Abstract
N6-Methyladenosine (m6A) is present at internal sites in mRNA isolated from all higher eukaryotes, but has not previously been detected in the mRNA of the yeast Saccharomyces cerevisiae. This nucleoside modification occurs only in a sequence- specific context that appears to be conserved across diverse species. The function of this modification is not fully established, but there is some indirect evidence that m6A may play a role in the efficiency of mRNA splicing, transport or translation. The S.cerevisiae gene IME4, which is important for induction of sporulation, is very similar to the human gene MT-A70, which has been shown to be a critical subunit of the human mRNA [N6-adenosine]-methyltransferase. This observation led to the hypothesis that yeast sporulation may be dependent upon methylation of yeast mRNA, mediated by Ime4p. In this study we show that induction of sporulation leads to the appearance of low levels of m6A in yeast mRNA and that this modification requires IME4. Moreover, single amino acid substitutions in the putative catalytic residues of Ime4p lead to severe sporulation defects in a strain whose sporulation ability is completely dependent on this protein. Collectively, these data suggest very strongly that the activation of sporulation by Ime4p is the result of its proposed methyltransferase activity and provide the most direct evidence to date of a physiologic role of m6A in a gene regulatory pathway.
INTRODUCTION
Post-transcriptional processing of mRNA in eukaryotes is a complicated multistep process. In recent years, research focusing on RNA molecular biology has shown that these post-transcriptional events play important roles in the overall regulation of gene expression. In addition to the obvious processing steps (such as capping, polyadenylation and splicing), post-transcriptional modification of certain nucleosides in mRNA also occurs, but has received substantially less attention. This has been due to the difficulty in determining what role, if any, modified nucleosides play in the regulation of gene expression.
Despite the huge diversity of nucleoside modifications found in rRNA, tRNA and snRNA, the modified nucleosides found in eukaryotic mRNA are few and are structurally simple. The N7-amino group is methylated on the ‘cap’ guanosine (m7G). The 2′-O-hydroxyl group on the ribose moiety is methylated only on the first two encoded nucleosides (Nm). If the first encoded nucleoside is an adenosine, it can be doubly methylated—at both the 2′-O-position on the ribose and the N6-amino position on the purine ring to form (N6-methyl-,2′-O-methyl-)-dimethyladenosine (m6Am). All of these methylated residues are part of the cap structure (1–3). The only other modified nucleoside found in mRNA is N6-methyladenosine (m6A), which is present at low levels throughout the body of the mRNA.
m6A is present in mRNA in all higher eukaryotes tested, including mammals, plants and insects (4–8), and in viruses that replicate in the nucleus [adenovirus, SV40, RSV, herpesviruses, influenza (9)]. It is present on average in one to three residues per typical mammalian mRNA molecule. It is present only within a defined sequence context that has been conserved across diverse species from plants to man (9). Despite its ubiquity and conserved sequence specificity, the functional significance of this modification remains a mystery. Point mutagenesis of m6A sites in viral RNAs had no apparent effect on viral replication or infectivity (10,11). Inhibition of mRNA methylation with agents such as cycloleucine and S-tubercidinylhomocysteine leads to accumulation of mRNA precursors in the nucleus (12–14), but the pleiotropic nature of these agents makes it impossible to ascertain whether the effects are mediated through inhibition of m6A formation, or by inhibition of methylation of some other moiety. More recently, Tuck et al. (15) have shown that m6A-deficient mRNA encoding dihydrofolate reductase is translated less efficiently than m6A-containing mRNA.
m6A has not previously been observed in the mRNA of the yeast Saccharomyces cerevisiae (16), even though this organism does contain a gene (IME4/SPO8) with striking sequence similarity to the mammalian gene encoding the N6-adenosine methyltransferase subunit, MT-A70 (17). This sequence similarity raised the possibility that yeast mRNA does contain m6A, but only under nutritional conditions conducive to high expression of the IME4 gene. IME4 mRNA levels are very low in exponentially-growing cells and are greatly elevated during sporulation and in stationary phase cells (18).
IME4 is a key gene in the highly regulated pathway that leads to meiosis and sporulation in S.cerevisiae (19). Sporulation occurs only in the diploid MATa/MATα cell type, and requires nitrogen starvation and a respirable carbon source. Under these conditions, expression and activation of the IME1 gene and its product lead to the initiation of a complex cascade of gene expression (20) that culminates in the formation of four haploid ascospores within the confines of the original mother cell. The IME4 gene is important for IME1 transcript accumulation as well as downstream events, and is essential for meiosis and sporulation in standard laboratory strains. The mechanism by which Ime4p exerts its effects is unknown, although the protein does not function as a transcriptional activator or repressor in tethering assays (B.Jursic, Y.Wang and M.Clancy, unpublished results).
The close sequence similarity between the IME4 gene and MT-A70 leads to the hypothesis that Ime4p activates meiosis and spore formation by a mechanism that involves the formation of m6A in yeast RNA, possibly mRNA. In order to test this hypothesis, we have measured the m6A content of total RNA and polyadenylated RNA isolated from vegetative yeast cells and sporulating yeast cells, and have observed an increase in m6A formation in both types of preparations. Moreover, inactivation of the IME4 gene leads to the loss of m6A in the mRNA of the mutant yeast incubated in sporulation medium. We also examined whether mutations in the catalytic MTase motif IV sequence of Ime4p lead to sporulation defects, as expected if the formation of m6A is important for IME4’s role in promoting sporulation. Indeed, such mutations do lead to severely defective sporulation. Taken together, these results suggest that the formation of m6A is important for RNA biogenesis during sporulation in yeast.
MATERIALS AND METHODS
Isolation of total RNA from yeast
RNA analyses used the rapidly-sporulating SK1 strain (Hanson SK1-can1; ATCC). The SK1-ime4 mutant diploid was constructed by disrupting IME4 in haploid strains AMP107 and AMP108, kindly provided by Aaron Mitchell. Plasmid pJS21 (19), containing LEU2 within the IME4 open reading frame, was used as a template for PCR using primers 901F (5′-ATCGTGAAACTGCGAGTG) and 1420R (5′-GTC TCTCTGGTCATTGAT), and the haploids were transformed with the resulting PCR product. Transformants were screened for the desired disruption using the same primers and isolates of opposite mating type were mated to form SK1-ime4 (MATa/MATα ime4:LEU2/ime4:LEU2 leu2/leu2 ura3/ura3 trp1/trp1 lys2:ho:LYS2/lys2:ho:LYS2).
