Abstract
In the course of macronuclear differentiation in spirotrichous ciliates massive DNA reorganization processes take place, which include splicing, cutting, rearranging and eliminating specific DNA sequences. In order to identify genes involved in these processes we took advantage of suppression subtractive hybridization. We have identified three transcripts that are exclusively expressed during macronuclear development in the ciliate Stylonychia lemnae. Two of the three differentially expressed mRNAs we have analyzed encode for novel proteins. One gene, mdp1 [macronuclear development protein 1 (MDP1)], encodes a homolog of the PIWI protein family. PIWI proteins are involved in germline differentiation processes and RNA silencing in worms, flies, mice, humans and in plants. Possible functions of the S.lemnae PIWI related protein MDP1 in the regulation of macronuclear development will be discussed.
INTRODUCTION
Programmed DNA rearrangements and subsequent elimination of specific germline DNA play crucial roles in the development and evolution of many organisms. Well studied systems include V(D)J recombination, which leads to antibody diversity (1), Trypanosome surface antigen variation to escape the host’s immune system (2), switching of mating types in yeast (3), programmed chromosome breakage and chromatin elimination events in Ascaris megalocephala (4) and Cyclops (5), and the elimination of complete chromosomes in Scaria coprophila (6) and some hagfish species during early embryogenesis (7). The most dramatic example of programmed DNA reorganization and elimination occurs in spirotrichous ciliates, e.g. Stylonychia, Oxytricha, Sterkiella and Euplotes (8).
The stichotrichous ciliate Stylonychia lemnae, like all ciliates, contains two types of nuclei: a transcriptionally inert germline micronucleus used in cell mating and a transcriptionally active somatic macronucleus used for vegetative cell growth. The DNA in the micronucleus is organized in chromosomes, with genes found singly or in groups separated by long regions of AT-rich spacer DNA. Micronuclear genes are interrupted by multiple, short, non-coding, AT-rich sequences, called internal eliminated sequences (IESs) (9). IESs interrupt macronuclear destined segments (MDSs). In 20–30% of genes the MDSs are scrambled, such that they are in a permuted order relative to the functional macronuclear gene (9). In contrast, the macronuclear DNA occurs as short molecules, ranging from 400 bp to ∼40 kb and averaging about 2.5 kb in length. The number of different DNA molecules found in a macronucleus is estimated to be ∼20 000 (8). Almost all sequenced macronuclear molecules encode a single gene and each DNA molecule is present in one to several thousand copies.
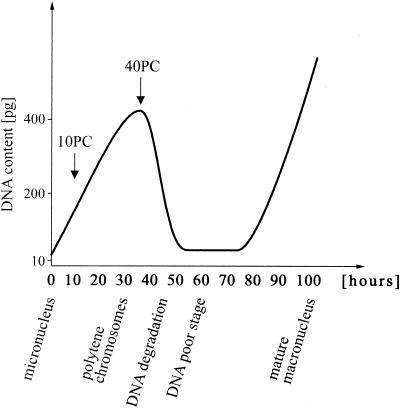
Macronuclear DNA derives from micronuclear DNA after sexual reproduction (conjugation), during a ∼100 h process called macronuclear development (10). In spirotrichous ciliates macronuclear development begins with several rounds of replication to yield polytene chromosomes with a maximum polytenization of 64C (8,10) (Fig. 1). During polytene chromosome formation IESs and transposon-like elements are eliminated and MDSs ligated to eventually form a functional gene (11). Destruction of the polytene chromosomes is coupled with elimination of up to 98% of the DNA, leading to the DNA poor stage (8). In the course of this DNA elimination event macronuclear DNA becomes specifically fragmented and telomeres are added de novo to each DNA molecule. Another four or five rounds of replication yield the mature macronucleus (8).
Figure 1.
Schematic drawing of differentiation processes taking place during macronuclear development in S.lemnae modified after Kraut et al. (38) and Ammermann et al. (14). At defined time points [arrows; 10 and 40 h post conjugation (PC)] RNA was isolated and further characterized.
