Abstract
Oligodeoxyribonucleotides containing CpG dinucleotides (CpG DNAs) are currently being evaluated as novel immunomodulators in clinical trials. Recently, we showed that an accessible 5′ end is required for immunostimulatory activity and blocking the 5′ end of CpG DNA by conjugation of certain ligands abrogates immunostimulatory activity. Based on these results, we designed and synthesized 3′–3′-linked CpG DNAs that contained two or more identical CpG DNA segments, referred to here as ‘immunomers’. The use of solid support bearing diDMT-glyceryl-linker permitted convenient synthesis of immunomers with both segments synthesized simultaneously, giving better yields and purity. The in vitro and in vivo studies suggest that as a result of accessibility to two 5′ ends for recognition, immunomers show an enhanced immunostimulatory activity compared with linear CpG DNAs. We also studied the suitability of a number of different linkers for attaching the two segments of immunomers. A C3-linker was found to be optimal for joining the two segments of immunomers. Incorporation of multiple linkers between the two segments of immunomers resulted in different cytokine profiles depending on the nature and number of linkers incorporated. Additionally, the length of immunomer also plays a significant role in inducing immune responses. An immunomer containing 11 nt in each segment showed the highest activity and an 11mer linear CpG DNA failed to stimulate an immune response. These results suggest that immunomers have several advantages over conventional linear CpG DNAs for immunomodulatory activity studies.
INTRODUCTION
The presence of CpG dinucleotides in certain sequence contexts in bacterial and synthetic oligonucleotides (CpG DNAs) are known to activate vertebrate innate immune system cells and B cells (1–4). The activation of immune cells by CpG DNA induces secretion of a myriad of cytokines, including IL-6, IL-12, TNF-α and IFN-γ, and stimulates expression of co-stimulatory surface molecules (5–8). The presence of a CpG dinucleotide and the sequences flanking it play a critical role in determining the immunostimulatory activity of CpG DNAs (3,9–11). The use of CpG DNAs as antitumor, anti-infectious, anti-asthmatic and anti-inflammatory agents, and as adjuvants in immunotherapy has been reported (12–14).
Evidence suggests that TLR9, a molecular pattern recognition receptor, recognizes CpG dinucleotides present in DNA and activates intracellular immune signaling pathways (15). Substitution of C and G nucleotides in a CpG dinucleotide with ribonucleotides or 2′-O-alkyl substituted ribonucleotides neutralizes immune responses, suggesting that the receptor has high specificity for deoxyribonucleotides in a CpG dinucleotide (16). Our laboratory has been studying sequence and structural changes in CpG DNA that might potentiate or neutralize immunostimulatory activities in a predictable fashion (17). In addition, these studies could lead to the understanding of the molecular basis of recognition between CpG DNA and its receptor(s). Our studies have shown a number of critical structural and functional group requirements of the pentose sugar (16,18–20), phosphate backbone (16,21) and nucleobases (22) for the immunostimulatory activity of CpG DNA. Moreover, we demonstrated the importance of functional groups on cytosine and guanine required for receptor recognition by substitution of natural cytosine and guanine with synthetic pyrimidine and purine bases in a CpG dinucleotide, thereby introducing synthetic motifs referred to as YpG and CpR motifs (23).
Recently, we showed that when two CpG DNA molecules were linked through their 5′ ends, immunostimulatory activity was abrogated compared with a 5′→3′ CpG DNA containing the same number of CpG dinucleotides (24,25). In contrast, an increased immunostimulatory activity was observed when they were linked through their 3′ ends. Based on these observations, we postulated that an accessible 5′-end of CpG DNA was required for recognition by receptor(s) and subsequent initiation of the immunostimulatory signaling cascade. In the present paper, we describe a number of 3′–3′-linked CpG DNAs, referred to here as ‘immunomers’, to establish the nature and length of linker required for tethering CpG DNAs and report the optimal length of the immunomers required for immunostimulatory activity.
MATERIALS AND METHODS
CpG DNA and immunomer synthesis and purification
CpG DNAs and immunomers were synthesized on a 1–2 µmol scale using β-cyanoethylphosphoramidite chemistry on a PerSeptive Biosystem’s 8909 Expedite DNA synthesizer as described earlier (24–27). The 3′-phosphoramidites of dA, dG, dC and T were obtained from PE Biosystems. The 5′-phosphoramidites of dA, dG, dC, and T were obtained from Glen Research Corporation. DiDMT protected glyceryl linker attached to CPG-solid-support was obtained from ChemGenes. Symmetric doubler (long linker, 3) phosphoramidite was obtained from Glen Research Corporation. Beaucage reagent was used as an oxidant to obtain the phosphorothioate backbone modification (28). After the synthesis, immunomers were deprotected using standard protocols, purified by HPLC and dialyzed against United States Pharmacopea quality sterile water for irrigation (Braun). The immunomers were lyophilized and dissolved again in distilled water and the concentrations were determined by measuring the UV absorbance at 260 nm (29). All the immunomers synthesized were characterized by CGE and MALDI–TOF mass spectrometry (Applied Biosystem’s Voyager-DE™ STR Biospectrometry™ Workstation) for purity and molecular mass (Table 1), respectively. The purity of full-length immunomers ranged from 89 to 95% with the rest being shorter by one or two nucleotides (n–1 and n–2) as determined by CGE and/or denaturing PAGE.
Table 1. Phosphorothioate immunomers and related CpG DNAs showing the number and position of linker.
| CpG DNA number | Sequence | Linker (X)a | Lengthb | Molecular Weight | |
|---|---|---|---|---|---|
| Calculated | Foundc | ||||
| 10 | 5′-CTATCTGACGTTCTCTGT-3′ | None | 18mer | 5704 | 5705 |
| 11 | (5′-CTATCTGACGTTCTCTGT)2-X-C-5′ | Glyceryl linker (2) | 18mer | 11945 | 11943 |
| 12 | (5′-CTATCTGACGTTCTCTGT)2-X-C-5′ | Long linker (3) | 18mer | 12158 | ND |
| 13 | (5′-CTATCTGACGTTCTCTGT)3-X | Long linker (3) | 18mer | 17635 | ND |
| 14 | 5′-CTGACGTTCTCTGT-3′ | None | 14mer | 4429 | 4428 |
| 15 | 5′-CTGACGTTCTCTGT-X-TGTCTCTTGCAGTC-5′ | Glyceryl linker (2) | 14mer | 9106 | ND |
| 16 | (5′-CTGACGTTCTCTGT)2-X-C-5′ | Long linker (3) | 14mer | 9609 | ND |
| 17 | 5′-TCTGACGTTCTCTGT-3′ | None | 15mer | 4749 | 4746 |
| 18 | 5′-TCTGACGTTCT-3′ | None | 11mer | 3458 | 3457 |
| 19 | 5′-GACGTTCT-3′ | None | 8mer | 2513 | 2512 |
| 20 | 5′-CTGACGTTCTCTG-X-GTCTCTTGCAGTC-5′ | Glyceryl linker (2) | 13mer | 8465 | 8455 |
| 21 | 5′-CTGACGTTCTCT-X-TCTCTTGCAGTC-5′ | Glyceryl linker (2) | 12mer | 7775 | 7772 |
| 22 | 5′-TCTGACGTTCT-X-TCTTGCAGTCT-5′ | Glyceryl linker (2) | 11mer | 7164 | 7166 |
| 23 | 5′-CTGACGTTCT-X-TCTTGCAGTC-5′ | Glyceryl linker (2) | 10mer | 6524 | 6522 |
| 24 | 5′-TGACGTTCT-X-TCTTGCAGT-5′ | Glyceryl linker (2) | 9mer | 5914 | 5913 |
| 25 | 5′-GACGTTCT-X-(T-5′)TCTTGCAG-5′ | Glyceryl linker (2) | 8mer | 5594 | 5593 |
| 26 | 5′-TCTGAGCTTCT-X-TCTTCGAGTCT-5′ | Glyceryl linker (2) | 11mer | 7164 | 7166 |
| 27 | 5′-CTGACGTTCT-X-TCTTGCAGTC-5′ | Phosphorothioate (1) | 10mer | 6354 | 6354 |
| 28 | 5′-CTGACGTTCT-X-TCTTGCAGTC-5′ | C2-linker (4) | 10mer | 6494 | 6519 |
| 29 | 5′-CTGACGTTCT-X-TCTTGCAGTC-5′ | C3-linker (5) | 10mer | 6508 | 6503 |
| 30 | 5′-CTGACGTTCT-X-TCTTGCAGTC-5′ | C4-linker (6) | 10mer | 6522 | 6515 |
| 31 | 5′-CTGACGTTCT-X-TCTTGCAGTC-5′ | C12-linker (7) | 10mer | 6634 | 6629 |
| 32 | 5′-CTGACGTTCT-X-TCTTGCAGTC-5′ | Spacer18 (8) | 10mer | 6714 | 6710 |
| 33 | 5′-CTGACGTTCT-X-TCTTGCAGTC-5′ | Abasic linker (9) | 10mer | 6550 | 6547 |
| 34 | 5′-CTGACGTTCT-XX-TCTTGCAGTC-5′ | C3-linker (5) | 10mer | 6662 | 6660 |
| 35 | 5′-CTGACGTTCT-XXX-TCTTGCAGTC-5′ | C3-linker (5) | 10mer | 6816 | 6814 |
| 36 | 5′-CTGACGTTCT-XX-TCTTGCAGTC-5′ | Abasic linker (9) | 10mer | 6746 | 6743 |
| 37 | 5′-CTGACGTTCT-XXX-TCTTGCAGTC-5′ | Abasic linker (9) | 10mer | 6942 | 6939 |
| 38 | 3′-C-X-(CTATCTGACGTTCTCTGT-3′)2 | Glyceryl linker (2) | 18mer | 11945 | ND |
| 39 | 5′-CTGACGTTCTCTGTCTTGTCCTGACGTTCTCTGT-3′ | None | 34mer | 10852 | ND |
aSee Figure 1A for chemical structures of linkers 1–9.
