Abstract
Oligonucleotide arrays are powerful tools to study changes in gene expression for whole genomes. These arrays can be synthesized by adapting photolithographic techniques used in microelectronics. Using this method, oligonucleotides are built base by base directly on the array surface by numerous cycles of photodeprotection and nucleotide addition. In this paper we examine strategies to reduce the number of synthesis cycles required to construct oligonucleotide arrays. By computer modeling oligonucleotide synthesis, we found that the number of required synthesis cycles could be significantly reduced by focusing upon how oligonucleotides are chosen from within genes and upon the order in which nucleotides are deposited on the array. The methods described here could provide a more efficient strategy to produce oligonucleotide arrays.
INTRODUCTION
The advent of genomics has facilitated a shift in molecular biology from studies of the expression of single genes to studies of whole-genome expression profiles. Genome-wide expression profiling is a powerful tool being applied in gene identification, drug discovery, pathological and toxicological mechanisms and clinical diagnosis. By simultaneously measuring the expression of thousands of genes, researchers can get a picture of the transcriptional profile of a whole genome in a given physiological condition. One of the leading technologies for expression profiling is oligo or gene chips. Oligo chips consist of oligonucleotides immobilized upon a support substrate, commonly silica. They have certain advantages over other technologies. Since all of the oligomers can be carefully designed, inter-feature variability is low. Also, oligo chips can be designed to contain several oligonucleotides representing each gene, allowing more quantitative analysis of expression levels.
One of the most successful methods used to make oligonucleotide chips is an adaptation of photolithographic techniques used in microelectronics (http://www.affymetrix.com). Initially, a specific mask is fabricated for each cycle of nucleotide addition that permits light to penetrate only at positions where nucleotides are to be added. A synthesis cycle consists of shining light through the mask onto the chip surface. The positions where light passes through the mask and reaches the chip are activated for synthesis by the removal of a photolabile protective group from the exposed end of the oligonucleotide. Thus, the pattern in which light penetrates the masks directs the base by base synthesis of oligonucleotides on a solid surface (1). After photodeprotection the chip is washed in a solution containing a single nucleotide (A, C, G or T) that binds to oligonucleotides at the deprotected positions. This method results in the in situ synthesis of oligonucleotides on an array surface. Light-directed chemical synthesis has been used to produce arrays with as many as 300 000 features (up to 1 000 000 on experimental products) with minimal cross-hybridization or inter-feature variability (2).
When using photolithography to make DNA arrays, the series of masks and the sequence in which nucleotides are added defines the oligonucleotide products and their locations. Because a separate photolithographic mask must be designed for each synthesis cycle it is advantageous to build oligo chips in as few deposition cycles as possible. To this end, we developed an algorithm to reduce the number of cycles required to build an array of oligonucleotides. If the length of the oligomer is N and the number of possible subunits of the oligomer is K, our goal was to build a set of oligomers in as many fewer than N × K steps as possible. The simplest strategy for the in situ synthesis of oligonucleotides upon an array surface is to first add A everywhere it is needed for the first base, then C, G and T. Using this strategy, a set of oligonucleotides of length N can be synthesized in a maximum of 4N steps (3). An array of 25mer oligonucleotides thus would take 100 cycles to build.
Our strategy reduced the number of required synthesis cycles by focusing upon two areas of improvement. First, we focused upon how to best select regions of each gene to be used for oligonucleotides. From within each gene we selected oligonucleotides that could be deposited most efficiently. Once the set of oligonucleotides had been selected they could be deposited on the array surface. The second part of our strategy was to determine a deposition order of nucleotide bases on the array surface with a minimum number of steps. We allowed the deposition order to vary so as to add the most common base at each point in the deposition process. During deposition we added bases at every available position and thus allowed oligonucleotides to be built at different rates. Thus, after four cycles, a given oligonucleotide could theoretically have no bases added and another have four bases. By simultaneously optimizing oligonucleotide selection and deposition we significantly reduced the number of deposition cycles required to synthesize an oligonucleotide array.
MATERIALS AND METHODS
Our strategy consists of two basic parts. Initially, we focused upon selecting those oligonucleotides from each gene that could be most efficiently deposited upon the array. Second, we determined an order of oligonucleotide deposition that could efficiently deposit these oligonucleotides. The source code used in modeling is freely available and can be obtained by emailing tolonen@mit.edu.