Nothing is known about the trans-acting factors that are involved in restructuring the micronuclear genome, leaving many fundamental questions about the mechanism, origin and evolutionary conservation of these processes among ciliates and eukaryotes in general. Studies in the distantly related ciliates Paramecium and Tetrahymena indicate epigenetic regulation of some of the DNA rearrangement events occurring during macronuclear development possibly through RNA ‘cross-talk’ from the old degenerating macronucleus (12).
In order to identify proteins responsible for these processes in S.lemnae, we have employed a suppression subtractive hybridization procedure to enrich for transcripts differentially expressed during macronuclear development. Here we describe three mRNAs that are transcribed exclusively during macronuclear development. Two genes encode novel polypeptides with no significant identity to sequences in GenBank. The other transcript encodes a homolog of the PIWI protein. Members of the PIWI group are involved in germline development and the RNA silencing pathway in evolutionarily diverse eukaryotes (13), suggesting that the process of macronuclear development in S.lemnae requires functions related to those involved in germline development and differentiation in a wide variety of eukaryotes.
MATERIALS AND METHODS
Accession numbers
cDNA sequences have been deposited in GenBank with the accession nos AY128524 for mdp1, AY128525 for mdp2 and AY128526 for mdp3.
Growth of Stylonychia lemnae and isolation of macronuclear DNA
Stylonychia lemnae was cultivated and macronuclear DNA was isolated as described previously (14,15).
RNA isolation
Total RNA was isolated from vegetative cells and from cells at various stages of macronuclear development (Fig. 1). The cells were filtered through 120 µm gauze and concentrated on 30 µm gauze. For total RNA isolation 750 µl Trizol LS-Reagent (Invitrogen) was added to 250 µl of the cell suspension and the RNA was isolated according to the manufacturer’s protocol. RNA concentration was determined by UV spectroscopy. Total RNA (300–500 µg) was isolated from a 1000 ml culture. When needed, poly(A)+ RNA was isolated with the PolyATract mRNA Isolation System (Promega). Poly(A)+ RNA yields were typically 6–10 µg per mg of total cellular RNA.
cDNA subtraction
Suppression subtractive hybridization was used to identify differentially expressed transcripts. The PCR Select cDNA Subtraction Kit (Clontech) allows comparison of two populations of mRNAs by obtaining clones of genes that are present in one RNA pool but not in the other (16–18). The mRNA populations of interest were reverse transcribed and cDNA synthesis was performed. The cDNA pool containing the specific transcripts is referred to as the tester, and the reference cDNA pool as the driver. Two micrograms of Poly(A)+ RNA was used for cDNA synthesis. RsaI digestion, adaptor ligation, subtractive hybridizations and subsequent PCR amplifications were performed according to the manufacturer’s protocol (Clontech PCR-Select cDNA Subtraction Kit User Manual PT1117–1). The resulting cDNAs were cloned into a TA vector (see below) for further analysis.
Cloning and sequencing
PCRs were either carried out with Biotherm Taq polymerase (Genecraft) or with the Advantage cDNA PCR Kit (Clontech). PCR products were cloned into the TA cloning vector pGEM-TEasy (Promega). Plasmid DNA templates for sequencing were prepared with the Miniprep Kit (Qiagen). Dye terminator DNA dideoxy cycle sequencing was carried out by MWG-Biotech AG (Ebersberg, Germany).
RT–PCR analysis
In order to determine the expression pattern of individual transcripts, RT–PCR analyses with total RNA derived from different stages during macronuclear development and, as a control, from total RNA derived from vegetatively growing cells were performed. DNase I digested total RNA (2.5 µg) was reverse transcribed with the RACE adaptor primer in a 20 µl reaction (Superscript II RT, Invitrogen). Of this cDNA, 2.5 µl was used for subsequent PCR amplification with individual primer pairs. Primers amplifying a region of the actin I gene were used as a control.