blength of each segment in 3′–3′- or 5′–5′-linked CpG DNA excluding linker.
cND, not determined.
Cell culture conditions and reagents
Spleen cells from 4–8 week old BALB/c mice were cultured in RPMI complete medium as described earlier (16,30). Murine macrophage-like cells, J774 (American Type Culture Collection, Rockville, MD), were cultured in Dulbecco’s modified Eagles medium supplemented with 10% (v/v) FCS and antibiotics (100 IU/ml of penicillin G/streptomycin). All other culture reagents were purchased from Mediatech (Gaithersburg, MD).
ELISAs for IL-12, IFN-γ, IL-10, TNF-α and IL-6
BALB/c mouse spleen or J774 cells were plated in 24-well dishes at a density of 5 × 106 or 1 × 106 cells/ml, respectively. The CpG DNA dissolved in TE buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA) was added to mouse spleen cell cultures to give final concentrations of 0.03, 0.1, 0.3, 1.0, 3.0 or 10.0 µg/ml. With J774 cell cultures, final concentrations of CpG DNA were 1.0, 3.0 or 10.0 µg/ml. The cells were then incubated at 37°C for 24 h and the supernatants were collected for ELISAs. The experiments with each CpG DNA were repeated two or three times using triplicates of each concentration. The secretion of IL-12, IFN-γ, IL-10, TNF-α and IL-6 was measured by sandwich ELISA. The required reagents, including cytokine antibodies and standards were purchased form BD PharMingen. ELISA plates (Costar) were incubated with appropriate antibodies at 5 µg/ml in PBSN buffer (PBS/0.05% sodium azide, pH 9.6) overnight at 4°C and then blocked with PBS/1% BSA at 37°C for 30 min. Cell culture supernatants and cytokine standards were appropriately diluted with PBS/1% BSA, added to the plates in triplicate, and incubated at 25°C for 2 h. Plates were washed and incubated with 1 µg/ml of appropriate biotinylated antibody and incubated at 25°C for 1.5 h. The plates were washed extensively with PBS/0.05% Tween-20 and then further incubated at 25°C for 1.5 h after the addition of streptavidine-conjugated peroxidase (Sigma). The plates were developed with Sure Blue™ (Kirkegaard and Perry) chromogenic reagent and the reaction was terminated by adding Stop Solution (Kirkegaard and Perry). The color change was measured on a Ceres 900 HDI Spectrophotometer (Bio-Tek Instruments) at 450 nm. The levels of IL-12, IFN-γ, IL-10, TNF-α and IL-6 in the cell culture supernatants were calculated from the standard curve constructed under the same experimental conditions for IL-12, IFN-γ, IL-10, TNF-α and IL-6, respectively.
Mouse splenomegaly assay
Female BALB/c mice (4–6 weeks, 19–21 g) were divided into groups of three mice. CpG DNAs were dissolved in sterile PBS and administered subcutaneously (SC) to mice at a dose of 5 mg/kg. After 4 days, mice were sacrificed and the spleens were harvested and weighed.
Preparation of J774 cell nuclear extracts and EMSA
For NF-κB activation studies, J774 cells were plated at a density of 5 × 106 cells/well in six-well plates and nuclear extracts were prepared as described earlier (31). Briefly, after stimulation of J774 cells for 1 h with CpG DNAs at a concentration of 10 µg/ml, cells were washed three times in ice-cold PBS, harvested, and suspended in 0.24 ml of buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT and protease inhibitor cocktail) (Boehringer-Mannheim) and allowed to swell on ice for 15 min. Then 16 µl of 10% Nonidet-P40 was added and the tubes were vortexed vigorously for 10 s. Nuclei were separated from cytosol by centrifugation at 500 g for 5 min and were resuspended in 60 µl of buffer B (20 mM HEPES pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA and protease inhibitor cocktail). After 30 min at 4°C, lysates were separated by centrifugation (14 000 g, 10 min) and supernatants containing nuclear proteins were transferred to new vials. Protein concentrations were determined (32) and the samples were either used immediately or stored frozen at –70°C.
The oligonucleotide probe d(AGTTGAGGGGACTTTCCCAGGC) containing the κB binding motif was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Invitrogen) and purified over two successive G-50 mini columns (Amersham-Pharmacia Biotech). Reaction mixtures (25 µl) containing 10 µg of nuclear protein, 5 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, 4% glycerol (v/v) and 0.08 mg/ml salmon sperm DNA were preincubated on ice for 15 min. Then, 1 × 106 c.p.m. of the probe was added and the binding reaction was allowed to proceed for 20 min at room temperature. DNA–protein complexes were then resolved in a 6% native polyacrylamide gel in TBE buffer (22.5 mM Tris, 22.5 mM boric acid and 0.5 mM EDTA, pH 8.3) at 140 V for 2–3 h. Gels were dried and exposed to Kodak Biomax MR film at –70°C. Films were scanned and the images were processed using Adobe imaging software.