A 5 ml culture of YPD medium (1% Bacto-yeast extract, 2% Bacto-peptone, 2% dextrose) was inoculated with a single colony of S.cerevisiae Hansen SK1-can1 (ATCC). The culture was incubated at 30°C with vigorous shaking until reaching OD595 = 0.5 (∼7 h). Fifty milliliters of methionine-free SD medium (0.67% Bacto-yeast nitrogen base without amino acids, 2% dextrose, 530 mg/ml complete drop-out medium minus methionine) was inoculated with 200 µl of the log-phase culture and incubated for 16 h at 30°C with vigorous shaking. For control cultures, 2 × 107 cells were centrifuged at 1000 g at room temperature and resuspended in 2 ml of SD-methionine-free medium. 350 µCi of l-[methyl-3H]methionine (84 Ci/mmol) (Amersham) was added and the culture was incubated for 60 min at 30°C with vigorous shaking. Cells were harvested by centrifugation at 1000 g for 3 min at room temperature. Total RNA was isolated from the cells after zymolase treatment for 20 min at 30°C, using the RNeasy Kit (Qiagen) according to the manufacturer’s instructions. For sporulating cultures, 2 × 107 cells were centrifuged at 1000 g at room temperature and resuspended in 2 ml of sporulation medium (3.0 g of potassium acetate, 0.2 g of raffinose in 1 l of water). The culture was incubated for 5 h at 30°C with vigorous shaking. 350 µCi of l-[methyl-3H]methionine was added and the culture was incubated for an additional hour at 30°C with vigorous shaking. Yeast cells were then harvested and RNA was prepared as above.
Real-time RT–PCR
Primers: IME1 sense primer, TGATGAATCCGCATC TACGTTCCAC; IME1 antisense primer, CGGAGGCGT TGTTATTATTGCTGG; IME4 sense primer, ATGAT GACATCCTAAGAGCACCGC; IME4 antisense primer, CTCCAAGCAGTCTACCCAGCAG. Reverse transcription reactions were performed as follows: ∼2 µg of total RNA was incubated with 100 ng of random hexamers and 1 µl of 10 mM dNTPs at 65°C for 5 min, then quick-chilled on ice for 1 min. RT–PCR was performed using the SUPERSCRIPT First-Strand Synthesis System for RT–PCR (GibcoBRL) according to the manufacturer’s instructions. Five microliters of a 1:50 dilution of the RT–PCR reaction was used per 20 µl reaction using the Cybr Green protocol (Roche Molecular Biochemicals). PCR conditions were 95°C for 10 s, 62°C for 10 s, 72°C for 18 s for 30 cycles. Lightcycler data analysis was performed using the manufacturer’s software package.
HPLC analysis
Labeled yeast RNA was digested with 10 µg of ribonuclease P1 (Calbiochem) and 0.125 U nucleotide pyrophosphatase (Sigma) in 5 mm sodium acetate pH 6.0, 1 mM MgCl2, in a final volume of 50 µl for 4 h at 37°C. The nucleotides were then treated with 11.4 U alkaline phosphatase in 6 mM ammonium acetate, in a final volume of 60 µl overnight at 37°C. The reaction was dried under vacuum and resuspended in 20 µl of dH2O. The sample was injected onto a Supelcosil LC-18-S column 25 cm × 2.1 mm column, and was eluted isocratically with 7.5% methanol/30 mM sodium phosphate, pH 5.3 at a flow rate of 0.5 ml/min. A260 was monitored, and 0.25 ml fractions were collected and analyzed for 3H c.p.m. using a Beckman liquid scintillation counter.
Mutagenesis
Mutations were constructed at amino acid positions 348 and 351 in Ime4p using primer pairs W351A-F/W351A-R (5′ GGT ATT GCA GAT CCT GCA GCG AAT ATC CAT ATG AAC CTA; 5′ GTA GGT TCA TAT GGA TAT TCG CTG CAG GAT CTG CAA TAA C) and D348A-F/D348A-R (5′ CGG TAG TTA TTG CAG CTC CTG CAT GGA ATA TCC; 5′ GGA TAT TCC ATG CAG GAG CTG CAA TAA CTA CCG). Primers were annealed to the pJS18 template (19) and extended with Pfu polymerase by the method of Weiner et al. (21). Following DpnI digestion of the parental strands, plasmids were recovered in Escherichia coli and sequenced. Segments containing the desired mutations were then swapped for their wild-type counterparts in IME4-CEN (20) to form yeast plasmids W351A-CEN and D348A-CEN. The vector in all cases was pRS316 (22). HA-tagged GAL1,10-driven alleles were obtained by cloning the full-length IME4 coding regions from wild-type and mutant plasmids into pYEF1U (23). PCR products using IME4-NotI-F and IME4-RI-R (5′-GGG GCG GCC GCG ATT AAC GAT AAA CTA GTA CAT and 5′-GGG GAA TTC TTA CTG AGC AAA ATA TAG GTT ATT TAG) were cloned into the NotI–EcoRI-cut vector in frame with the N-terminal HA tag encoded by the vector. All constructs were fully sequenced.
Sporulation assays
Transformation of yeast strains MC309 (MATa/MATα ime4:LEU2/ ime4:LEU2 leu2/leu2 trp1/trp1 ura3/ura3 his3/his3 ho:HIS3/ho:HIS3 spr3:lacZ/spr3:lacZ) (19) and BJ5465 (24) was accomplished using the rapid transformation method of Gietz et al. (25), selecting on SD medium (supplemented as required by the strain) for the URA3 gene present on the vectors. For sporulation experiments, cells were grown with vigorous aeration in supplemented PSP2 (26) without phthalate buffer, collected by centrifugation, washed, and suspended in supplemented SPM (1% potassium acetate) to a final cell concentration of 3–5 × 107 cells/ml. Nuclear divisions and ascospore formation were monitored in the sporulating cultures by 4′,6-diamidino-2,3-phenylindole staining (DAPI; Sigma) (27) and bright field microscopy, respectively. Samples (0.3 ml) for DAPI staining were mixed with 0.6 ml of 95% ethanol for fixation and storage. For visualization of the meiotic nuclei, the cells were pelleted and resuspended in 0.1 µg/ml DAPI in 10% glycerol, 50 mM sodium phosphate buffer pH 7.5 and observed immediately using a Nikon or Olympus microscope equipped for epifluorescence.
RESULTS
Conservation of a family of MT-A70-like methyltransferases in diverse species
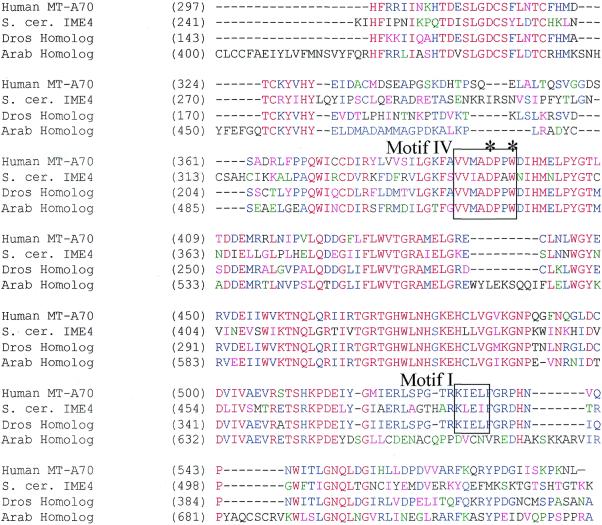
Database analysis and protein sequence alignments have revealed that there is a striking homology between mammalian MT-A70, S.cerevisiae IME4, a putative Drosophila gene (CG5933 gene product) and a putative Arabidopsis gene (g15236910, putative methyltransferase). It was not surprising to identify plant and fly homologs of MT-A70, because earlier work had shown that plants contain m6A in their mRNA in the same sequence context as in mammals (28). Likewise, Drosophila mRNA contains m6A, although the sequence context has not been investigated (8). In contrast, it was surprising to find that the most homologous gene to MT-A70 was IME4, because previous experiments had shown that S.cerevisiae mRNA did not contain m6A. Alignment of the mammalian gene and the homologous yeast, fly and plant genes is shown in Figure 1.