Rapid amplification of cDNA ends (RACE)
To determine the ends of the transcripts RACE reactions were performed (19). Following reverse transcription (SuperScript II RT, Invitrogen) of total RNA with the RACE adaptor primer 5′-GACTCGAGTCGACATCGA(T)17-3′, a PCR with a gene specific primer and the RACE primer was performed (3′ RACE). The 5′ end was amplified by reverse transcription (SuperScript II RT, Invitrogen) of total RNA with a gene specific primer. The cDNA was purified with the QIAquick PCR Purification Kit (Qiagen), the 3′ end of the target cDNA was A-tailed (terminal dideoxynucleotidyl transferase; Amersham Biosciences), and a PCR with the RACE adaptor primer and a nested gene specific primer was performed. The resulting PCR products were then cloned and sequenced.
Virtual northerns
Total RNA (3 µg) was reverse transcribed and second strand cDNA synthesis was carried out with Escherichia coli DNA polymerase I, E.coli RNase H and E.coli DNA Ligase (SuperScript™ Choice System for cDNA Synthesis, Invitrogen). For agarose gel electrophoresis equal volumes from the same cDNA preparation were used for each of the four gels. The gels were blotted onto Hybond N+ nitrocellulose membranes (Amersham Biosciences) and hybridized with probes generated from the differentially expressed cDNA clones by the PCR DIG Probe Synthesis Kit (Roche Molecular Biochemicals). Primers used for probe generation were the Clontech primers, nested PCR primer 1 and nested PCR primer 2R (PCR-Select cDNA Subtraction Kit, see above). Prehybridizations and hybridizations were carried out with Church hybridization buffer (20) at 68°C. Probe concentration was set to 25–35 ng/ml hybridization buffer. After hybridization the membranes were washed according to standard procedures (20). Specific cDNAs were detected with NBT/BCIP using the Dig DNA Detection Kit (Roche Molecular Biochemicals).
Determination of open reading frames
Polarity of the transcripts was determined by 3′ RACE. The first AUG on the sense strand marks the origin of the only significant open reading frame (ORF) on all of the transcripts analyzed in this study. ORFs were translated taking into account that UAA and UGA encode glutamine in stichotrichous ciliates (21).
Protein sequence analysis
Protein search was carried out with SWISS-PROT/TrEMBL (22). PSI-BLAST similarity search was performed at NCBI (23). Multiple sequence alignments were carried out with the ClustalX application (software version 1.81). Determination of the theoretical isoelectric point (pI) and molecular weight (MW) were carried out with protein identification and analysis tools on the ExPASy server (24).
RESULTS
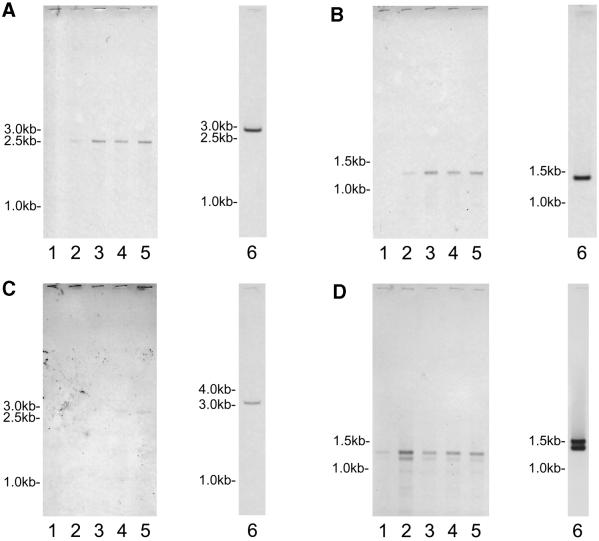