RESULTS
Design and synthesis of immunomers
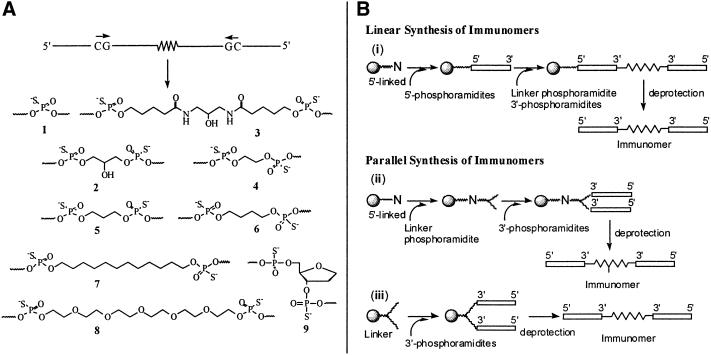
An 18mer CpG DNA sequence 10, which has a ‘GACGTT’ hexameric motif, was used in the present study (Table 1). This sequence has been shown to induce immunostimulatory activity in mice in earlier studies (21,22). The immunomers used in the present study contain two identical segments that are tethered through their 3′ends via a non-nucleoside linker or a phosphorothioate linkage (see Fig. 1A for linkers used in the study). Since they contain two identical sequence segments (Fig. 1A), they can be synthesized on an automated DNA synthesizer using two different synthetic strategies, referred to here as linear and parallel synthesis (Fig. 1B). In a linear synthetic strategy, as reported earlier, synthesis is started from one end of the immunomer using a solid support linked 5′-nucleoside and nucleoside-5′-β-cyanoethylphosphoramidites (24–27). When the synthesis of first oligonucleotide segment is complete, it is coupled with a DMT-protected-β-cyanoethylphosphoramidite derivatized linker and the synthesis is continued using nucleoside-3′-β-cyanoethylphosphoramidites to construct the second segment. The second oligonucleotide segment could be identical or different from the first segment in terms of its length, base composition and/or chemical modifications incorporated. In the present study, however, we used identical sequences and modifications in both the segments of immunomers 27–37 (Table 1), which are synthesized using linear synthesis strategy (route i, Fig. 1B).
Figure 1.
(A) A line drawing of immunomer showing CpG dinucleotide directionality and different linkers used to tether two CpG DNAs for synthesis of immunomers. (B) Schemes showing (i) linear synthesis of CpG DNA and immunomers, and parallel synthesis of immunomers using (ii) diDMT protected brancher phosphoramidite or (iii) solid support attached diDMT protected brancher. N stands for any nucleoside.
Alternately, immunomers containing identical sequence segments can be synthesized using a parallel synthesis strategy as shown in Figure 1B (routes ii and iii). This strategy requires a solid support derivatized with a linker having two reactive sites protected with DMT (route iii, Fig. 1B) or its corresponding phosphoramidite (route ii, Fig. 1B). This enables simultaneous synthesis of two identical sequence segments using nucleoside-3′- (or 5′-)β-cyanoethylphosphoramidites. Parallel synthesis of immunomers has certain advantages over linear synthesis: (i) it permits incorporation of two or more identical sequence segments; (ii) the segments are synthesized concurrently, thereby halving the number of synthetic steps and the time required; and (iii) with fewer synthetic cycles, the yield and purity of the final product improves. Immunomers 11–13, 15, 16 and 20–26 (Table 1) were synthesized using the parallel synthesis strategy. Out of the immunomers synthesized by parallel synthesis strategy, immunomers 11–13 and 16 were synthesized using 5′-nucleoside attached solid support and diDMT-protected linker phosphoramidite (route ii, Fig. 1B). Note that immunomer 13 contains three identical oligonucleotide segments. The remaining immunomers, 15 and 20–26 were synthesized using solid support bearing diDMT glyceryl linker (route iii, Fig 1B).
Effect of a linker on immunostimulatory activity of immunomers
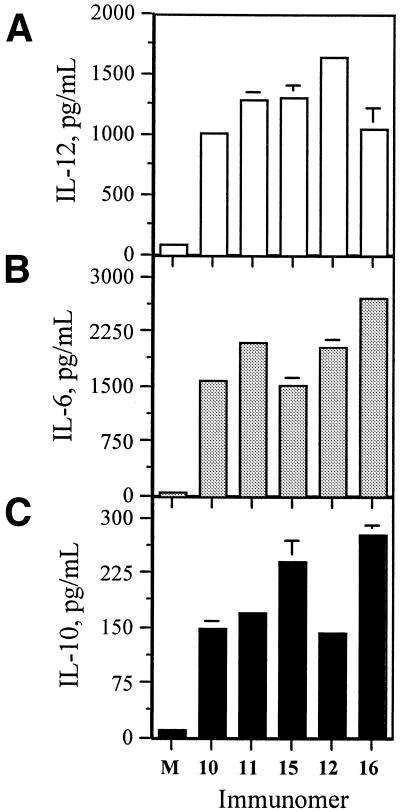
Previously, we showed that immunomers having the two segments 3′–3′-linked through a phosphorothioate group (1) are more immunostimulatory than those 5′–5′-linked (24,25). To examine the effect of different length linkers, we synthesized two immunomers 11 and 12 incorporating linkers 2 and 3 (Table 1), respectively. Their immunostimulatory activities were determined by induction of cytokines IL-12, IL-6 and IL-10 secretion in BALB/c mouse spleen cell cultures. Both immunomers 11 and 12 induced higher levels of IL-12 and IL-6, but similar levels of IL-10, compared with its parent CpG DNA 10 at a concentration of 1.0 µg/ml (Fig. 2). These results suggest that neither of these linkers interfered with activity of immunomers. Immunomer 13, which has three identical 18mer segments branching from the same linker (a ‘Y’-shaped molecule), produced similar or slightly lower levels of cytokine secretion compared with CpG DNA 10 (data not shown), suggesting that more than two segments do not enhance activity. We, therefore, used immunomers containing two identical sequence segments in all subsequent studies.
Figure 2.
(A) IL-12, (B) IL-6 and (C) IL-10 secretion in BALB/c mouse spleen cell cultures induced by immunomers at a concentration of 1.0 µg/ml. M represents control in the absence of CpG DNA.
To examine whether each segment of immunomer should have the same length as that of CpG DNA 10 (18 nt), we synthesized immunomers 15 and 16 (Table 1) with linkers 2 and 3, respectively, but with each sequence segment containing 14 nt, and studied immunostimulatory activity in BALB/c mouse spleen cell cultures. As shown in Figure 2, immunomer 15 induced similar levels of IL-12 and lower levels of IL-6 compared with immunomer 11 at a concentration of 1.0 µg/ml. In contrast, truncated immunomer 16, which contained a longer linker 3, induced lower IL-12 and higher IL-6 compared with immunomer 12 at the same concentration (Fig. 2A and B). Both immunomers 15 and 16 produced higher levels of IL-10 compared with 11 and 12, respectively (Fig. 2C). As these results suggested that immunomer length affects its activity, we carried out a systematic study to find the optimal length.
Effect of length of immunomer on immunostimulatory activity
Immunomers 11 and 12, with segments of 18 nt, were compared with 20–25 (Table 1) having shorter segments of 8–14 nt. Immunomers 20–25 contain glyceryl-linker 2 that, earlier, increased the activity of 15, with 14mer segments, relative to CpG DNA 14 (data not shown). Immunomers 15 and 20–25 were tested for their immunostimulatory properties in BALB/c mouse spleen cell cultures.
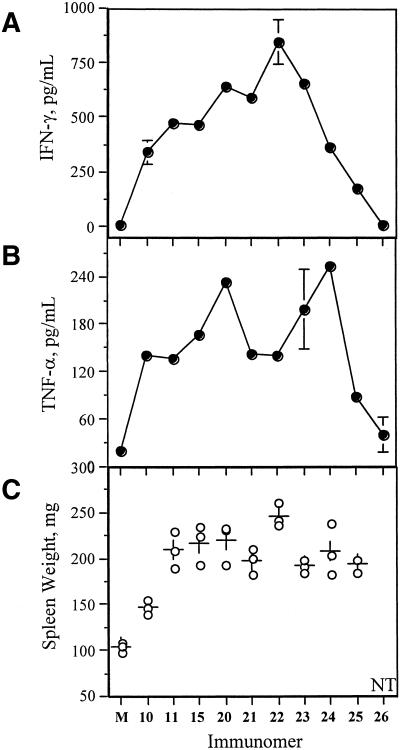
Table 2 shows the concentration-dependent induction of IL-12 and IL-6 secretion by immunomers 15 and 20–24 in BALB/c mouse spleen cell cultures. In general, a trend of increased secretion of IL-12 and IL-6 was observed as immunomer length reduced from 14 nt (15) to 11 nt (22) in repeat experiments. However, immunomer 20 showed a lower activity in this particular assay compared with 15. Further reduction, to 10 nt or less, resulted in a decreased cytokine secretion compared with 11mer immunomer 22, but maintained a similar or higher activity compared with 18mer parent CpG DNA 10 (Table 2). Similar results were observed with IFN-γ secretion in BALB/c mouse spleen cell cultures (Fig. 3A). However, a different profile was found for TNF-α secretion (Fig. 3B). TNF-α secretion increased as chain-lengths decreased from 18mer (11) to 13mer (20), dropped as the length decreased further to 12mer (21) and 11mer (22), then rose again as the length continued to decrease in immunomers 23 and 24 (Fig. 3B). The control immunomer 26, which has a GpC dinucleotide, induced minimal levels of cytokines in the same assays (Fig. 3 and data not shown), suggesting that a CpG dinucleotide is required for immunostimulatory activity.