Figure 1.
Conserved protein sequences of putative mRNA N6-adenosine methyltransferases. Amino acid sequences of human MT-A70, S.cerevisiae IME4 and closely related proteins from Arabidopsis and Drosophila that were identified by database searches were aligned using AlignX (a component of VectorNTI Suite 6.0 software). The subsequences shown roughly correspond to the C-terminal halves of the respective proteins, as these regions were most highly conserved. Regions of similarity were also present in alignments of the N-terminal portions, but are not shown. Methyltransferase consensus motifs are shown in boxes. The positions that correspond to Ime4p residues D348 and W351, which are predicted catalytic residues, are identified by asterisks. Red letters represent consensus residues derived from a completely conserved amino acid at a given position. Residues that are identical in two or three of the sequences are shown in blue. Residues that are highly similar to a consensus at a given position are shown in pink. Weak similarities are depicted in green. Overall sequence identity is ∼40% across all four proteins (excluding gaps). Sequence similarity is almost 95% across this region (when calculated for at least three of the four proteins at each position). GenBank accession numbers for the sequences are: AF014837 (human MT-A70), D23721 (S.cerevisiae SPO8/IME4), g15236910 (Arabidopsis thaliana homolog), AE003746 (translation of subsequence beginning at position 1654, also CG5933 gene product, Drosophila melanogaster homolog).
These proteins contain examples of some of the consensus methyltransferase motifs that have been derived from mutational and structural studies of bacterial DNA methyltransferases, including the universally conserved motif IV catalytic residues and a proposed motif I (AdoMet binding) element, also indicated (29,30). These conserved motifs are numbered based on the cytosine-directed DNA methyltransferases, which share significant structural similarity with the adenosine, protein and small-molecule-directed enzymes. In light of the high degree of homology of MT-A70 with these putative methyltransferases from other species, we set out to determine whether Ime4p behaves as an mRNA N6-adenosine methyltransferase component.
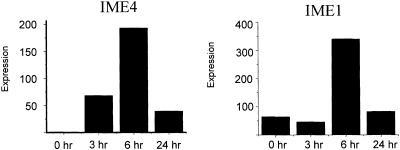
Time course of induction of IME4 and IME1 expression in response to starvation conditions
Yeast growing in log phase were transferred to sporulation medium and RNA was prepared at 0, 3, 6 and 24 h. IME4 and IME1 mRNA levels were measured using real-time RT–PCR. As seen in Figure 2, mRNA transcripts for both genes increased to significant levels by 6 h and had decreased substantially to near baseline levels by 24 h after transfer to sporulation medium. These results are consistent with those of Shah and Clancy (19) and with the sporulation microarray time course of Chu et al. (20) Subsequent experiments in which m6A levels in RNA were measured were all performed at the 6 h time point, where IME4 expression was maximal.
Figure 2.
Induction of IME4 and IME1 mRNA levels. Yeast were grown vegetatively as described, and then switched to sporulation medium. Cells were harvested and RNA was prepared at the indicated times. Levels of IME4 and IME1 mRNA were measured by real-time RT–PCR using a LightCycler. Relative levels of expression are indicated.
HPLC analysis of modified nucleosides
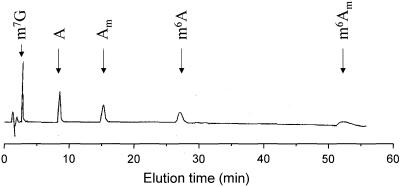
Modified nucleoside standards were used to optimize a HPLC protocol using a Supelcosil LC-18-S column for the resolution of m6A from other commonly modified nucleosides present in yeast total RNA. The protocol that provided both good resolution and ease of use was an isocratic elution using 7.5% methanol/30 mM sodium phosphate, pH 5.3. The absorbance profile from a typical chromatogram of adenosine analog standards is shown in Figure 3. m6A elutes at ∼27 min, and is well resolved from other methylated adenosine nucleosides. In addition to the standards shown in this figure, other modified nucleoside standards that were evaluated include: pseudouridine, 3-methylcytidine, 5-methylcytidine, 1-methyladenosine, unmodified guanosine, uridine and cytidine, and 2′-O-methylated guanosine, uridine and cytidine. All of these nucleosides eluted well before m6A.
Figure 3.
Absorbance profile of an HPLC separation of adenosine and methylated adenosine nucleosides. N7-Methylguanosine and methylated adenosine analogs were separated by isocratic elution from a Supelcosil LC-18-S column (250 × 2.1 mm). The mobile phase was 7.5% methanol/30 mM sodium phosphate. Additional standards were also run, all of which were well-resolved from m6A; their elution times are indicated on subsequent figures. m7G, N7-methyl guanosine; A, adenosine; Am, 2′-O-methyladenosine; m6A, N6-methyladenosine; m6Am, (N6-,2′-O-)-dimethyladenosine.