Suppression subtractive hybridization was performed using cDNA created from RNA isolated 10 and 40 h after initiation of macronuclear development as testers and vegetative cDNA as driver. Following the hybridization procedure, the resulting cDNAs were cloned and plated. Twelve colonies from both the 10 and 40 h suppression subtractive hybridization procedures were sequenced. Six of the S.lemnae sequences originated from known genes (i.e. several histones, cyclin B and PABP) that are not transcribed exclusively during macronuclear development as confirmed by RT–PCR. Presumably these genes are upregulated during macronuclear development when chromatin remodeling and DNA replication to yield polytene chromosomes occurs. These sequences were not further analyzed in this study. Ten clones were identical in sequence, and the other eight were unique. Primer pairs were designed from each sequence and PCR was performed on cDNA isolated from 0, 10, 20, 30, 40, 50 and 60 h after initiation of macronuclear development and on cDNA isolated from vegetative cells. Based on these PCR results, four of the nine sequences represented genes that are transcribed during macronuclear development but not during vegetative growth. 5′ and 3′ RACE was performed in order to clone and sequence the entire cDNA molecules. Sequence analysis revealed that two of the four transcripts are identical. Thus, the procedure yielded a total of three differentially expressed genes. These genes have been named macronuclear development protein (mdp)1, 2 and 3 (the corresponding proteins are referred to as MDP1, 2 and 3). mdp1 and mdp2 were isolated from the 10 h subtraction; mdp3 was isolated from the 40 h subtraction. While mdp1 and mdp2 are highly abundant transcripts showing hybridization intensities almost identical to the actin transcripts in stages after conjugation, mdp3 mRNA is expressed at least at a 10-fold lower rate as determined by virtual northern analysis (Fig. 2).
Figure 2.
Detection of stage-specific RNAs and the corresponding macronuclear genes. Total RNA of vegetative cells and different stages during macronuclear development was isolated, first and second strand cDNA synthesis was performed (see Materials and Methods). For agarose gel electrophoresis equal volumes from the same cDNA preparation were used for each of the four gels. Lane 1, cDNA derived from vegetative cells; lanes 2–5, cDNA derived from exconjugants, 0, 20, 30 or 40 h PC, respectively. The gels were blotted and hybridized with Dig labeled probes generated by PCR amplification of the differentially expressed clones mdp1 (A), mdp2 (B) and mdp3 (C). To check the integrity of the different cDNAs, actin I was used as a control (D). Macronuclear DNA was isolated and separated by agarose gel electrophoresis. The gels were blotted and hybridized with the corresponding Dig labeled probes (lane 6).
To determine the time course of mdp expression, cDNAs from different time points during macronuclear differentiation and from vegetative cells were synthesized and equal amounts of the same preperation were loaded onto four different agarose gels and blotted onto nylon membranes. To demonstrate the integrity of these cDNA preparations one of these membranes was hybridized with an S.lemnae actin gene probe.
MDP1
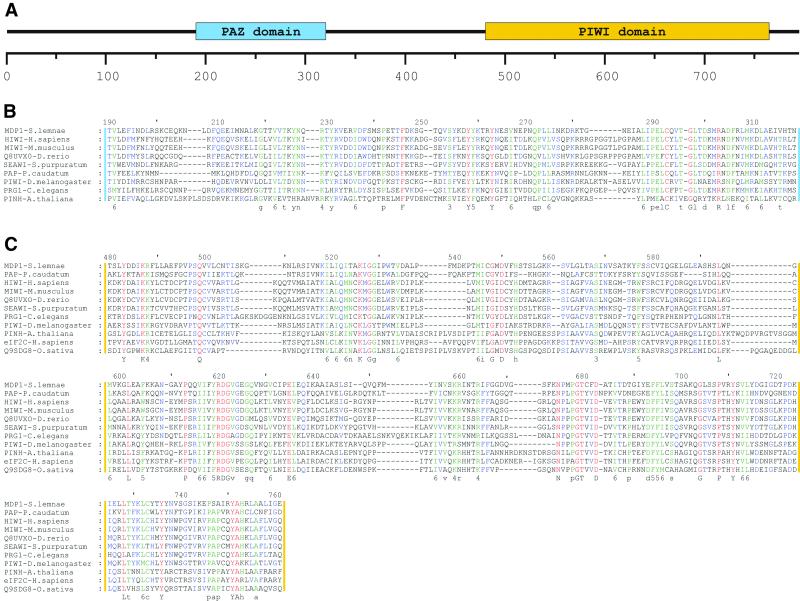