Table 2. Concentration-dependent cytokine secretion in BALB/c mouse spleen cell cultures by immunomers.
| CpG DNA | IL-12 (pg/ml) ± SD | IL-6 (pg/ml) ± SD | ||||||
|---|---|---|---|---|---|---|---|---|
| 0.1 µg/ml | 0.3 µg/ml | 1.0 µg/ml | 3.0 µg/ml | 0.1 µg/ml | 0.3 µg/ml | 1.0 µg/ml | 3.0 µg/ml | |
| 10 | 70 ± 13 | 444 ± 33 | 1142 ± 100 | 2570 ± 46 | 150 ± 8 | 211 ± 26 | 3979 ± 514 | 21491 ± 1951 |
| 15 | 189 ± 24 | 1404 ± 189 | 2972 ± 275 | 2910 ± 191 | 314 ± 56 | 456 ± 21 | 2550 ± 356 | 20923 ± 103 |
| 20 | 70 ± 8 | 1025 ± 36 | 2135 ± 96 | 2155 ± 128 | 211 ± 51 | 533 ± 4 | 2696 ± 138 | 17918 ± 1495 |
| 21 | 217 ± 56 | 1397 ± 42 | 2443 ± 48 | 2921 ± 64 | 183 ± 22 | 452 ± 15 | 3812 ± 96 | 23894 ± 713 |
| 22 | 504 ± 58 | 1609 ± 132 | 2428 ± 226 | 2743 ± 269 | 233 ± 5 | 1965 ± 1 | 9307 ± 95 | 26825 ± 1239 |
| 23 | 103 ± 22 | 1417 ± 76 | 2385 ± 168 | 3482 ± 81 | 198 ± 57 | 376 ± 82 | 7537 ± 964 | 24827 ± 1917 |
| 24 | 34 ± 3 | 135 ± 18 | 1351 ± 106 | 3282 ± 115 | NT | |||
| Culture medium | 71 ± 1 | 155 ± 28 | ||||||
NT, not tested.
Figure 3.
(A) IFN-γ and (B) TNF-α secretion in BALB/c mouse spleen cell cultures induced by immunomers at a concentration of 1.0 µg/ml. M represents control in the absence of CpG DNA. (C) Splenomegaly in BALB/c mice at a dose of 5 mg/kg dose of immunomer administered SC. Spleen weight of each mouse is represented by open circles. Plus sign indicates average spleen weight. M indicates control mice administered with vehicle (PBS). NT stands for not tested.
We also tested immunomers 10, 11, 15, and 20–25 for their ability to induce spleen enlargement in BALB/c mice. Following subcutaneous administration at a dose of 5 mg/kg, spleen weights were measured after 4 days as described in the Materials and Methods. Increases in spleen weight in excess of that produced by vehicle (PBS) are considered the result of immunostimulatory activity of immunomers (16,29). The splenomegaly data are presented in Figure 3C. CpG DNA 10 caused a 40% increase in spleen weight compared with control mice treated with vehicle. Immunomers containing glyceryl-linker (2) and different chain lengths are more potent immunostimulators and caused 83–135% increases in spleen weights. These splenomegaly assays also suggest that immunomer 22 with 11mer segments is optimal for inducing higher immunostimulatory activity as determined by in vitro assays.
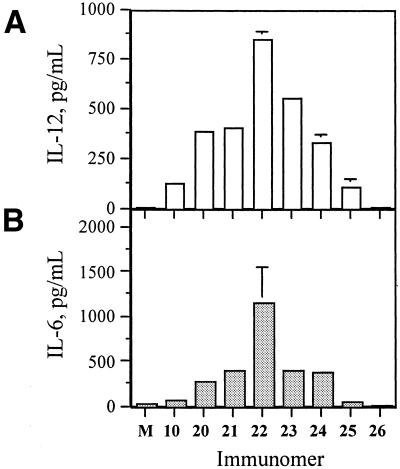
In addition, we also examined the ability of immunomers 20–26 to induce IL-12 and IL-6 secretion in macrophage- like J774 cell cultures. All immunomers tested induced a concentration-dependent cytokine secretion (data not shown). As shown in Figure 4, immunomer 22 induced the highest levels of IL-12 and IL-6 secretion at 3 µg/ml concentration in J774 cell cultures further confirming the results obtained from BALB/c mouse spleen cell culture assays. Importantly, shorter immunomers with 9- and 8-nt segments also showed immunostimulatory activity in BALB/c mouse spleen and J774 cell culture and splenomegaly assays compared with parent linear CpG DNA 10, whereas the linear CpG DNAs of the same length completely failed to show activity in these assays.
Figure 4.
(A) IL-12 and (B) IL-6 secretion in J774 cell cultures induced by immunomers at a concentration of 3.0 µg/ml. M represents control in the absence of CpG DNA.
Effect of nature and length of linker on immunostimulatory activity
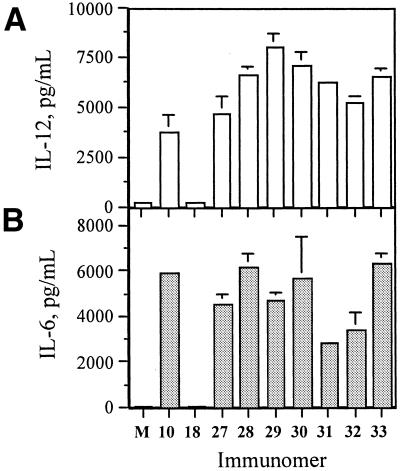
After establishing the optimal length for immunomers, we used linkers 1 and 4–9 (Fig. 1A) for tethering the two segments of immunomers to compare immunostimulatory activity. All immunomers (27–33), had 10-nt long segments (Table 1) and induced concentration-dependent cytokine secretion in BALB/c mouse spleen cell cultures (Table 3). The levels of IL-12 and IL-6 induced by immunomers 10, 18 and 27–33 in spleen cell cultures at a concentration of 1.0 µg/ml are shown in Figure 5 for comparison. CpG DNA 18, which is an 11mer, failed to induce cytokine secretion in these assays suggesting CpG DNAs as short as 11mers do not induce immune responses. All seven immunomers with different linker groups (27–33) induced increased cytokine secretion. As shown in Figure 5A, IL-12 secretion increased on going from immunomer 27 with no linker, to a C2-linker containing 28 and then to a C3-linker (5) containing 29. Other immmunomers 30–32, which have longer linkers [C4- (6), C12- (7) and hexaethylene glycol-linkers (8)], induced lower IL-12 secretion compared with 28 and 29. Importantly, all seven immunomers induced higher IL-12 than did CpG DNA 10. Immunomer 33, which has a more constrained abasic linker (9), but maintains a similar distance between the two phosphates as in the C3-linker, induced comparable levels of IL-12 secretion as 29. All seven immunomers induced similar or lower IL-6 secretion compared with CpG DNA 10 (Fig. 5B). These results suggest that a C3-linker is optimal for immunomer construction.