Analysis of methylated nucleosides in total RNA prepared from control or sporulating yeast cultures
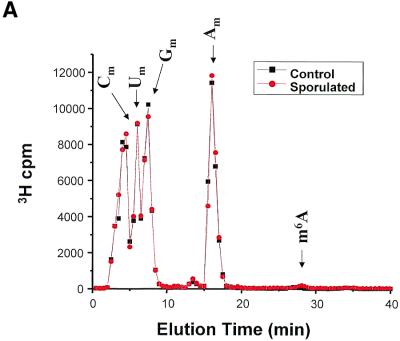
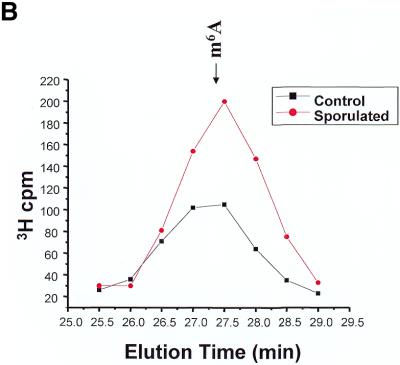
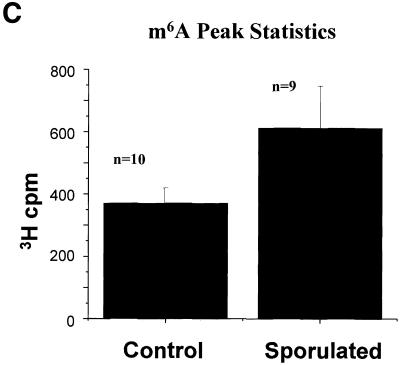
Yeast cells growing in methionine-free SD medium or in sporulation medium for 5 h were labeled using l-[methyl-3H]methionine for 1 h. RNA was isolated using standard techniques: 3H incorporation was measured, and the RNA was then treated with ribonuclease P1, nucleotide pyrophospha tase and alkaline phosphatase. This treatment would be expected to yield primarily mononucleosides. Any resistant polynucleotides would not interfere with the detection of m6A, as these charged moieties should elute very quickly from the Supelcosil LC-18-S column. Figure 4 summarizes the chromatographic data from 10 control experiments and nine sporulation experiments. In each experiment, non-sporulating and sporulating RNA samples were chromatographed sequentially. In all cases, roughly equivalent amounts of radioactivity were loaded. In order to determine whether a real difference in m6A levels was observed, the mean data for all experiments were calculated and are plotted in Figure 4A. No difference was seen in the amount of radioactivity in the peaks corresponding to the predominant methylated nucleosides (Cm, Um, Gm and Am). Most of the less common methylated nucleosides would be expected to elute prior to 10 min (based on our standardization runs, data not shown), and would be indistinguishable from the Cm, Um and Gm peaks. The m6A peak, as expected, represents a tiny fraction of the total 3H c.p.m., but is easily distinguished from the predominant methylated nucleosides due to its relatively long retention time. There was in fact an increase in the amount of m6A present in the RNA prepared from the sporulating yeast cells as compared with the non-sporulating cells (Fig. 4B and C). Although the absolute amount of the modified nucleoside is small compared with the others, the relative difference in the amount of m6A is significant—1.6-fold.
Figure 4.
Methylated nucleoside content of total RNA isolated from sporulating and non-sporulating yeast. (A) Yeast grown in methionine-deficient medium were labeled with [3H-methyl-]methionine. Total RNA was prepared, digested with ribonuclease P1 and nucleoside pyrophosphatase, and dephosphorylated with alkaline phosphatase. The sample was then chromatographed as described in the text. The positions of the predominant 2′-O-methylribonucleosides (Nm) are shown, as is the position of the much smaller m6A peak. The data shown represent the mean of nine separate experiments (control) and 10 experiments (sporulated). Each sample consisted of ∼10 µg of total RNA, containing ∼100 000 c.p.m. of incorporated 3H. All data were standardized to account for differences in the total counts loaded onto the column. (B) The same data are replotted on an expanded scale to better illustrate the m6A peak. (C) The radioactivity in the m6A peak is represented graphically. Error bars represent the standard deviation from the mean for each group of samples.
m6A is present in one residue in each of the large and small subunits of rRNA (31). Because the vast majority of methylated nucleosides (primarily ribose-methylated) occur in rRNA, by loading an equal number of counts in each sample, the data are essentially normalized for newly synthesized rRNA. m6A is also present in a few tRNAs (32) and in the snRNA U6 (33,34). In all of these cases, the nucleoside modification appears to be stoichiometric; i.e. every mature rRNA subunit contains exactly one m6A residue and each potential m6A site in the snRNA and tRNA is in fact methylated. This is in contrast to m6A sites in mammalian mRNA, where potential m6A sites are not methylated on every mRNA molecule (14). Furthermore (also in mam malian systems), the N6-adenosine methyltransferase(s) that methylate rRNA and snRNA are different from the mRNA N6-adenosine methyltransferase that is the homolog of IME4 (34,35). It is likely that the baseline level of m6A seen in the control cells is from ribosomal, transfer and small nuclear RNAs for two reasons: first, by analogy to this distribution of m6A in other organisms, and, secondly, based upon previous work that shows that mRNA prepared from yeast grown under normal (non-sporulating) conditions does not contain m6A (16). We hypothesize that the increment in m6A levels in sporulating cells is the result of N6-adenosine methylation in mRNA.
Analysis of methylated nucleosides in polyadenylated RNA prepared from sporulating yeast cultures
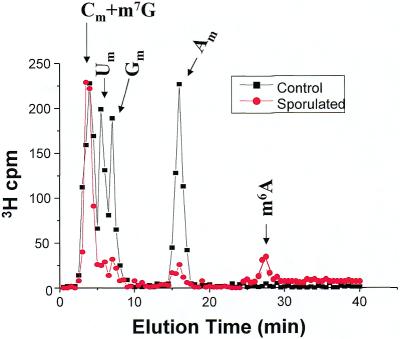
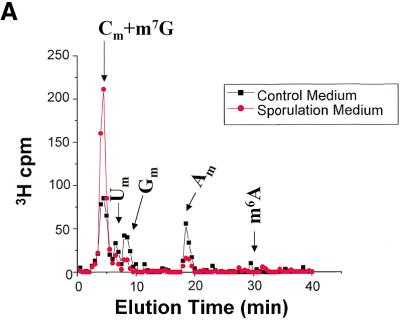
Yeast cells were again grown in sporulation medium, labeled with [3H-methyl]methionine, RNA was isolated, and polyadenylated RNA was prepared by oligo-dT selection. The yeast culture and total RNA preparation had to be scaled up 10-fold in order to recover enough labeled polyadenylated RNA for HPLC analysis. The polyadenylated RNA was treated with ribonuclease, pyrophosphatase and phosphatase exactly as was done for the total RNA, and nucleosides were analyzed by HPLC. The chromatographic profile of the [3H-methyl]-labeled nucleosides is shown in Figure 5. A peak of radioactivity, albeit in low amounts, is clearly present in the m6A fractions. No m6A peak was detected in polyadenylated RNA prepared from cells growing under non-sporulating conditions. The ratio of the m6A peak to the Am peak provides evidence that the m6A seen in mRNA prepared from sporulating yeast is not simply carry-over from contaminating rRNA. In total RNA, the m6A peak is approximately 0.01 times the height of the Am peak, whereas in the polyadenylated samples the m6A peak and the Am peak are roughly equivalent. These results support the hypothesis that induction of IME4, by the appropriate genetic and nutritional signals, leads to N6-adenosine methylation in some or all polyadenylated RNA.
Figure 5.
Methylated nucleoside content of polyadenylated RNA isolated from sporulating and non-sporulating yeast. Total RNA was prepared as before, but the preparations were scaled up 10-fold to yield ∼100 µg of 3H- labeled RNA. From this, ∼1 µg of polyadenylated RNA was isolated, containing ∼1000 c.p.m. 3H. The samples were treated with ribonuclease, pyrophosphatase, phosphatase, and then chromatographed. All data were standardized to account for differences in the total counts loaded onto the column. The curves represent the mean values from three independent experiments.