The mdp1 transcript is 2500 bases and is contained on a macronuclear molecule of ∼2.8 kb (Fig. 2A). Of the 24 colonies analyzed in this report, 11 originated from this transcript. Ten were identical in sequence, while one came from an adjacent RsaI fragment. Restriction digestion (RsaI) was a prerequisite for achieving an optimal suppression subtractive hybridization. Transcription of mdp1 is first detected at the beginning of macronuclear development and remains constant for 40 h (Fig. 2A). The ORF begins 46 bases downstream of the 5′ end of the transcript. The ORF is 2376 bases and encodes a polypeptide of 791 amino acids. The predicted MW is 89 kDa and the protein has a pI of 9.01. PSI-BLAST indicated MDP1 contains the recently characterized PAZ and PIWI domains. MDP1 shares highest similarity with PIWI from Danio rerio (GenBank accession no. AF336369) with an e-value of e–104. Over a 794 amino acid region these two proteins share 29% identity and 51% similarity. Multiple sequence alignments of the PAZ and the PIWI domain from MDP1 and several highly similar proteins from other species are shown in Figure 3. The alignment of the PIWI domain reveals a high conservation of these domains over a long period of eukaryotic evolution (Fig. 3C).
Figure 3.
Conserved domains and protein alignments. (A) Schematic drawing of the localization of the PAZ and the PIW domain within MDP1. Multiple sequence alignments of the PAZ and the PIWI domain of different organisms are shown in (B) and (C). Identical amino acids are shaded in red, an 80% similarity is indicated in blue and a 60% similarity is shown in green. The amino acid similarity groups are numbered: 1, DN; 2, EQ; 3, ST; 4, KR; 5, FYW; and 6, LIVM. Upper case letters indicate complete identity, lower case letters indicate 80% identity. Proteins, organisms and accession numbers: HIWI from H.sapiens (gi18098558), MIWI from M.musculus (Q9JMB7), Q8UVX0 from D.rerio (Q8UVX0), SEAWI from Strongylocentrotus purpuratus (gi12007640), PAP from P.caudatum (gi6630673), PIWI from D.melanogaster (gi4165140), PRG1 from C.elegans (D2030.6), PRG2 from C.elegans (C01G5.2), PINH/ZLL from A.thaliana (Q9XGW1), eIF2C from H.sapiens (gi6912352) and Q9SDG8 from Oryza sativa. (B) Alignment within the PAZ domain. (C) Alignment within the PIWI domain.
MDP2
The mdp2 transcript is 1263 bases and is located on a macronuclear molecule of ∼1.5 kb (Fig. 2B). Transcription of MDP2 begins immediately after initiation of macronuclear development, and remains relatively constant for the next 40 h (Fig. 2B). There is only one significant ORF on this transcript, starting with the first AUG 32 bases downstream of the 5′ end. This ORF is 1026 bases and terminates with the UGA stop codon. The 341 amino acid polypeptide has a predicted MW of 40 kDa and a pI of 5.94. PSI-BLAST search revealed no known sequence motifs. MDP2 shares strongest similarity with an unknown protein from Arabidopsis thaliana (GenBank accession no. ACO16827) with an e-value of 4e–05. Over a 120 amino acid region the two polypeptides share 31% identity and 49% similarity. Other secondary structure prediction and amino acid analyses tools did not reveal any insight into the nature of MDP2. TargetP analysis indicates an 83% likelihood that MDP2 is localized in the nucleus (25).
