Abstract
We describe a rapid and inexpensive method to monitor the kinetics of small RNA-cleaving deoxyribozymes, based on the exogenous fluorophore ethidium bromide. Ethidium binds preferentially to double-stranded nucleic acids, and its fluorescence emission increases dramatically upon intercalation. Thus, ethidium can be used in single-turnover experiments to measure both annealing of the deoxyribozyme to its substrate and release of the products. Under conditions in which dissociation of the product is fast compared with cleavage, the apparent rate of product release reflects the cleavage step. The method was developed for characterizing the so-called 8-17 catalytic DNA, but its general applicability in the deoxyribozyme field was verified using the 10-23 RNA-cleaving construct. Catalysis by both deoxyribozymes was not inhibited in the presence of substoichiometric amounts of ethidium, and the rates obtained through the ethidium assay were virtually identical to the rates determined using radiolabeled substrates. In contrast, the assay cannot be applied to the large, structured ribozymes, and its use to study the kinetics of the small hammerhead ribozyme was hampered by the presence on the catalyst of at least one high-affinity ethidium binding site.
INTRODUCTION
In recent years, application of in vitro selection protocols has led to the discovery of many catalytic DNA motifs that, by analogy with the better known ribozymes, have been termed deoxyribozymes (reviewed in 1–4). Especially interesting from an applicative viewpoint has been the identification of several small RNA-cleaving deoxyribozymes (5–14), which can be designed to recognize and cut almost any given target sequence, thanks to their substrate-binding ‘arms’ that are variable in composition and length. These catalysts are being increasingly employed in applications that range from molecular therapy (reviewed in 15–17) to the development of molecular-scale computational devices (18) and from analytical biotechnology (19,20) to the manipulation and analysis of RNA (21,22).
In connection with the manifold applications of RNA-cleaving deoxyribozymes, there is currently a demand for techniques that allow a rapid and high-throughput kinetic characterization of these catalysts. Indeed, whereas all experimental works on DNA enzymes involve the measurement of RNA cleavage reactions, monitoring such reactions in real time and on a large scale faces some inherent difficulties.
The most established method for measuring the kinetics of deoxyribozyme reactions is based on the use of radiolabeled substrates. This method entails the cumbersome sampling, electrophoretic separation, and quantitation of the reaction products, as well as the continuous loss of substrate due to decay of the radioactive label. Moreover, the technique is not appropriate for studying reactions occurring with half times substantially <1 min.
A possible alternative to the use of radioactive substrates would be to resort to measurements of intramolecular fluorescence resonance energy transfer (FRET) in dye-labeled oligonucleotide substrates; an approach successfully applied in the ribozyme field (23–25). FRET assays are continuous and suitable for high-throughput analyses (26). However, the synthesis of RNA substrates containing appropriate fluorophores at both ends is rather expensive, and the possibility that the bulky fluorescent groups might interfere with the kinetic behavior of the catalysts is always a concern (24).
Herein, we report a continuous assay in which an exogenous fluorophore, ethidium bromide, is exploited to measure the rates of kinetic steps in the reaction catalyzed by the RNA-cleaving 8-17 deoxyribozyme (see Fig. 1). This method allows the real-time kinetic analysis of the deoxyribozyme reaction, yielding reaction rates that are essentially identical to the rates obtained through the use of radioactive substrates. The assay has a general applicability, which has been verified using another RNA-cleaving construct, the 10-23 catalytic DNA.
Figure 1.
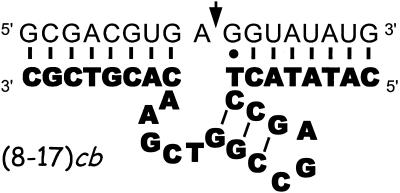
Primary and secondary structure of the 8-17 deoxyribozyme complexed to its substrate (7). The DNA enzyme (bold letters) binds its RNA substrate (not bold letters) through two substrate-recognizing arms, each involving Watson–Crick base pairing. Under the conditions adopted in this study, the thermodynamic dissociation constant of the (8-17)cb substrate is <5 nM (unpublished results). Once bound, the substrate is cleaved at the site indicated by the arrow.
Overall, the results presented in this paper help delineate the advantages and the limitations of this ‘exogenous fluorophore’ approach, which appears to be, in many cases, a convenient alternative or a useful complement to more traditional methods for assaying the activity of RNA-cleaving DNAzymes.