Oligonucleotide selection
First, we determined a candidate set of unique 25mer oligonucleotides to be deposited on the array. As the input to our program, we arbitrarily selected the second chromosome of Arabidopsis thaliana (ftp://ncbi.nlm.nih.gov/ genbank/genomes/A_thaliana/CHR_II/). This chromosome is 19.6 Mb and contains 4036 genes. In this paper we modeled the deposition of the first 1000 genes on the chromosome that were >300 bp. However, our strategy could be applied to any number of genes in any genome. For each gene we chose five non-overlapping 25mer oligonucleotides to be deposited on the array. To define the source for each oligonucleotide we parsed the 3′ 300 bp into five 60 bp regions. Thus, each 60 bp region consisted of a total of 35 potential 25mers. We subjected each potential oligonucleotide to a series of simple tests for biological suitability. The tests required that each oligonucleotide be unique in the genome, have a GC content between 25 and 75% and have no region of self-complementarity of five or more bases at either end. In our data set, 2.7% of the 60 bp gene regions contained no suitable oligonucleotides. From the set of oligonucleotides that passed the tests, we then selected one oligonucleotide from each region. Thus, for 1000 genes, we selected a total of 5000 oligonucleotides that were evenly distributed across the 3′ region of each gene.
Modeling oligonucleotide construction
Once we had selected a complete set of oligonucleotides, the next step in our method was to evaluate how many deposition cycles were required to build each oligonucleotide in situ on an array surface. Broadly, our deposition strategy was to maximize the number of bases added at each step of the oligonucleotide synthesis. A position was defined as available if it was the next undeposited base in the oligonucleotide sequence. During each deposition cycle, we assumed that a specific base could be added only once at an available position. For example, even if the next two bases to be added to an oligonucleotide were CC, we added only one C at a time.
For each step of oligonucleotide construction, we identified the first available base in each oligonucleotide in the data set. We calculated the frequency of each base at this position and selected the most common base for deposition. This base was deposited for each oligonucleotide in which this base occupied the first position. In each of these oligonucleotides, we then incremented the next available position by one base. One loop of our program was analogous to one cycle of oligonucleotide deposition. The deposition subroutine continued to loop until we had calculated the total number of steps required to synthesize each oligonucleotide.
Optimizing oligonucleotide selection
The goal of this section was to see if selecting alternative oligonucleotides from the same gene region could streamline the deposition process. We investigated two strategies to optimize oligonucleotide selection, iterative re-selection and pooling of candidate oligonucleotides. Our iterative re- selection strategy identified those oligonucleotides that took the most steps to build, replaced them with an equivalent oligonucleotide from the same section of the same gene and tested if the new set of oligonucleotides could be deposited more efficiently. We viewed this process as analogous to an ‘oligonucleotide natural selection’ to weed out unfit oligonucleotides and replace them with potentially more fit substitutes. After completing an iteration of the deposition process, we knew the number of steps required to deposit each oligonucleotide. We identified the 75th percentile as the number of steps to produce 75% of the oligonucleotides. For example, if 75% of the oligonucleotides were deposited in 50 steps, we focused upon all oligonucleotides that took 51 or more steps to deposit. We then replaced all oligonucleotides above the 75th percentile with alternative oligonucleotides from the same gene region. We replaced oligonucleotides by going back to the input sequence and re-selecting an oligonucleotide that started one position downstream. If that oligonucleotide passed our biological suitability criteria it was used instead of the original oligonucleotide in the next iteration of the deposition process. If the replacement failed our suitability criteria, then we again replaced this oligonucleotide with one from one base downstream. Our goal was to converge upon a set of oligonucleotides that could be most efficiently deposited by repeated oligonucleotide re-selection.
Our second method of oligonucleotide optimization was to initially include all possible 25mer oligonucleotides in the data set passed to the deposition subroutine and then to select the oligonucleotide that is deposited in the fewest steps for each gene region. Thus, all 35 25mers from each gene region were initially included in the data set. When a single oligonucleotide was completed from a given gene region it was selected and the remaining oligonucleotides were deleted from the data set. After completing the deposition subroutine we had selected the oligonucleotide from each 60 bp region that could be deposited in the fewest steps. This method circumvented the need to iterate the oligonucleotide selection process.