An additional, unanticipated finding is also evident in this data. The amounts of the 2′-O-methylyated nucleosides, Um, Gm and Am, are substantially and consistently reduced in the polyadenylated RNA fraction from sporulated cells as compared with the control cells. It is likely that the amount of Cm is also reduced, although the Cm peak is not distinguishable from the m7G peak. This result suggests that in addition to the appearance of m6A in the mRNA of yeast during sporulation, there is a significant decrease in the 2′-O-methylation of other nucleosides. However, Sripati et al. (16) reported that there are no other methylated nucleosides in yeast mRNA, so the significance of this finding is not known. One possible explanation is that yeast mRNA cap nucloesides are in fact methylated at the 2′-O position, and this methylation is reduced under sporulating conditions. An alternative explanation is that these peaks represent contamination with rRNA. We feel that this is not likely due to the remarkable consistency in the relative peak heights across experiments.
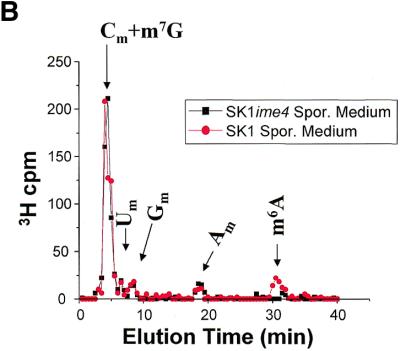
To assess whether the appearance of m6A depended on IME4, a similar experiment was done using SK1 cells containing an ime4 null mutation. SK1-ime4 cells were labeled with l-[methyl-3H]methionine exactly as in the experiments above, in methionine-free SD medium and in sporulation medium. mRNA was isolated and prepared as before, and analyzed for m6A content by HPLC. As shown in Figure 6A, no m6A peak was detected in the mRNA from the SK1-ime4 cells either in control medium or in sporulation medium. Interestingly, the changes in the peaks corresponding to the 2′-O-methyl nucleosides were observed just as they were for the wild-type SK1 cells (Fig. 6B). This experiment indicates that IME4 is necessary for N6-adenosine methylation in the mRNA of sporulating yeast, and also indicates that the putative changes in cap methylation are independent of Ime4p activity, and of the loss of m6A.
Figure 6.
Methylated nucleoside content of polyadenylated RNA isolated from SK1-ime4 (null mutant). (A) Comparison of methylated nucleosides from SK1-ime4 polyadenylated RNA prepared from the null-mutant yeast labeled in control medium and sporulation medium. (B) Comparison of methylated nucleosides in polyadenylated RNA prepared from from SK1-ime4 cells and wild-type SK1 cells, both labeled in sporulation medium. In this experiment, the m6A had a slightly longer retention time than in the other experiments due to regeneration of the HPLC column in between the experiments. The m6A retention time was verified with unlabeled standards.
Mutagenesis of motif IV
While previous work demonstrated that the IME4 gene is essential for sporulation in standard laboratory strains (19), it was possible that the proposed methyltransferase activity of the protein was not important for this process. To address this issue, we created mutations in the highly conserved motif IV sequence, found at amino acids 348–351 in Ime4p. Examples of the consensus sequence (N/D/S, P/I, P, Y/F/W) are found in all N6 adenosine-directed methyltransferases. These residues form part of the active sites of these enzymes (29,30,36–42) and mutations strongly reduce or eliminate catalytic activity (43–46). We expected, therefore, that the analogous mutations would reduce or eliminate any methyltransferase activity of Ime4p. Thus, the conserved acidic and aromatic amino acids at positions 348 and 351 (aspartate and tryptophan, respectively) were changed to alanine to form the D348A and W351A alleles. The mutant alleles were transferred to the CEN-based plasmid vector pRS316, forming W351A-CEN and D348A-CEN.
The activity of the mutant alleles in promoting sporulation was assessed. We chose a standard laboratory strain background (S288c) for these experiments, because unlike the rapidly sporulating SK1 cells used above, these strains are completely dependent on IME4 for sporulation. SK1-ime4 cells, in contrast, are impaired in sporulation but can complete the process to some extent (47), a property that could obscure any effects of the W351A and D348A mutations. Thus, plasmids containing mutant and wild-type alleles were transferred into MC309, an ime4-deficient yeast strain, and the ability of the plasmids to complement the sporulation deficiency of this strain was assessed. The production of full-length stable proteins was verified by western blotting (data not shown) following cloning the mutant and wild-type alleles in frame with the HA tag sequence in vector pYeF1-U (22).
Table 1 shows the results of an experiment in which the kinetics of spore formation were compared in MC309-ime4 null mutant cells carrying plasmid-borne mutant D348A or W351A alleles versus wild-type and vector-only controls. Cells carrying the wild-type allele began to form visible spores between 8 and 12 h after shift to sporulation as assessed by bright-field microscopy. By 24 h, ∼50% of the cells had sporulated, with final levels achieved by 48 h. The mutant alleles, in contrast, were severely compromised in their ability to promote sporulation of the ime4-deficient strain. Cultures of cells containing the D348A mutation achieved only 5% sporulation, ∼10-fold less than for the wild-type. The W351A allele was even more impaired, with <1% of the cells forming microscopically-visible asci even after prolonged incubation in sporulation medium (144 h).
Table 1. Sporulation kinetics of strains containing motif IV mutations.
| Time after shift to sporulation medium (h) | |||||||
|---|---|---|---|---|---|---|---|
| |
3 |
8 |
12 |
29 |
48 |
72 |
144 |
| Gene on plasmida | |||||||
| Wild-type | 0.0 | 0.0 | 13.5 | 50.5 | 58.2 | 58.2 | 61.5 |
| None | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| D348A | 0.0 | 0.0 | 0.0 | 4.3 | 5.1 | 5.1 | 4.9 |
| W351A | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.1 | 0.0 |
Transformants of the ime4-deficient MC309 strain were grown in pre-sporulation medium and shifted to sporulation medium. The percentage of the cells that had formed visible spores was determined at intervals by bright-field microscopy. At least 600 cells were counted for each determination shown. For the W351A and D348A mutants, at least 900 cells were counted.
aWild-type and mutant genes were carried on the CEN-URA3 vector, pRS316.
We also used DAPI staining and epifluorescence microscopy to examine the timing and extent of the meiotic divisions in these cells (Table 2). As expected from the kinetics of spore formation, binucleate and tetranucleate cells began to appear between 5 and 8 h after the shift in the culture carrying the wild-type IME4 allele, and cells that had completed both meiotic divisions comprised more than 50% of the population by 24 h. In contrast, cultures carrying the mutant alleles produced only a small fraction of tetranucleate cells, even after 48 h (∼7.5 and 0.5% for the D348A and W351A alleles, respectively.) Thus, the low fraction of asci appearing in these cultures reflects the failure of the cells to undertake the meiotic divisions. We conclude that the motif IV element is very important for IME4 function in promoting sporulation in yeast, a result that suggests a direct role for m6A in this process.