Table 3. Effect of linkers on cytokine secretion in BALB/c mouse spleen cell cultures.
| CpG DNA | IL-12 (pg/ml) ± SD | IL-6 (pg/ml) ± SD | ||||||
|---|---|---|---|---|---|---|---|---|
| 0.1 µg/ml | 0.3 µg/ml | 1.0 µg/ml | 3.0 µg/ml | 0.1 µg/ml | 0.3 µg/ml | 1.0 µg/ml | 3.0 µg/ml | |
| 10 | 310 ± 1 | 1962 ± 78 | 3780 ± 805 | 6084 ± 1390 | 181 ± 57 | 582 ± 143 | 5981 ± 138 | 13879 ± 612 |
| 18 | 203 ± 76 | 148 ± 26 | 310 ± 35 | 1229 ± 45 | 43 ± 6 | 35 ± 4 | 71 ± 3 | 2562 ± 157 |
| 27 | 1826 ± 230 | 4770 ± 301 | 4698 ± 881 | 6052 ± 372 | 257 ± 44 | 1184 ± 111 | 4551 ± 389 | 10613 ± 507 |
| 28 | 1419 ± 455 | 5113 ± 870 | 6697 ± 346 | 7558 ± 1139 | 233 ± 24 | 1385 ± 75 | 6233 ± 549 | 16011 ± 924 |
| 29 | 4118 ± 1099 | 5768 ± 1250 | 8042 ± 632 | 7414 ± 457 | 421 ± 30 | 806 ± 131 | 4725 ± 291 | 15093 ± 636 |
| 30 | 930 ± 28 | 7083 ± 297 | 7135 ± 651 | 9045 ± 421 | 127 ± 39 | 1570 ± 767 | 5718 ± 1746 | 14559 ± 1774 |
| 31 | 2177 ± 254 | 5735 ± 308 | 6254 ± 190 | 7443 ± 424 | 779 ± 64 | 747 ± 130 | 2840 ± 140 | 15311 ± 2459 |
| 32 | 365 ± 109 | 4731 ± 857 | 5254 ± 279 | 7283 ± 905 | 69 ± 19 | 480 ± 57 | 3414 ± 717 | 13954 ± 151 |
| 33 | 778 ± 136 | 4854 ± 498 | 6563 ± 358 | 8803 ± 666 | 294 ± 107 | 587 ± 56 | 6359 ± 372 | 17006 ± 1274 |
| Culture medium | 256 ± 42 | 82 ± –17 | ||||||
Figure 5.
(A) IL-12 and (B) IL-6 secretion in BALB/c mouse spleen cell cultures induced by immunomers at a concentration of 1.0 µg/ml. M represents control in the absence of CpG DNA.
Effect of multiple linkers in immunomers on immunostimulatory activity
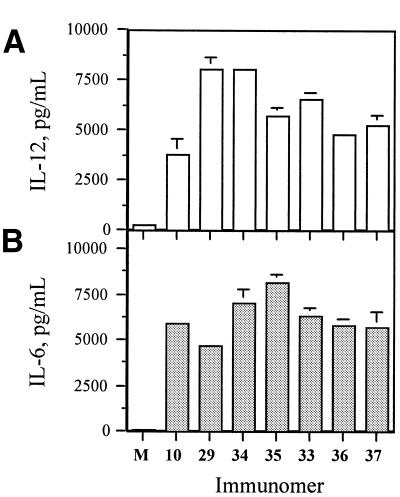
Since the use of longer linkers showed reduced immunostimulatory activity, we tested whether incorporation of multiple linkers would be preferable. We synthesized immunomers 34 and 35 with two and three copies of C3-linker (5), respectively, and 36 and 37 with two and three copies of the abasic linker (9), respectively (Table 1). All four immunomers induced concentration-dependent IL-12 and IL-6 secretion in BALB/c mouse spleen cell cultures (Table 4). When compared at 1.0 µg/ml concentration, the presence of two and three C3-linkers in immunomers 34 and 35 maintained or reduced IL-12 secretion but increased IL-6 secretion considerably compared with one C3-linker containing 29 (Fig. 6). These results suggest that it may be possible to modulate cytokine secretion by incorporation of an appropriate number of C3-linkers in immunomers.
Table 4. Effect of multiple linkers on cytokine secretion in BALB/c mouse spleen cell cultures.
| CpG DNA | IL-12 (pg/ml) ± SD | IL-6 (pg/ml) ± SD | ||||||
|---|---|---|---|---|---|---|---|---|
| 0.1 µg/ml | 0.3 µg/ml | 1.0 µg/ml | 3.0 µg/ml | 0.1 µg/ml | 0.3 µg/ml | 1.0 µg/ml | 3.0 µg/ml | |
| 10 | 310 ± 1 | 1962 ± 78 | 3780 ± 805 | 6084 ± 1390 | 181 ± 57 | 582 ± 143 | 5981 ± 138 | 13879 ± 612 |
| 29 | 4118 ± 1099 | 5768 ± 1250 | 8042 ± 632 | 7414 ± 457 | 421 ± 30 | 806 ± 131 | 4725 ± 291 | 15093 ± 636 |
| 34 | 4005 ± 174 | 6488 ± 1123 | 8112 ± 270 | 6858 ± 251 | 276 ± 26 | 2581 ± 285 | 7033 ± 743 | 15958 ± 2371 |
| 35 | 3917 ± 677 | 6156 ± 728 | 5806 ± 365 | 6782 ± 539 | 660 ± 144 | 1908 ± 268 | 8223 ± 378 | 12319 ± 468 |
| 33 | 778 ± 136 | 4854 ± 498 | 6563 ± 358 | 8803 ± 666 | 294 ± 107 | 587 ± 56 | 6359 ± 372 | 17006 ± 1274 |
| 36 | 1812 ± 150 | 4879 ± 432 | 4874 ± 170 | 6126 ± 253 | 331 ± 18 | 1348 ± 128 | 5823 ± 330 | 12888 ± 1186 |
| 37 | 2994 ± 46 | 5293 ± 544 | 5341 ± 456 | 5602 ± 91 | 408 ± 8 | 1310 ± 82 | 5764 ± 748 | 12470 ± 1373 |
| Culture medium | 256 ± 42 | 82 ± –17 | ||||||
Figure 6.
(A) IL-12 and (B) IL-6 secretion in BALB/c mouse spleen cell cultures induced by immunomers at a concentration of 1.0 µg/ml. M represents control in the absence of CpG DNA.
The incorporation of multiple abasic linkers in immunomers (36 and 37) reduced both IL-12 and IL-6 secretion compared with 33, which contained a single abasic linker (Fig. 6).
5′ end of CpG DNA is required for activation of NF-κB and subsequent immunostimulatory effects
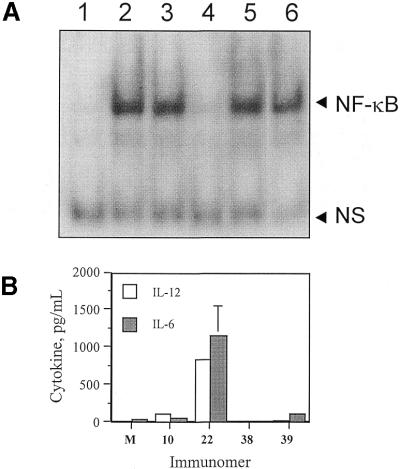
CpG DNA has been shown to activate a number of signaling pathways, including the NF-κB pathway, which plays a critical role in up-regulation of cytokine gene expression (33). We studied the activation of NK-κB in J774 cell nuclear extracts by immunomers 10, 22, 38, and 39 (Table 1) using EMSA. Figure 7A shows the results obtained. CpG DNA 10 activated NF-κB as indicated by the presence of a complex in lane 3 as is the case with LPS treated cells in lane 2 (Fig. 7A). Immunomer 22, which has two 5′-accessible ends, also activated NF-κB as indicated by the presence of the appropriate complex in lane 5. In contrast, 5′–5′-linked immunomer 38 failed to activate the transcription factor NF-κB in J774 cells (lane 4, Fig. 7A). A 34mer linear CpG DNA (39) with two CpG dinucleotides separated by 18 nt also activated the transcription factor NF-κB (see complex band in lane 6, Fig. 7A). This suggests that the enhanced activity of immunomer(s) (22) is not as a result of the presence of multiple CpG dinucleotides, but of the presence of accessible 5′ ends.