MDP3
The mdp3 transcript is ∼2.5 kb and is contained on a macronuclear molecule of ∼3200 kb. This rare transcript is first detected 30 h after initiation of macronuclear development (Fig. 2C). The first 1038 bases of the transcript were sequenced. The sequence of the 3′ end could not be obtained through TA cloning, unidirectional cloning or direct sequencing. The 5′ untranslated region is 45 bases. Of the ORF, 993 bases, encoding 331 amino acids, were translated. This region of the protein is highly basic, with a pI of 9.17. No significant similarity could be determined by PSI-BLAST. The lowest e-value obtained was 0.42 with a polypeptide similar to Wasp family 1 in Mus musculus (GenBank accession no. XM_142051). These two polypeptides are 32% identical and 46% similar over a 75 amino acid region.
DISCUSSION
During macronuclear development in S.lemnae, extensive cutting, splicing and rearranging of DNA occurs, resulting in elimination of up to 98% of the germline DNA (8). Until now only some aspects of the nature of cis-acting sequences involved in the process of macronuclear development in S.lemnae have been studied in detail. A consensus inverted repeat in both subtelomeric regions was suggested to be necessary for processing of macronuclear destined sequences during development (26,27). Nothing is known about the proteins involved in these processes. Using a suppression subtractive hybridization procedure we have identified three mRNAs differentially expressed during macronuclear development.
Two of the three cDNAs sequenced, mdp2 and mdp3, encode polypeptides with no significant similarity to proteins in GenBank. The third gene, mdp1, identified in nearly 50% of the isolated cDNA clones, is a homolog of the PIWI protein family, characterized by a highly basic amino acid composition (pI of MDP1, 9.01) and the highly conserved PIWI and PAZ domains. PIWI was discovered in Drosophila melanogaster, where it is required for asymmetric division of germline stem cells (28,29). Subsequently, PIWI homologs have been identified in plants (A.thaliana), worms (Caenorhabditis elegans), mammals (M.musculus and Homo sapiens) and ciliates (Paramecium caudatum), but not in yeast. In many of these organisms over 20 paralogs have been identified by similarity searches (30). PAP, a PIWI homolog in P.caudatum, is transcribed at a very low level during vegetative growth, and is extensively upregulated during macronuclear development (31). While the definitive function of PIWI homologs vary, approximately all twenty studied at the genetic level play crucial roles in germline-specific genetic suppression mechanisms and/or the double-stranded RNA silencing pathway.
Work in several ciliates on epigenetic regulation of macronuclear development, the identification of proteins involved in macronuclear development and studies on PIWI in other eukaryotes combined with our results suggest a crucial role of RNA mediated events during macronuclear development in S.lemnae. (i) Studies by Meyer and co-workers (12,32) in Paramecium indicate that homology-dependent epigenetic effects play an important role in specifying proper excision of IESs through the action of RNA molecules cross-talking between macronuclear and micronuclear genomes. (ii) Chalker and Yao (33) found that IESs in Tetrahymena are transcribed bidirectionally during macronuclear development. Inhibition of transcription prevents IES excision. (iii) Pdd1p from Tetrahymena is differentially expressed during macronuclear development, co-localizes with IESs and is necessary for IES excision. It contains three chromo (chromatin organizer modifier) domains (34,35). Chromodomain proteins do not bind DNA, but a recent study in Drosophila shows that members of this family are capable of binding RNA (36). A homolog of Pdd1p was identified in S.lemnae based on immunochemical methods (37). (iv) An RNA proofreading mechanism has been postulated for DNA processing in S.lemnae (26). (v) Homologs of PIWI, an evolutionarily conserved protein family involved in germline differentiation and RNA silencing, have now been identified in the two ciliate species, P.caudatum and S.lemnae. The very specific transcription of mdp1 during macronuclear differentiation together with the observations discussed above strongly suggest that this S.lemnae homolog of PIWI is involved in the extraordinary genome modifications occurring during macronuclear development.