Table 2. Meiotic divisions in strains containing motif IV mutations.
| Time after shift to sporulation medium (h) | ||||||||
|---|---|---|---|---|---|---|---|---|
| 2 | 5 | 8 | 11 | 13.5 | 24 | 30 | 48 | |
| Gene on plasmida | ||||||||
| None | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Wild-type | 0.0 | 0.0 | 23.0 | 38.3 | 38.4 | 54.6 | 54.6 | 54.8 |
| D348A | 0.0 | 0.0 | 0.4 | 2.8 | 2.8 | 6.0 | 6.4 | 7.5 |
| W351A | 0.0 | 0.0 | 0.3 | 0.6 | 0.8 | 1.0 | 0.3 | 0.3 |
Transformants of the ime4-deficient MC309 strain were grown pre-sporulation medium and shifted to sporulation medium. Each culture was sampled at the hourly intervals shown for DAPI staining and epifluorescence microscopy. The percentage of cells containing two or more nuclear masses was determined by counting at least 600 cells for each time point shown. For the D348A and W351A strains, at least 800 cells were counted.
aWild-type and mutant genes were carried on the CEN-URA3 vector, pRS316.
DISCUSSION
This work brings together two disparate research problems: control of yeast differentiation and nucleoside modification. Saccharomyces cerevisiae sporulation (meiosis) is controlled by a complicated cascade of genes that are highly regulated by genetic and environmental signals. IME4 is expressed throughout sporulation, and its activity is important at early times for accumulation of the IME1 and IME2 mRNAs, and later for unknown aspects of spore formation (47, M.Clancy, unpublished results). The mechanism whereby IME4 influences activation of sporulation has been previously unknown, but, unlike many of the regulatory genes in the cascade, it does not appear to function as a transcription factor (B.Jursic, Y.Wang and M.Clancy, unpublished results).
The work presented here strongly suggests that IME4 encodes an mRNA N6-adenosine methyltransferase and that the formation of m6A serves an important function in promoting sporulation in yeast. These experiments were motivated by the high degree of sequence similarity between MT-A70 and IME4, including regions encompassing consensus methyltransferase motifs that were identified by comparison with prokaryotic N6-adenine-specific DNA methyltransferases enzymes (36–42). These similarities led to the hypothesis that Ime4p may function like MT-A70, as an S-adenosylmethionine-binding component of an RNA- directed methyltransferase.
In accord with this idea, preparations of total RNA and polyadenylated RNA isolated from sporulating cells contained higher levels of m6A than did those from vegetative cells. As expected, m6A was present in total RNA prepared from yeast cells growing in log phase. Upon switching the MATa/MATα diploid yeast to sporulation medium, IME4 expression was induced, and a concomitant 1.6-fold rise in the amount of m6A in total RNA was observed. Consistent with the hypothesis that Ime4p is an mRNA m6A-methyltransferase subunit, m6A was now present in the polyadenylated RNA fraction, where it had been undetectable in polyadenylated RNA prepared from vegetative cells. These experiments support the hypothesis that Ime4p is an essential component of the methyltransferase responsible for this increase. Furthermore, SK1 cells that lack a functional IME4 gene do not have detectable levels of m6A in their mRNA when incubated in sporulation medium. The high degree of homology of IME4 to MT-A70, the temporal correlation of IME4 induction and the appearance of m6A in mRNA, and the results of the IME4 mutation experiments strongly support the hypothesis that Ime4p is directly responsible for yeast mRNA methylation. These experiments do not rule out the possibility that other RNA species may also be hypermethylated when IME4 is induced. Indeed, m6A is present in both the large and small rRNA subunits and in tRNA in all eukaryotes and many prokaryotes, and is present in a few snRNA in higher eukaryotes (31,33). The methyltransferases that are responsible for N6-adenosine methylation in these RNA species have not been isolated, but several lines of evidence suggest that IME4 is not involved. First, rRNA, tRNA and snRNA methylation is constitutive, but IME4 is expressed at high levels only in sporulating and stationary phase cells. Additionally, we have shown that in vitro, the human IME4 homolog, mRNA m6A-MT, is not able to methylate human rRNA- or U6 snRNA-like substrates (34, J.Bokar and M.E.Shambaugh, unpublished results).
The putative active-site mutations in methyltransferase motif IV of IME4 led to a severe reduction in sporulation, strongly supporting the hypothesis that the methyltransferase activity of this protein and the formation of m6A are important for sporulation in yeast. We observe, however, that the motif IV mutations permit some level of sporulation in these cells, albeit at markedly reduced levels, whereas the null mutant strain does not sporulate at all. One possibility is that the individual D348A mutation and, to a lesser extent, the W351A mutation, still have residual catalytic activity, allowing a low level of m6A to be formed that is below the limit of detection of m6A in our HPLC assay. Another explanation is that IME4 has additional functions distinct from the methyltransferase catalytic activity. This would be reminiscent of a similar study involving the dimethyladenosine residue near the 3′ end of the 18S rRNA. The Dim1 protein that is responsible for this modification is essential for viability, while the modification itself is not essential; rather, the inviability of dim1 cells is the result of failure in ribosomal precursor processing (48).
How might mRNA methylation in yeast, catalyzed by Ime4p, lead to induction of the sporulation pathway? Recent microarray studies show that more than 500 mRNAs are present at elevated levels in sporulating cells, including some encoding proteins involved in general aspects of mRNA biogenesis, as well as many that promote sporulation-specific processes (20). We suggest that IME4 activity governs part of this global response to nutritional and cell-type signals by facilitating a general process such as splicing, 3′-end formation, nuclear pre-mRNA turnover, export or localization of mature mRNAs, or translation efficiency.
The function of m6A in mRNA in higher eukaryotes has remained elusive, despite numerous lines of investigation. Several groups have used two general approaches in an effort to define the role of m6A in mRNA biogenesis and function in mammalian cells and their viruses. Mutations of the major methylation sites within the Rous sarcoma virus src and env genes have been tested (10,11). Mutations of several of these sites, which rendered them incapable of being methylated, did not lead to any differences in steady-state levels of src mRNA in mutant versus wild-type virus in infected CEF cells. No differences in the levels of src mRNA in the nuclear fraction were detected either, and no unusual spliced products were observed. Furthermore, there was no difference in the translation of the src protein, packaging of RNA into virions or infectivity of mutant virus compared with wild-type. Several explanations for the lack of an effect of these methylation mutations have been proposed. If methylation affects the kinetics of processing or transport, it may be necessary to study only newly synthesized mRNA to observe the effect. Alternatively, multiple methylation sites may be redundant for function. A second series of experiments explored m6A function by using methylation inhibitors. Cycloleucine treatment of CHO cells resulted in a 96% decrease in 3H-methyl incorporation into polyadenylated RNA, and the decay of this hypomethylated polyadenylated RNA from the nucleus was prolonged in pulse-chase experiments (12). Cycloleucine treatment of cells infected with B77 avian sarcoma virus resulted in a shift in the ratio of subgenomic env RNA to genome-length RNA (49). The methylation inhibitor, S-tubercidinylhomocysteine, caused a significant delay in the cytoplasmic appearance of newly synthesized polyadenylated RNA in HeLa cells (13). Another inhibitor, neplanocin, led to nuclear accumulation of an unspliced pre-mRNA in CHO cells stably transfected with a bovine prolactin minigene (14). Taken together, these experiments support a model in which methylation of specific internal adenosine residues in mRNA somehow affects the efficiency of processing and/or transport of nuclear pre-mRNA to mature cytoplasmic mRNA. The major limitation of these experiments is that they all used methylation inhibitors that function by altering the equilibrium of the S-adenosylhomocysteine/S-adenosylmethionine pool, and therefore are likely to have pleiotropic effects on a wide range of methyltransferases. More recently, Tuck et al. (15) have shown that m6A-deficient mRNA encoding dihydrofolate reductase is translated less efficiently than m6A-containing mRNA. In these experiments, the mRNA was made m6A-deficient by treatment of cells with the non-selective methylation inhibitor, cycloleucine. The translation efficiency was determined in vitro using a rabbit reticulocyte lysate assay. This observation has not been confirmed in an in vivo system.