Figure 7.
(A) Activation of NF-κB in J774 cells by different CpG DNAs. J774 cells were stimulated with LPS (lane 2), CpG DNAs 10 (lane 3), 38 (lane 4), 22 (lane 5) and 39 (lane 6), nuclear extracts were prepared and analyzed on a native polyacrylamide gel as described in Materials and Methods. Lane 1 represents medium treated control. NS stands for non- specific band. NF-κB band is indicated with an arrow. (B) Induction of IL-12 and IL-6 secretion by immunomers 10, 22, 38 and 39 in J774 cell cultures. M represents control in the absence of CpG DNA.
Immunomer 38, lacking an accessible 5′ end, induced 11 ± 1 and 7 ± 5 pg/ml of IL-12 and IL-6 secretion, respectively, in J774 cell culture assays at 3 µg/ml concentration (Fig. 7B). This result further confirms the inability of 38 to activate NF-κB in this cell culture system. CpG DNA 39, which has two CpG dinucleotides, induced 35 ± 14 and 125 ± 10 pg/ml of IL-12 and IL-6, respectively, at a concentration of 3 µg/ml in J774 cell cultures.
DISCUSSION
CpG DNAs have been shown as potential immune adjuvants (34,35). Their use as antiallergens (36,37) and for the treatment of infectious diseases (38,39) and cancer (40,41) has been demonstrated. Our earlier studies demonstrated that an accessible 5′ end is required for immunostimulatory activity and the blocking of the 5′ end by attaching two CpG DNAs via their 5′ ends abrogates immunostimulatory activity (24,25). In the present studies, we examined the use of linkers between CpG DNA strands and the required length of immunomers for optimal immunostimulatory activity. The use of linkers for tethering immunomers permitted development of a fast and convenient parallel synthesis approach for producing immunomers. The use of the parallel synthesis approach resulted in higher yields of the immunomer products with improved purity.
The present results support our earlier finding that a 3′–3′-linked immunomer is a more potent immunostimulator than linear CpG DNA containing one or two CpG dinucleotides (24,25). In addition, these results also provided further insight into the immunological effects of CpG DNA and possible molecular basis for recognition of CpG DNA by its receptor. Several studies showed that the presence of multiple CpG dinucleotides increased immunostimulatory activity (42). Immunomers used here inherently contain at least twice as many CpG dinucleotides per molecule as the parent CpG DNA 10. The higher immunostimulatory activity of immunomer 22 compared with CpG DNA 10 could be as a result of the increased number of CpG dinucleotides per molecule. However, the different activities of 3′–3′- and 5′–5′- immunomers (with two or three branches) and 34mer CpG DNA 39, containing two or more CpG dinucleotides, in J774 cell cultures for NF-κB activation and cytokine secretion cannot be explained by the multiple CpG dinucleotides in immunomers. Immunomers 13, 38 and CpG DNA 39 (34mer) also contained two or more CpG dinucleotides but showed lower or no activation of NF-κB and less or no induction of cytokine secretion in J774 cell cultures. Taken together, enhanced activity of immunomers results from the 3′–3′-linked immunomer structure rather than the number of CpG dinucleotides present.
Upon internalization, CpG DNA is recognized by TLR9 with initiation of the signaling cascade that activates NF-κB and expression of cytokines (43,44). However, the molecular mechanism of recognition between CpG DNA and TLR9 is not known. The present results, at least to some extent, suggest that TLR9 reads the CpG DNA sequences from the 5′ to the 3′ end. Blocking the 5′ end by 5′–5′-linking of two CpG DNAs abrogates recognition of CpG DNA by TLR9 and subsequent events such as NF-κB activation and cytokine secretion. The blocking of the immunostimulatory activity also depends on the size of the blocking group conjugated at the 5′ end of the CpG DNA as shown recently (25). Immunomers having two free 5′ ends might be recognized by TLR9 at either or both 5′ ends. The increased immunostimulatory activity of immunomers, however, suggests that they are recognized by the receptors at both 5′ ends. The failure of 5′–5′-linked CpG DNA to activate NF-κB and induce cytokine secretion suggests that the receptor is unable to recognize the molecule lacking an accessible 5′ end even though it has two appropriate CpG dinucleotides.
In the case of CpG DNA 39, though it has two CpG dinucleotides, it is no more immunostimulatory than a CpG DNA with only a single copy. It appears that the recognition of one of the CpG dinucleotides by the receptor may preclude recognition of the second CpG dinucleotide by a second receptor, because the molecule has a single accessible 5′ end.
The immunostimulatory activity of different immunomers may be affected by the impact of 3′–3′- and 5′–5′-linkages on cellular uptake. Our recent cellular uptake studies using the 5′- or 3′-end fluorescein-labeled CpG DNAs in J774 cell cultures suggested that the uptake is not affected by whether the DNA molecule has a free 3′ or 5′ end (25). Therefore, it is reasonable to assume both 3′–3′- and 5′–5′-linked immunomers are taken up by cells similarly and differences in immunostimulatory activity result from structural effects on receptor recognition and/or binding.
Degradation of oligonucleotides in vivo occurs primarily by 3′-exonucleases acting from the 3′ ends (45–47). Blocking groups, ligands or sequences forming secondary structures at the 3′ end of oligonucleotides increase metabolic stability (48–52). Ligation of two CpG DNAs through their 3′ ends could enhance nuclease stability and, thereby, immunostimulatory activity in the present study. If so, 5′–5′-linked immunomers should have similar or higher nuclease stability compared with CpG DNA 10 as oligonucleotides are also degraded from the 5′ end though to a lesser extent. However, 5′–5′-linked immunomer 38 failed to induce immunostimulatory activity despite expected equal, if not higher, nuclease stability compared with parent CpG DNA 10. Therefore, the data strongly argue that the differences in immunostimulatory activity of 3′–3′- and 5′–5′-linked DNAs do not arise from differences in nuclease stability or their cellular uptake but from the accessibility to the 5′ end for receptor recognition and/or binding.
An immunomer with 11-nt segments shows increased immunostimulatory activity. Further reduction in length of immunomers lowered immunostimulatory activity compared with 11mer immunomer, but still maintained similar or higher activity than linear CpG DNA 10 in in vitro and in vivo assays. These results suggest that the receptor requires a certain length of nucleotides for recognition.
CpG DNA stimulates secretion of several cytokines by B cells, macrophages and dendritic cells (5–8). Recently it has been shown that CpG DNAs containing different backbone and sequences elicit different cytokine secretion properties, perhaps through targeting different immune cell populations (53–55). Although the mechanism is not clear, in the present study, we found altered cytokine secretion profiles by incorporating one or more copies of several linkers but without changing the backbone or sequence of immunomers. For example, incorporation of a single C3-linker (immunomer 29) induced higher levels of IL-12 secretion, which is a Th1 type cytokine. Such an immunomer might be useful for cancer and asthma therapies or as an adjuvant with vaccines, antibodies and allergens. In contrast, when lower levels of IL-12 and higher levels of IL-6 are required, incorporation of two or more C3-linkers is appropriate.
Taken together, the present results suggest that 3′–3′-linked CpG DNAs are potent immunostimulators because of the presence of two 5′-accessible ends for receptor recognition. Additionally, the 3′–3′-linkage provides increased stability against nucleases (26,27,51). Linking of CpG DNAs through their 5′–5′ ends blocks the immunostimulatory activity, though they possess required CpG dinucleotides in appropriate sequence context, nuclease stability and cellular uptake properties. Our preliminary results indicate potent immunostimulatory activity of immunomers containing synthetic YpG and CpR motifs instead of CpG. Additional immunomodulatory moieties (18–22) reported can also be incorporated in immunomers to modulate cytokine secretion profiles. We are currently examining the immunomers containing synthetic YpG and CpR motifs in human specific sequences for immunostimulatory responses.
