehler,B.L. (1972) Loss of genes coding for ribosomal RNA in ageing brain cells. Nature, 240, 412–414. [DOI] [PubMed] [Google Scholar]
- 24.Strehler B.L. and Chang,M.P. (1979) Loss of hybridizable ribosomal DNA from human post-mitotic tissues during aging: II. Age-dependent loss in human cerebral cortex–hippocampal and somatosensory cortex comparison. Mech. Ageing Dev., 11, 379–382. [DOI] [PubMed] [Google Scholar]
- 25.Strehler B.L., Chang,M.P. and Johnson,L.K. (1979) Loss of hybridizable ribosomal DNA from human post-mitotic tissues during aging: I. Age-dependent loss in human myocardium. Mech. Ageing Dev., 11, 371–378. [DOI] [PubMed] [Google Scholar]
- 26.Gacy A.M., Goellner,G., Juranic,N., Macura,S. and McMurray,C.T. (1995) Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell, 81, 533–540. [DOI] [PubMed] [Google Scholar]
- 27.Petruska J., Arnheim,N. and Goodman,M.F. (1996) Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res., 24, 1992–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson C.E. and Sinden,R.R. (1998) In Wells,.R.D. and Warren,S.T. (eds), Genetic Instabilities and Hereditary Neurological Diseases, 1st Edn. Academic Press, New York, pp. 585–621.
- 29.Lilley D.M. (2000) Structures of helical junctions in nucleic acids. Q. Rev. Biophys., 33, 109–159. [DOI] [PubMed] [Google Scholar]
- 30.Lilley D.M. and White,M.F. (2001) The junction-resolving enzymes. Nature Rev. Mol. Cell Biol., 2, 433–443. [DOI] [PubMed] [Google Scholar]
- 31.Pearson C.E. and Sinden,R.R. (1996) Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry, 35, 5041–5053. [DOI] [PubMed] [Google Scholar]
- 32.Pearson C.E., Eichler,E.E., Lorenzetti,D., Kramer,S.F., Zoghbi,H.Y., Nelson,D.L. and Sinden,R.R. (1998) Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry, 37, 2701–2708. [DOI] [PubMed] [Google Scholar]
- 33.Pearson C.E., Wang,Y.H., Griffith,J.D. and Sinden,R.R. (1998) Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res., 26, 816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szybalski W., Kim,S.C., Hasan,N. and Podhajska,A.J. (1991) Class-IIS restriction enzymes—a review. Gene, 100, 13–26. [DOI] [PubMed] [Google Scholar]
- 35.Winkle S.A., Aloyo,M.C., Morales,N., Zambrano,T.Y. and Sheardy,R.D. (1991) Enhanced reactivity of a B-Z junction for cleavage by the restriction enzyme MboI. Biochemistry, 30, 10601–10606. [DOI] [PubMed] [Google Scholar]
- 36.Podhajska A.J. and Szybalski,W. (1985) Conversion of the FokI endonuclease to a universal restriction enzyme: cleavage of phage M13mp7 DNA at predetermined sites. Gene, 40, 175–182. [DOI] [PubMed] [Google Scholar]
- 37.Murchie A.I., Portugal,J. and Lilley,D.M. (1991) Cleavage of a four-way DNA junction by a restriction enzyme spanning the point of strand exchange. EMBO J., 10, 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong D.M. and Wetmur,J.G. (1994) Some class-IIS restriction endonucleases can cleave across a three-way DNA junction. Gene, 150, 63–66. [DOI] [PubMed] [Google Scholar]
- 39.Hartenstine M.J., Goodman,M.F. and Petruska,J. (2000) Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J. Biol. Chem., 275, 18382–18390. [DOI] [PubMed] [Google Scholar]
- 40.Yu A., Dill,J. and Mitas,M. (1995) The purine-rich trinucleotide repeat sequences d(CAG)15 and d(GAC)15 form hairpins. Nucleic Acids Res., 23, 4055–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadowski P.D. (1971) Bacteriophage T7 endonuclease. I. Properties of the enzyme purified from T7 phage-infected Escherichia coli B. J. Biol. Chem., 246, 209–216. [PubMed] [Google Scholar]
- 42.de Massy B., Weisberg,R.A. and Studier,F.W. (1987) Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J. Mol. Biol., 193, 359–376. [DOI] [PubMed] [Google Scholar]