The most well characterized changes in mRNA biogenesis during sporulation involve splicing (50–53). Sporulation induces the expression of the novel splicing factor encoded by the MER1 gene, as well as the expression of a relatively large number of intron-containing pre-mRNAs, many containing non-canonical splice junctions (54). It is possible that methylation by Ime4p facilitates recognition of particular pre-mRNAs by components of the splicing or export machinery, thereby contributing to the regulation of expression of these mRNAs during sporulation. It is also possible that methylation of the pre-mRNA facilitates 3′-end maturation. Indeed, the best-studied methylation substrate in mammalian systems is the single site in the RNA encoded by a bovine prolactin minigene, which occurs 1 nt upstream of the polyA site. In yeast, 3′-end formation can respond to nutritional conditions (53), and some genes encoding components of the polyadenylation cleavage and specificity factor are induced during sporulation, including NAB4/HRP1 and CLP1 (20).
It is tempting to speculate that Ime4p methylates specific internal adenosine residues in a subset of mRNAs produced in sporulating cells, possibly including the IME1, IME2 and NDT80 mRNAs among many others. Methylation of these transcripts may alter their expression by modifying their splicing, stability, translation efficiency or compartmentalization, ultimately leading to high level accumulation of RNAs corresponding to IME1 and other sporulation-specific genes. These exciting hypotheses are directly testable, and will need to be addressed in future studies. However, the discovery that m6A is induced in the mRNA of sporulating yeast, along with the finding that mutations within the putative catalytic methyltransferase motifs in IME4 severely attenuate sporulation, provide the strongest evidence yet that this nucleoside modification has a critical role in a gene regulatory pathway.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Sandra Ferguson (Roche Molecular Biochemicals) for providing the RNA Amplification Kit SYBR Green I and Control Kit RNA. We also thank Dr Daniel Gelperin for his assistance with sporulation of Hansen SK1-can1 cells and Drs Aaron Mitchell and Elizabeth Jones for strains. This work was supported by National Science Foundation grant MCB-9983555 (to M.J.C.) and National Cancer Institute grant CA77472 (to J.A.B.).
REFERENCES
- 1.Rottman F.M., Shatkin,A.J. and Perry,R.P. (1974) Sequences containing methylated nucleotides at the 5′-termini of messenger RNAs: possible implications for processing. Cell, 3, 197–199. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A.K. (1980) 5′-Terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev., 44, 175–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy R., Singh,R. and Shimba,S. (1992) Methylated CAP structures in eukaryotic RNAs: structure, synthesis and function. Pharmacol. Ther., 54, 249–267. [DOI] [PubMed] [Google Scholar]
- 4.Desrosiers R.C., Friderici,K.H. and Rottman,F.M. (1975) Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5′ terminus. Biochemistry, 14, 4367–4374. [DOI] [PubMed] [Google Scholar]
- 5.Perry R.P., Kelley,D.E., Friderici,K. and Rottman,F. (1975) The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell, 4, 387–394. [DOI] [PubMed] [Google Scholar]
- 6.Wei C.M., Gershowitz,A. and Moss,B. (1976) 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry, 15, 397–401. [DOI] [PubMed] [Google Scholar]
- 7.Nichols J.L. (1979) ‘Cap’ structures in maize poly(A)-containing RNA. Biochim. Biophys. Acta, 563, 490–495. [DOI] [PubMed] [Google Scholar]
- 8.Levis R. and Penman,S. (1978) 5′-Terminal structures of poly (A)+ cytoplasmic messenger RNA and of poly (A)+ and poly (A)– heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster. J. Mol. Biol., 120, 487–515. [DOI] [PubMed] [Google Scholar]
- 9.Bokar J.A. and Rottman,F.M. (1998) Biosynthesis and functions of modified nucleosides in eukaryotic mRNA. In Grosjean,H. and Benne,R. (eds), Modifications and Editing of RNA. ASM Press, Washington, DC, pp. 183–200.