REFERENCES
- 1.Tokunaga T., Yamamoto,H., Shimada,S., Abe,H., Fukuda,T., Fujisawa,Y., Furutani,Y., Yano,O., Kataoka,T., Sudo,T., Makiguchi,N. and Suganuma,T. (1984) Antitumor activity of deoxyribonucleic acid fraction from Mycobacterium bovis BCG.I. Isolation, physicochemical characterization, and antitumor activity. J. Natl Cancer Inst., 72, 955–962. [PubMed] [Google Scholar]
- 2.Messina J.P., Gilkeson,G.S. and Pisetsky,D.S. (1991) Stimulation of in vitro murine lymphocyte proliferation by bacterial DNA. J. Immunol., 147, 1759–1764. [PubMed] [Google Scholar]
- 3.Krieg A.M., Yi,A.K., Matson,S., Waldschmidt,T.J., Bishop,G.A., Teasdale,R., Koretzky,G.A. and Klinman,D.M. (1995) CpG motifs in bacterial DNA trigger direct B-cell activation. Nature, 374, 546–549. [DOI] [PubMed] [Google Scholar]
- 4.Sato Y., Roman,M., Tighe,H., Lee,D., Corr,M., Nguyen,M.D., Silverman,G.J., Lotz,M., Carson,D.A. and Raz,E. (1996) Immunostimulatory DNA sequences necessary for effective intradermal gene immunization. Science, 273, 352–354. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto S., Kuramoto,E., Shimada,S. and Tokunaga,T. (1988) In vitro augmentation of natural killer cell activity and production of interferon-α/β and -γ with deoxyribonucleic acid fraction from Mycobacterium bovis BCG. Jpn. J. Cancer Res., 79, 866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halpern M.D., Kurlander,R.J. and Pisetsky,D.S. (1996) Bacterial DNA induces murine interferon-γ production by stimulation of interleukin-12 and tumor necrosis factor-α. Cell. Immunol., 167, 72–78. [DOI] [PubMed] [Google Scholar]
- 7.Klinman D.M., Yi,A.K., Beaucage,S.L., Conover,J. and Krieg,A.M. (1996) CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon-γ. Proc. Natl Acad. Sci. USA, 93, 2879–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Q., Temsamani,J., Zhou,R.Z. and Agrawal,S. (1997) Pattern and kinetics of cytokine production following administration of phosphorothioate oligonucleotides in mice. Antisense Nucleic Acid Drug Dev., 7, 495–502. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto S., Yamamoto,T., Kataoka,T., Kuramoto,E., Yano,O. and Tokunaga,T. (1992) Unique palindromic sequences in synthetic oligonucleotides are required to induce IFN and augment IFN-mediated natural killer activity. J. Immunol., 148, 4072–4076. [PubMed] [Google Scholar]
- 10.Tokunaga T., Yano,O., Kuramoto,E., Kimura,Y., Yamamoto,T., Kataoka,T. and Yamamoto,S. (1992) Synthetic oligonucleotides with particular base sequences from the cDNA encoding proteins of Mycobacterium bovis BCG induce interferons and activate natural killer cells. Microbiol. Immunol., 36, 55–66. [DOI] [PubMed] [Google Scholar]
- 11.Liang H., Nishioka,Y., Reich,C.F., Pisetsky,D.S. and Lipsky,P.E. (1996) Activation of human B cells by phosphorothioate oligodeoxynucleotides. J. Clin. Invest., 98, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurunathan S., Klinman,D.M. and Seder,R.A. (2000) DNA vaccines: immunology, application, and optimization. Annu. Rev. Immunol., 18, 927–974. [DOI] [PubMed] [Google Scholar]
- 13.Krieg A.M. (2002) CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol., 20, 709–760. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal S. and Kandimalla,E.R. (2002) Medicinal chemistry and therapeutic potential of CpG DNA. Trends Mol. Med., 8, 114–121. [DOI] [PubMed] [Google Scholar]
- 15.Hemmi H., Takeuchi,O., Kawai,T., Kaisho,T., Sato,S., Sanjo,H., Matsumoto,M., Hoshino,K., Wagner,H., Takeda,K. and Akira,S. (2000) A Toll like receptor recognizes bacterial DNA. Nature, 408, 740–745. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Q., Temsamani,J., Iadarola,P.L., Jiang,Z. and Agrawal,S. (1996) Effect of different chemically modified oligodeoxynucleotides on immune stimulation. Biochem. Pharmacol., 51, 173–182. [DOI] [PubMed] [Google Scholar]
- 17.Kandimalla E.R., Yu,D. and Agrawal,S. (2002) Towards optimal design of second-generation immunomodulatory oligonucleotides. Curr. Opin. Mol. Ther., 4, 122–129. [PubMed] [Google Scholar]
- 18.Zhao Q., Yu,D. and Agrawal,S. (1999) Site of chemical modifications in CpG containing phosphorothioate oligodeoxynucleotide modulates its immunostimulatory activity. Bioorg. Med. Chem. Lett., 9, 3453–3458. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Q., Yu,D. and Agrawal,S. (2000) Immunostimulatory activity of CpG containing phosphorothioate oligodeoxynucleotide is modulated by modification of a single deoxynucleoside. Bioorg. Med. Chem. Lett., 10, 1051–1054. [DOI] [PubMed] [Google Scholar]
- 20.Yu D., Kandimalla,E.R., Zhao,Q., Cong,Y. and Agrawal,S. (2002) Immunostimulatory properties of phosphorothioate CpG DNA containing both 3′–5′- and 2′–5′-internucleotide linkages. Nucleic Acids Res., 30, 1613–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu D., Kandimalla,E.R., Zhao,Q., Cong,Y. and Agrawal,S. (2001) Immunostimulatory activity of CpG oligonucleotides containing non-ionic methylphosphonate linkages. Bioorg. Med. Chem., 9, 2803–2808. [DOI] [PubMed] [Google Scholar]
- 22.Yu D., Kandimalla,E.R., Zhao,Q., Cong,Y. and Agrawal,S. (2001) Modulation of immunostimulatory activity of CpG oligonucleotides by site-specific deletion of nucleobases. Bioorg. Med. Chem. Lett., 11, 2263–2267. [DOI] [PubMed] [Google Scholar]
- 23.Kandimalla E.R., Yu,D., Zhao,Q. and Agrawal,S. (2001) Effect of chemical modifications of cytosine and guanine in a CpG-motif of oligonucleotides: structure-immunostimulatory activity relationships. Bioorg. Med. Chem., 9, 807–813. [DOI] [PubMed] [Google Scholar]
- 24.Yu D., Zhao,Q., Kandimalla,E.R. and Agrawal,S. (2000) Accessible 5′-end of CpG-containing phosphorothioate oligodeoxynucleotides is essential for immunostimulatory activity. Bioorg. Med. Chem. Lett., 10, 2585–2588. [DOI] [PubMed] [Google Scholar]
- 25.Kandimalla E.R., Bhagat,L., Yu,D., Cong,Y., Tang,J. and Agrawal,S. (2002) Conjugation of ligands at the 5′-end of CpG DNA affects immunostimulatory activity. Bioconjug. Chem., 13, 966–974. [DOI] [PubMed] [Google Scholar]
- 26.Kandimalla E.R., Agrawal,S., Venkataraman,S. and Sasisekharan,V. (1995) Single strand targeted triplex formation: parallel-stranded DNA hairpin duplexes for targeting pyrimidine strands. J. Am. Chem. Soc., 117, 6416–6417. [Google Scholar]