RESULTS
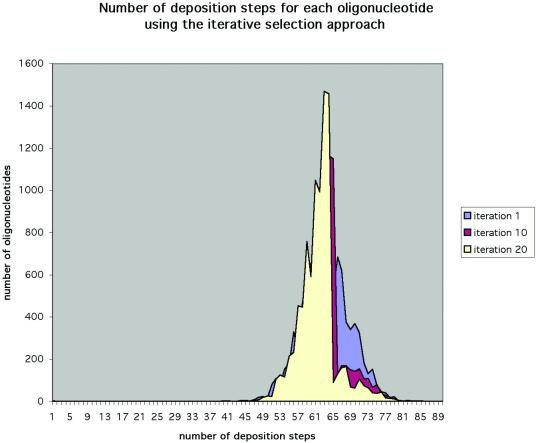
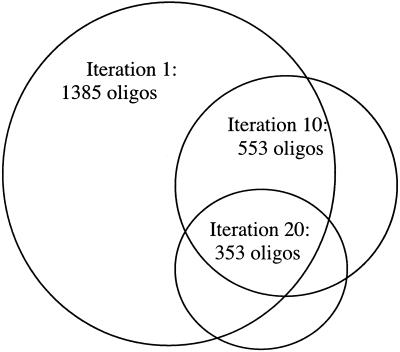
Our oligonucleotide selection and deposition strategy demonstrated that oligonucleotides can be synthesized in situ upon an array in many fewer than 4N steps. In our trial data set, we deposited all oligonucleotides in 83 steps. To further reduce the number of required steps, we investigated the effect of iterative replacement of the most costly oligonucleotides. We observed that across iterations the distribution became compressed and the mean number of steps decreased (Fig. 1). However, even when the oligonucleotide selection process was iterated 20 times, the number of steps required to complete the deposition process was not reduced. In fact, it increased by two cycles. While in the upper tail the distribution became reduced in size, we were unable to eliminate those oligonucleotides that required the most steps to build from the data set. In light of this result, we identified the gene regions that contained oligonucleotides above the 75th percentile. Because in the upper tail the distribution diminished in successive iterations, the number of oligonucleotides above the 75th percentile became smaller. It became clear that the oligonucleotides above the 75th percentile were coming from the same gene regions across iterations. Figure 2 is a Venn diagram showing that the most costly oligonucleotides came from the same gene regions across iterations. For example, of the 353 oligonucleotides above the 75th percentile in iteration 20, 263 were from the same gene regions represented in iteration 1.
Figure 1.
Distribution of the number of steps required to build each oligonucleotide across iterations. Data from iterations 1, 10 and 20 are shown. As the number of iterations increased, the upper tail of the distribution became compressed. However, the number of cycles required to build the entire oligonucleotide set did not decrease.
Figure 2.
The oligonucleotides requiring the most deposition cycles were from the same gene regions across iterations. This diagram shows overlap in the gene regions that contained oligonucleotides above the 75% percentile. Common oligonucleotides: iterations 1 and 10 share 421 common gene regions; iterations 1 and 20 share 263 gene regions; iterations 10 and 20 share 241 gene regions.
As an alternative means to select more efficient oligonucleotides, we investigated a pooling approach in which the initial data set consisted of all potential oligonucleotides from each gene region. We passed this complete data set to our deposition subroutine and when a single oligonucleotide from a given gene region was completed, it was selected and the remaining oligonucleotides from that gene region were deleted from the data set. We found that this strategy produced significant improvements (Fig. 3). Using this strategy, the entire set of oligonucleotides could be deposited in 73 steps. A summary comparing the results of these two strategies is shown in Table 1.
Figure 3.
Distribution of the number of steps required to build each oligonucleotide using the oligonucleotide pooling strategy.
Table 1. Summary of the synthesis cycles required to deposit oligonucle otides using the iterative and pooling strategies.
| Deposition strategy | Median cycles | Maximum cycles |
|---|---|---|
| 1 iteration | 60 | 83 |
| 10 iterations | 60 | 85 |
| 20 iterations | 59 | 85 |
| Pool | 54 | 73 |
Iterative results are shown for the first, tenth and twentieth iterations. For each strategy, the number of cycles required to deposit 50% (median) of oligonucleotides and the number of cycles to deposit all the oligonucleotides (maximum) are shown.
DISCUSSION
Our results demonstrate that both oligonucleotide selection and nucleotide deposition order are important steps towards minimizing the number of steps required to construct oligonucleotides in situ upon an array surface. From within a specific gene region, selecting one oligonucleotide versus another can have a significant impact upon the number of deposition steps required. Further, the opportunistic deposition of bases in which the most common next base is added and oligonucleotides may grow at different rates will almost always result in fewer deposition steps than when all oligonucleotides are built at the same rate. Our strategy minimized the number of required deposition steps by attempting to simultaneously optimize oligonucleotide selection and deposition. Because the photolithographic synthesis of oligonucleotides requires expensive reagents and a custom mask for each step of synthesis, our methods could reduce the time and money required to synthesize these arrays.
Our oligonucleotide selection program required that each oligonucleotide pass a set of criteria for biological suitability before it was accepted into the data set. Our criteria included uniqueness in the genome, moderate CG content, no self-complementarity and availability of a unique mismatch oligonucleotide. However, our process of oligonucleotide selection was by no means rigorous. We did not explicitly test whether the melting temperatures of the oligonucleotides were similar. Also, cross-hybridization might be better prevented by searching the genome for regions of significant local alignment rather than perfect matches.