- 10.Kane S.E. and Beemon,K. (1987) Inhibition of methylation at two internal N6 methyladenosine sites caused by GAC to GAU mutations. J. Biol. Chem., 262, 3422–3427. [PubMed] [Google Scholar]
- 11.Csepany T., Lin,A., Baldick,C.J. and Beemon,K. (1990) Sequence specificity of mRNA N6-adenosine methyltransferse. J. Biol. Chem., 265, 20117–20122. [PubMed] [Google Scholar]
- 12.Bachellerie J.P., Amalric,F. and Caboche,M. (1978) Biosynthesis and utilization of extensively undermethylated poly (A)+ RNA in CHO cells during a cycloleucine treatment. Nucleic Acids Res., 5, 2927–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camper S.A., Albers,R.J., Coward,J.K and Rottman,F.M. (1984) Effect of undermethylation on mRNA cytoplasmic appearance and half-life. Mol. Cell. Biol., 4, 538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll S.M., Narayan,P. and Rottman,F.M. (1990) N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol. Cell. Biol., 10, 4456–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuck M.T., Wiehl,P.E. and Pan,T. (1999) Inhibition of 6-methyladenine formation decreases the translation efficiency of dihydrofolate reductase transcripts. Int. J. Biochem. Cell Biol., 31, 837–851. [DOI] [PubMed] [Google Scholar]
- 16.Sripati C.E., Groner,Y. and Warner,J.R. (1976) Methylated, blocked 5′ termini of yeast mRNA. J. Biol. Chem., 251, 2898–2904. [PubMed] [Google Scholar]
- 17.Bokar J.A., Shambaugh,M.E., Polayes,D., Matera,A.G. and Rottman,F.M. (1997) Purification and cDNA cloning of the Adomet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA, 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 18.Gasch A.P., Spellman,P.T., Kao,C.M., Carmel-Harel,O., Eisen,M.B., Storz,G., Botstein,D. and Brown,P.O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell, 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah J.C. and Clancy,M.J. (1992) IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 21.Weiner M.P, Costa,G.L., Schoettlin,W., Cline,J., Mathur,E. and Bauer,J.C. (1994) Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene, 15, 119–125. [DOI] [PubMed] [Google Scholar]
- 22.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cullen C. and Minvielle-Sebastia,L. (1994) Multipurpose vectors designed for the fast generation of N- or C-terminal epitope-tagged proteins. Yeast, 10, 105–112. [DOI] [PubMed] [Google Scholar]
- 24.Jones E.W. (1991) Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol., 194, 428–453. [DOI] [PubMed] [Google Scholar]
- 25.Gietz R.D., Schiestl,R.H., Willems,A.R. and Wood,R.A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast, 1, 355–360. [DOI] [PubMed] [Google Scholar]
- 26.Kassir Y. and Simchen,G. (1991) Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol., 194, 94–110. [DOI] [PubMed] [Google Scholar]
- 27.Williamson D.H. and Fennel,D.J. (1975) The use of fluorescent DNA-binding agent for detecting and separating yeast mitochondrial DNA. Methods Cell Biol., 12, 335–351. [DOI] [PubMed] [Google Scholar]
- 28.Nichols J.L. and Welder,L. (1981) Nucleotides adjacent to N6 methyladenosine in maize zea-mays poly adenylated containing RNA. Plant Sci. Lett., 21, 75. [Google Scholar]
- 29.Klimasauskas S., Timinskas,A., Menkevicius,S., Butkiene,D., Butkus,V. and Janulaitis,A. (1989) Sequence motifs of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res., 17, 9823–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timinskas A., Butkus,V. and Janulaitus,A. (1995) Sequence motifs characteristic for DNA [cytosine-N4] and DNA [adenine-N6] methyltransferases. Classification of all DNA methyltransferases. Gene, 157, 3–11. [DOI] [PubMed] [Google Scholar]
- 31.Maden B.E. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- 32.Bjork G.R., Ericson,J.U., Gustafsson,C.E.D., Hagervall,T.G., Jonsson,Y.H. and Wikstrom,P.M. (1987) Transfer RNA Modification. Annu. Rev. Biochem., 56, 263–287. [DOI] [PubMed] [Google Scholar]
- 33.Reddy R. and Busch,H. (1988) SnRNP proteins. In Birnstiel,M.L. (ed.), Small Nuclear RNAs: RNA Sequences, Structure and Modifications. Springer-Verlag, Berlin, pp. 1–37.
- 34.Shimba S., Bokar,J.A., Rottman,F. and Reddy,R. (1995) Accurate and efficient N6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res., 13, 2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bokar J.A., Rath-Shambaugh,M.E., Ludwiczak,R.L., Narayan,P. and Rottman,F.M. (1994) Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. J. Biol. Chem., 269, 17697–17704. [PubMed] [Google Scholar]
- 36.Roth M., Helm-Kruse,S., Friedrich,T. and Jeltsch,A. (1998) Functional roles of conserved amino acid residues in DNA methyltransferases investigated by site-directed mutagenesis of the EcoRV adenine-N6-methyltransferase. J. Biol. Chem., 273, 17333–17342. [DOI] [PubMed] [Google Scholar]
- 37.Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- 38.Narva K.E., Van Etten,J.L., Slatko,B.E. and Benner,J.S. (1988) The amino acid seuence of the eukaryotic DNA [N6-adenine]methyltransferase, M CviBIII, has regions of similarity with the prokaryotic isoschizomer M Taq I and other DNA [N6-adenine] methyltransferases. Gene, 74, 253–259. [DOI] [PubMed] [Google Scholar]
- 39.Labahn J., Granzin,J., Schluckebier,G., Robinson,D.P., Jack,W.E., Schildkraut,I. and Saenger,W. (1994) Three-dimensional structure of the adenine-specific DNA methyltransferase M-Taq I in complex with the cofactor S-adenosylmethionine. Proc. Natl Acad. Sci. USA, 91, 10957–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong W., O’Gara,M., Blumenthal,R.M. and Cheng,X. (1997) Structure of PvUII DNA-(cytosine N4)methyltransferase, an example of domain peremutation and protein assignment. Nucleic Acids Res., 25, 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schluckebier G., O’Gara,M., Saenger,W. and Cheng,X. (1997) Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol., 247, 16–20. [DOI] [PubMed] [Google Scholar]
- 42.Scavetta R.D., Thomas,C.B., Walsh,W.A., Szegedi,S., Joachimiak,A., Gumport,R.I. and Churchill,M. (2000) Structure of RsrI methyltransferse, a member of the N6-adenine beta class of DNA methyltransferases. Nucleic Acids Res., 28, 3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holz B., Dank,N., Eickhoff,J.E., Lipps,G., Krauss,G. and Weinhold,E. (1999) Identification of the binding site for the extrahelical target base in N6-adenine DNA methyltransferases by photo-cross-linking with duplex oligodeoxyribonucleotides containing 5-iodouracil at the target position. J. Biol. Chem., 274, 15066–15072. [DOI] [PubMed] [Google Scholar]
- 44.Willcock D.F., Dryden,D.T.F. and Murray,N.E. (1994) A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J., 13, 3902–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pues H., Bleimling,N., Holz,B., Wolcke,J. and Weinhold,E. (1999) Functional roles of the conserved aromatic amino acid residues at position 108 (motif IV) and position 196 (motif VIII) in base flipping and catalysis by the N6-adenine DNA methyltransferase from Thermus aquaticus. Biochemistry, 38, 1426–1434. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad I. and Rao,D.N. (1996) Functional analysis of conserved motifs in EcoP151 DNA methyltransferase. J. Mol. Biol., 259, 229–240. [DOI] [PubMed] [Google Scholar]
- 47.Su S. and Mitchell,A.P. (1993) Molecular characterization of the meiotic regulatory gene RIM1. Nucleic Acids Res., 21, 3789–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lafontaine D., Vandenhaute,J. and Tollervey,D. (1995) The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev., 9, 2470–2481. [DOI] [PubMed] [Google Scholar]
- 49.Stoltzfus C.M. and Dane,R.W. (1982) Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J. Virol., 42, 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leu J.Y. and Roeder,G.S. (1999) Splicing of the meiosis-specific HOP2 transcript utilizes a unique 5′ splice site. Mol. Cell. Biol., 19, 7933–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakagawa T. and Ogawa,H. (1999) The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J., 18, 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spignola M. and Ares,M. (2000) A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol. Cell, 2, 329–338. [DOI] [PubMed] [Google Scholar]
- 53.Engebrecht J., Voelkel-Meiman,K. and Roeder,G.S. (1991) Meiosis-specific RNA splicing in yeast. Cell, 66, 1257–1268. [DOI] [PubMed] [Google Scholar]
- 54.Davis C.A., Grate,L., Spingola,M. and Eres,M. (2000) Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res., 28, 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sparks K.A. and Dieckmann,C.L. (1998) Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res., 26, 4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]