- 27.Chaix C., Iyer,R.P. and Agrawal,S. (1996) 3′–3′-Linked oligonucleotides synthesis and stability studies. Bioorg. Med. Chem. Lett., 6, 827–832. [DOI] [PubMed] [Google Scholar]
- 28.Iyer R.P., Egan,W., Regan,J.B. and Beaucage,S.L. (1990) 3H-1,2 benzodithiole-3-one1,1-dioxide as an improved sulfurizing reagent in the solid-phase synthesis of oligodeoxyribonucleoside phosphorothioates. J. Am. Chem. Soc., 112, 1253–1254. [Google Scholar]
- 29.Puglisi J.D. and Tinoco,I.,Jr (1989) Absorbance melting curves of RNA. Methods Enzymol., 180, 304–325. [DOI] [PubMed] [Google Scholar]
- 30.Branda R.F., Moore,A.L., Mathews,L., McCormack,J.J. and Zon,G. (1993) Immune stimulation by an antisense oligomer complementrary to the rev gene of HIV-1. Biochem. Pharmacol., 45, 2037–2043. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber E., Matthias,P., Muller,M.M. and Schaffner,W. (1989) Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res., 17, 6419–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 33.Stacey K.J., Sweet,M.J. and Hume,D.A. (1996) Macrophages ingest and are activated by bacterial DNA. J. Immunol., 157, 2116–2122. [PubMed] [Google Scholar]
- 34.O’Hagan D.T., MacKichan,M.L. and Singh,M. (2001) Recent developments in adjuvants for vaccines against infectious diseases. Biomol. Eng., 18, 69–85. [DOI] [PubMed] [Google Scholar]
- 35.Leitner W.W., Hammerl,P. and Thalhamer,J. (2001) Nucleic acid for the treatment of cancer: genetic vaccines and DNA adjuvants. Curr. Pharm. Des., 7, 1641–1667. [DOI] [PubMed] [Google Scholar]
- 36.Spiegelberg H.L. and Raz,E. (2002) DNA-based approaches to the treatment of allergies. Curr. Opin. Mol. Ther., 4, 64–71. [PubMed] [Google Scholar]
- 37.Kline J.N. and Ballas,Z.K. (2002) DNA immunomodulation of asthma. Clin. Allergy Immunol., 16, 551–564. [PubMed] [Google Scholar]
- 38.Lewis E.J., Agrawal,S., Bishop,J., Chadwick,J., Cristensen,N.D., Cuthill,S., Dunford,P., Field,A.K., Francis,J., Gibson,V., Greenham,A.K., Kelly,F., Kilkushie,R., Kreider,J.W., Mills,J.S., Mulqueen,M., Roberts,N.A., Roberts,P. and Szymkowski,D.E. (2000) Non-specific antiviral activity of antisense molecules targeted to the E1 region of human papillomavirus. Antiviral Res., 48, 187–196. [DOI] [PubMed] [Google Scholar]
- 39.Masihi K.N. (2001) Fighting infection using immunomodulatory agents. Expert Opin. Biol. Ther., 1, 641–653. [DOI] [PubMed] [Google Scholar]
- 40.Warren T.L. and Weiner,G.J. (2002) Synergism between cytosine-guanine oligodeoxynucleotides and monoclonal antibody in the treatment of lymphoma. Semin. Oncol., 29 (1 Suppl. 2), 93–97. [PubMed] [Google Scholar]
- 41.Hafner M., Zawatzky,R., Hirtreiter,C., Buurman,W.A., Echtenacher,B., Hehlgans,T. and Mannel,D.N. (2002) Antimetastatic effect of CpG DNA mediated by type I IFN. Cancer Res., 61, 5523–5528. [PubMed] [Google Scholar]
- 42.Hartmann G., Weeratna,R.D., Ballas,Z.K., Payette,P., Blackwell,S., Suparto,I., Rasmussen,W.L., Waldschmidt,M., Sajuthi,D., Purcell,R.H., Davis,H.L. and Krieg,A.M. (2000) Delineation of a CpG phosphorothioate oligodeoxynucleotide for activating primate immune responses in vitro and in vivo. J. Immunol., 164, 1617–1624. [DOI] [PubMed] [Google Scholar]
- 43.Chuang T.H., Lee,J., Kline,L., Mathison,J.C. and Ulevitch,R.J. (2002) Toll-like receptor 9 mediates CpG-DNA signaling. J. Leukoc. Biol., 71, 538–544. [PubMed] [Google Scholar]
- 44.Wagner H. (2002) Interactions between bacterial CpG-DNA and TLR9 bridge innate and adaptive immunity. Curr. Opin. Microbiol., 5, 62–69. [DOI] [PubMed] [Google Scholar]
- 45.Temsamani J., Roskey,A., Chaix,C. and Agrawal,S. (1997) In vivo metabolic profile of a phosphorothioate oligodeoxyribonucleotide. Antisense Nucleic Acid Drug Dev., 7, 159–165. [DOI] [PubMed] [Google Scholar]
- 46.Temsamani J., Kubert,M. and Agrawal,S. (1995) Sequence identity of the n-1 product of a synthetic oligonucleotide. Nucleic Acids Res., 23, 1841–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agrawal S., Temsamani,J. and Tang,J.Y. (1991) Pharmacokinetics, biodistribution, and stability of oligodeoxynucleotide phosphorothioates in mice. Proc. Natl Acad. Sci. USA, 88, 7595–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Temsamani J., Tang,J.Y. and Agrawal,S. (1992) Capped oligodeoxynucleotide phosphorothioates. Pharmacokinetics and stability in mice. Ann. N. Y. Acad. Sci., 660, 318–320. [DOI] [PubMed] [Google Scholar]
- 49.Temsamani J., Tang,J.Y., Padmapriya,A., Kubert,M. and Agrawal,S. (1993) Pharmacokinetics, biodistribution, and stability of capped oligodeoxynucleotide phosphorothioates in mice. Antisense Res. Dev., 3, 277–284. [DOI] [PubMed] [Google Scholar]
- 50.Levis J.T. and Miller,P.S. (1994) Properties of exonuclease-resistant, psoralen- conjugated oligodeoxyribonucleotides in vitro and in cell culture. Antisense Res Dev., 4, 231–241. [DOI] [PubMed] [Google Scholar]
- 51.Yu D., Zhu,F.G., Bhagat,L.,Wang,H., Kandimalla,E.R., Zhang,R. and Agrawal,S. (2002) Potent CpG oligonucleotides containing phosphodiester linkages: in vitro and in vivo immunostimulatory properties. Biochem. Biophys. Res. Commun., 297, 83–90. [DOI] [PubMed] [Google Scholar]
- 52.Pandolfi D., Rauzi,F. and Capobianco,M.L. (1999) Evaluation of different types of end-capping modifications on the stability of oligonucleotides toward 3′- and 5′-exonucleases. Nucl. Nucl., 18, 2051–2069. [DOI] [PubMed] [Google Scholar]
- 53.Iho S., Yamamoto,T., Takahashi,T. and Yamamoto,S. (1999) Oligodeoxynucleotides containing palindrome sequences with internal 5′-CpG-3′ act directly on human NK and activated T cells to induce IFN-γ production in vitro. J. Immunol., 163, 3642–3652. [PubMed] [Google Scholar]
- 54.Krug A., Rothenfusser,S., Hornung,V., Jahrsdorfer,B., Blackwell,S., Ballas,Z.K., Endres,S., Krieg,A.M. and Hartmann,G. (2001) Identification of CpG oligonucleotide sequences with high induction of IFN-α/β in plasmacytoid dendritic cells. Eur. J. Immunol., 31, 2154–2163. [DOI] [PubMed] [Google Scholar]
- 55.Verthelyi D., Ishii,K., Gursel,M., Takeshita,F. and Klinman,D.M. (2001) Human peripheral blood cells differentially recognize and respond to two distinct CpG motifs. J. Immunol., 166, 2372–2377. [DOI] [PubMed] [Google Scholar]