In conclusion, our experiments are a first step towards the identification of trans-acting factors involved in programmed DNA reorganization and elimination in stichotrichous ciliates. The finding that a PIWI homolog is exclusively expressed during macronuclear differentiation suggests that this protein plays a crucial role in these processes and provides indirect evidence that RNA mediated events are involved in restructuring the micronuclear genome.
NOTE ADDED IN PROOF
While this manuscript was in the publication process an immediate early online publication in Cell was published on August 13, 2002, with the title ‘Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena’ by K. Mochizuki et al.
Acknowledgments
ACKNOWLEDGEMENTS
We thank H. J. Wieden for his help with protein alignments and S. Feiler for technical assistance with the S.lemnae cultures. This work was supported by the Deutsche Forschungs gemeinschaft, the Alfried Krupp von Bohlen und Halbach Foundation and the National Science Foundation (subcontract no. 128-6114-1). D.J.H. was a recipient of a scholarship from the Fulbright foundation. C.P.F. was supported by the Werner Richard–Dr Carl Doerken-Foundation.
DDBJ/EMBL/GenBank accession nos AY128524–AY128526
REFERENCES
- 1.Harriman W., Volk,H., Defranoux,N. and Wabl,M. (1993) Immunoglobulin class switch recombination. Annu. Rev. Immunol., 11, 361–384. [DOI] [PubMed] [Google Scholar]
- 2.Borst P. (1991) Molecular genetics of antigenic variation. Immunol. Today, 12, A29–A33. [DOI] [PubMed] [Google Scholar]
- 3.Haber J.E. (1998) Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet., 32, 561–599. [DOI] [PubMed] [Google Scholar]
- 4.Tobler H. (1986) The differentiation of germ and somatic cell lines in nematodes. Results Probl. Cell Differ., 13, 1–69. [DOI] [PubMed] [Google Scholar]
- 5.Beermann S. (1977) The diminution of Heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda). Chromosoma, 60, 297–344. [DOI] [PubMed] [Google Scholar]
- 6.Gerbi S.A. (1986) Unusual chromosome movements in sciarid flies. Results Probl. Cell Differ., 13, 71–104. [DOI] [PubMed] [Google Scholar]
- 7.Kubota S., Kuro-o,M., Mizuno,S. and Kohno,S. (1993) Germ line-restricted, highly repeated DNA sequences and their chromosomal localization in a Japanese hagfish (Eptatretus okinoseanus). Chromosoma, 102, 163–173. [DOI] [PubMed] [Google Scholar]
- 8.Prescott D.M. (1994) The DNA of ciliated protozoa. Microbiol. Rev., 58, 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prescott D.M. (2000) Genome gymnastics: unique modes of DNA evolution and processing in ciliates. Nature Rev. Genet., 1, 191–198. [DOI] [PubMed] [Google Scholar]
- 10.Ammermann D. (1971) Morphology and development of the macronuclei of the ciliates Stylonychia mytilus and Euplotes aediculatus. Chromosoma, 33, 209–238. [DOI] [PubMed] [Google Scholar]
- 11.Klobutcher L.A. and Jahn,C.L. (1991) Developmentally controlled genomic rearrangements in ciliated protozoa. Curr. Opin. Genet. Dev., 1, 397–403. [DOI] [PubMed] [Google Scholar]
- 12.Meyer E. and Garnier,O. (2002) Non-Mendelian inheritance and homology-dependent effects in ciliates. Adv. Genet., 46, 305–337. [DOI] [PubMed] [Google Scholar]
- 13.Pal-Bhadra M., Bhadra,U. and Birchler,J.A. (2002) RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell, 9, 315–327. [DOI] [PubMed] [Google Scholar]
- 14.Ammermann D., Steinbruck,G., von Berger,L. and Hennig,W. (1974) The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma, 45, 401–429. [DOI] [PubMed] [Google Scholar]
- 15.Elsevier S.M., Lipps,H.J. and Steinbruck,G. (1978) Histone genes in macronuclear DNA of the ciliate Stylonychia mytilus. Chromosoma, 69, 291–306. [DOI] [PubMed] [Google Scholar]