- 43.Lu M., Guo,Q., Studier,F.W. and Kallenbach,N.R. (1991) Resolution of branched DNA substrates by T7 endonuclease I and its inhibition. J. Biol. Chem., 266, 2531–2536. [PubMed] [Google Scholar]
- 44.Dickie P., McFadden,G. and Morgan,A.R. (1987) The site-specific cleavage of synthetic Holliday junction analogs and related branched DNA structures by bacteriophage T7 endonuclease I. J. Biol. Chem., 262, 14826–14836. [PubMed] [Google Scholar]
- 45.Mashal R.D., Koontz,J. and Sklar,J. (1995) Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nature Genet., 9, 177–183. [DOI] [PubMed] [Google Scholar]
- 46.Lohman T.M. and Ferrari,M.E. (1994) Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem., 63, 527–570. [DOI] [PubMed] [Google Scholar]
- 47.Laskowski M. Sr (1980) Purification and properties of the mung bean nuclease. Methods Enzymol., 65, 263–276. [DOI] [PubMed] [Google Scholar]
- 48.Kabotyanski E.B., Zhu,C., Kallick,D.A. and Roth,D.B. (1995) Hairpin opening by single-strand-specific nucleases. Nucleic Acids Res., 23, 3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baldwin R.L. (1968) Kinetics of helix formation and slippage of the dAT copolymer. In Pullman,B. (ed.), Symposium on Molecular Associations in Biology. Academic Press, New York, pp. 145–162.
- 50.Olivera B.M. and Lehman,I.R. (1968) Enzymic joining of polynucleotides. 3. The polydeoxyadenylate-polydeoxythymidylate homopolymer pair. J. Mol. Biol., 36, 261–274. [DOI] [PubMed] [Google Scholar]
- 51.Leontis N.B., Kwok,W. and Newman,J.S. (1991) Stability and structure of three-way DNA junctions containing unpaired nucleotides. Nucleic Acids Res., 19, 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welch J.B., Walter,F. and Lilley,D.M. (1995) Two inequivalent folding isomers of the three-way DNA junction with unpaired bases: sequence-dependence of the folded conformation. J. Mol. Biol., 251, 507–519. [DOI] [PubMed] [Google Scholar]
- 53.Smith G.P. (1976) Evolution of repeated DNA sequences by unequal crossover. Science, 191, 528–535. [DOI] [PubMed] [Google Scholar]
- 54.Richard G.F., Goellner,G.M., McMurray,G.T. and Haber,J.E. (2000) Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J., 19, 2381–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kennedy L. and Shelbourne,P.F. (2000) Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Hum. Mol. Genet., 9, 2539–2544. [DOI] [PubMed] [Google Scholar]
- 56.Rosche W.A., Jaworski,A., Kang,S., Kramer,S.F., Larson,J.E., Geidroc,D.P., Wells,R.D. and Sinden,R.R. (1996) Single-stranded DNA-binding protein enhances the stability of CTG triplet repeats in Escherichia coli. J. Bacteriol., 178, 5042–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manley K., Shirley,T.L., Flaherty,L. and Messer,A. (1999) Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature Genet., 23, 471–473. [DOI] [PubMed] [Google Scholar]
- 58.Kovtun I.V. and McMurray,C.T. (2001) Trinucleotide expansion in haploid germ cells by gap repair. Nature Genet., 27, 407–411. [DOI] [PubMed] [Google Scholar]
- 59.van Den Broek W.J., Nelen,M.R., Wansink,D.G., Coerwinkel,M.M., te Riele,H., Groenen,P.J. and Wieringa,B. (2002) Somatic expansion behaviour of the (CTG)(n) repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet., 11, 191–198. [DOI] [PubMed] [Google Scholar]
- 60.Moore H., Greenwell,P.W., Liu,C.P., Arnheim,N. and Petes,T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bill C.A., Taghian,D.G., Duran,W.A. and Nickoloff,J.A. (2001) Repair bias of large loop mismatches during recombination in mammalian cells depends on loop length and structure. Mutat. Res., 485, 255–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.