Our deposition strategy of adding the most common base at each position can be thought of as similar to a chess game. At each stage in the game we selected the move that provided the greatest marginal benefit. However, an algorithm that could predict a few steps into the future might be a more optimal deposition solution. It is easy to see that the number of pathways for N steps into the future increases at 4N and rapidly becomes computationally prohibitive. However, we thought that if we calculated all the possibilities for a few steps ahead that this might yield some improvement. To this end, we tested two look-ahead strategies. First, we calculated all the possibilities for four moves ahead and chose the best path for these four moves. Second, we calculated the best path for the next four steps, executed a single move, and then re-evaluated the next move based upon the next four steps. Unfortunately, neither strategy yielded an improvement.
We found that strategies relating to oligonucleotide selection can result in a more efficient deposition. By replacing all the oligonucleotides above the 75th percentile, we hoped to gradually eliminate the most costly oligonucleotides from the data set. We examined how the distribution of synthesis steps required for each oligonucleotide changed as the number of iterations increased (Fig. 1). We found that reiteration compressed the distribution and reduced the mean, but it did not reduce the number of cycles needed to deposit the entire data set. We believe that this is due to certain genes that have a small pool of available oligonucleotides. Thus, even if the process is reiterated, costly oligonucleotides from these genes cannot be removed from the data set. In light of these results, we investigated a different strategy in which all the available oligonucleotides were pooled into the initial data set and passed to the deposition subroutine. When a single oligonucleotide from a given gene region was completed, it was selected and the remaining oligonucleotides from that gene region were deleted. We found that this strategy significantly reduced the number of required deposition steps (Fig. 3). Perhaps this is because it is less constrained by those genes with fewer available oligonucleotides.
Our deposition strategy allowed the oligonucleotides to be built at different rates. Thus, at any point in the deposition process the length of an oligonucleotide could be different from that of its neighbors. Hubbell et al. (4) wrote that it is usually desirable for the synthesis of adjacent probes to vary in as few synthesis cycles as possible. They explained that an undesirable ‘delta edge’ is produced when a monomer is added to a synthesis region but not to an adjacent region. To avoid delta edges, it may be important to distribute the oligonucleotides on the chip surface so that adjacent probes are built at similar rates.
With regard to oligonucleotide selection, there might be an unavoidable conflict between choosing oligonucleotides to minimize cross-hybridization and to lower the number of steps required for deposition. Oligonucleotide probes will more efficiently hybridize with only a single mRNA transcript if they represent regions of the genome that are specific to that gene. On the other hand, a set of oligonucleotides can be built in fewer steps if the oligonucleotides are more similar to each other and thus represent areas that are more conserved among genes. In our oligonucleotide selection procedure, we tested to ensure that each oligonucleotide was unique in the genome. However, the re-selection of oligonucleotides likely selected for oligonucleotides that were more similar to the rest of the data set. Thus, our method might result in increased cross-hybridization on the chip.
In conclusion, the optimal set of oligonucleotides can be deposited on an array in a minimum number of steps while retaining the ability to quantify the abundance of each transcript. Our process produces a set of oligonucleotides that can be deposited in many fewer than 4N steps. In the future, we would like to explore whether this process builds a chip that can effectively monitor changes in global mRNA expression.
Acknowledgments
ACKNOWLEDGEMENTS
We thank G. M. Church and S. Kasif for their helpful suggestions. We would also like to thank A. Derti for his insights and programming expertise.
REFERENCES
- 1.Fodor S.P., Read,J.L., Pirrung,M.C., Stryer,L., Lu,A.T. and Solas,D. (1991) Light-directed, spatially addressable parallel chemical synthesis. Science, 251, 767–773. [DOI] [PubMed] [Google Scholar]
- 2.Lipshutz R.J., Fodor,S.P., Gingeras,T.R. and Lockhart,D.J. (1998) High density synthetic oligonucleotide arrays. Nature Genet., 21 (suppl.), 20–24. [DOI] [PubMed] [Google Scholar]
- 3.Chee M., Yang,R., Hubbell,E., Berno,A., Huang,X.C., Stern,D., Winkler,J., Lockhart,D.J., Morris,M.S. and Fodor,S.P. (1996) Accessing genetic information with high-density DNA arrays. Science, 274, 610–614. [DOI] [PubMed] [Google Scholar]
- 4.Hubbell E.A., Morris,M.S. and Winkler,J.L. (1999) Computer-aided engineering system for design of sequence arrays and lithographic masks. US patent 5,856,101.