- 16.Attardi L.D., Reczek,E.E., Cosmas,C., Demicco,E.G., McCurrach,M.E., Lowe,S.W. and Jacks,T. (2000) PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev., 14, 704–718. [PMC free article] [PubMed] [Google Scholar]
- 17.Dessens J.T., Margos,G., Rodriguez,M.C. and Sinden,R.E. (2000) Identification of differentially regulated genes of Plasmodium by suppression subtractive hybridization. Parasitol. Today, 16, 354–356. [DOI] [PubMed] [Google Scholar]
- 18.Diatchenko L., Lukyanov,S., Lau,Y.F. and Siebert,P.D. (1999) Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol., 303, 349–380. [DOI] [PubMed] [Google Scholar]
- 19.Frohman M.A., Dush,M.K. and Martin,G.R. (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA, 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Hoffman D.C., Anderson,R.C., DuBois,M.L. and Prescott,D.M. (1995) Macronuclear gene-sized molecules of hypotrichs. Nucleic Acids Res., 23, 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 24.Wilkins M.R., Gasteiger,E., Bairoch,A., Sanchez,J.C., Williams,K.L., Appel,R.D. and Hochstrasser,D.F. (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol., 112, 531–552. [DOI] [PubMed] [Google Scholar]
- 25.Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- 26.Jonsson F., Steinbruck,G. and Lipps,H.J. (2001) Both subtelomeric regions are required and sufficient for specific DNA fragmentation during macronuclear development in Stylonychia lemnae. Genome Biol., 2, RESEARCH0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsson F., Wen,J.P., Fetzer,C.P. and Lipps,H.J. (1999) A subtelomeric DNA sequence is required for correct processing of the macronuclear DNA sequences during macronuclear development in the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res., 27, 2832–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox D.N., Chao,A., Baker,J., Chang,L., Qiao,D. and Lin,H. (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev., 12, 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin H. and Spradling,A.C. (1997) A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development, 124, 2463–2476. [DOI] [PubMed] [Google Scholar]
- 30.Grishok A., Pasquinelli,A.E., Conte,D., Li,N., Parrish,S., Ha,I., Baillie,D.L., Fire,A., Ruvkun,G. and Mello,C.C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- 31.Obara S., Iwataki,K. and Mikami,K. (2000) Identification of an Aubergine/Piwi homologue in the ciliated protozoan Paramecium caudatum. Proc. Jpn Acad. series B, 76, 57–62. [Google Scholar]
- 32.Duharcourt S., Butler,A. and Meyer,E. (1995) Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev., 9, 2065–2077. [DOI] [PubMed] [Google Scholar]
- 33.Chalker D.L. and Yao,M.C. (2001) Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev., 15, 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coyne R.S., Nikiforov,M.A., Smothers,J.F., Allis,C.D. and Yao,M.C. (1999) Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell, 4, 865–872. [DOI] [PubMed] [Google Scholar]
- 35.Madireddi M.T., Coyne,R.S., Smothers,J.F., Mickey,K.M., Yao,M.C. and Allis,C.D. (1996) Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell, 87, 75–84. [DOI] [PubMed] [Google Scholar]
- 36.Akhtar A., Zink,D. and Becker,P.B. (2000) Chromodomains are protein-RNA interaction modules. Nature, 407, 405–409. [DOI] [PubMed] [Google Scholar]
- 37.Maercker C., Kortwig,H., Nikiforov,M.A., Allis,C.D. and Lipps,H.J. (1999) A nuclear protein involved in apoptotic-like DNA degradation in Stylonychia: implications for similar mechanisms in differentiating and starved cells. Mol. Biol. Cell, 10, 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraut H., Lipps,H.J. and Prescott,D.M. (1986) The genome of hypotrichous ciliates. Int. Rev. Cytol., 99, 1–28. [DOI] [PubMed] [Google Scholar]