MATERIALS AND METHODS
Materials
The DNA oligonucleotides and DNA–RNA mixed oligonucleotides used in this study were purchased from MWG Biotech (Ebersberg, Germany) or from Eurogentech (Seraing, Belgium). One of the hammerhead constructs used, the J34 ribozyme, was a generous gift by Prof. Yi Lu (University of Illinois, Urbana-Champaign). All other RNA oligonucleotides were prepared by T7 RNA polymerase transcription (27); when internally labeled RNAs were required, the transcription mixture contained [α-32P]UTP. Transcripts were separated on a denaturing 20% polyacrylamide gel; the bands were cut from the gel and eluted in TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5). 32P-5′-end labeling of chemically synthesized oligonucleotides was accomplished with T4 polynucleotide kinase. Concentrations of radioactive oligonucleotides were determined from specific activities; concentrations of non-radioactive oligonucleotides were determined spectrophotometrically, using extinction coefficients calculated based on published values for nearest-neighbor pairs (28).
Ethidium bromide and 4,6-diamidino-2-phenyl indole (DAPI) were from Fluka; the concentration of ethidium was calculated using an ε480 of 5850 M–1cm–1 (29). Since ethidium is a mutagen and suspect carcinogen, all solutions containing the dye were handled wearing personal protective equipment (gloves, lab coat and goggles).
MgCl2, CaCl2 and MnCl2 (>99.99%) were from Aldrich. MnCl2 solutions were used immediately after preparation to minimize metal oxidation and formation of insoluble hydroxides.
1,4-Piperazinediethane sulfonic acid (PIPES) and N-(2-hydroxyethyl)piperazine-N′-2-ethane sulfonic acid (HEPES) were purchased from Fluka. Unless otherwise stated, measurements were carried out in 50 mM PIPES–NaOH, pH 7.4. The buffer solution also contained 75 mM NaCl, for a calculated total ionic strength of 0.21 M.
Constructs used: deoxyribozyme, substrates and inhibitors
The 8-17 deoxyribozyme used in this study, termed (8-17)cb, is a variant of the previously described (8-17)c construct (9); in that earlier work (8-17)cb was identified as variant A12T/A15.0. The catalytic core of (8-17)cb contains a ‘TCGAA’ unpaired region (see Fig. 1), instead of the ‘ACGA’ turn that was present in the ‘canonical’ (8-17)c DNA enzyme. This change renders (8-17)cb more active than its parent construct in the presence of Mg2+ and Mn2+, while reducing its preference for Ca2+ versus Mg2+ as a metal ion cofactor (9).
For the experiments involving the 8-17 construct, two substrates and one substrate analog were used (underlined, ribonucleotide; bold, deoxyribonucleotide):
rS 5′-GCG ACG UGA GGU AUA UG-3′
dS 5′-GCG ACG TGA GGT ATA TG-3′
dI 5′-GCG ACG TGA GGT ATA TG-3′
rS is the all-RNA substrate of (8-17)cb, whereas dS is a mostly-DNA substrate, containing a single ribonucleotide at the cleavage site. 8-17 constructs can cleave both kinds of substrate, but cleavage of dS occurs at a faster rate (9,10). dI is a non-cleavable dS analog, which was used for control experiments.
Conventional fluorescence measurements
Fluorescence emission spectra and continuous fluorometric assays of slow deoxyribozyme reactions were conducted on a thermostated Perkin Elmer LS-50B Luminescence Spectro meter. Experiments were carried out using either a standard quartz cuvette (total sample volume 700 µl) or a microcuvette (Hellma), which allowed reduction of the sample volume to 150 µl.
Reaction kinetics, unless otherwise stated, were performed at 25°C in the presence of 0.5 µM ethidium. Fluorescence changes were followed to completion, and the resulting time-courses were fit to the appropriate kinetic equation using Sigma Plot (SPSS Inc.).
Stopped-flow fluorescence measurements
Rapid kinetic experiments were performed on a SX.18MV stopped-flow apparatus from Applied Photophysics. To measure the rates of substrate cleavage, reactions were performed by mixing the deoxyribozyme (present in one syringe) with an equimolar amount of the substrate (in the other syringe). Both syringes contained equal concentrations of MnCl2 and of ethidium bromide.
To determine the association rates of (8-17)cb with dI, the DNAzyme and its substrate analog were placed in two separated syringes, containing 0.5 µM ethidium and, when required, 3 mM divalent metal ion chloride (MgCl2, CaCl2 or MnCl2). Association was measured under two regimes. In the first regime, equimolar amounts of the two nucleic acids were mixed in the stopped-flow. In this case, formation with time of the complex between the deoxyribozyme and its substrate analog proceeded according to equation 1 (30):
[ES] = [E]0 ([E]0 kt)/([E]0 kt + 1)1
where [ES] is the concentration of the complex between the deoxyribozyme and dI, [E]0 is the initial concentration of deoxyribozyme, and k is the second-order kinetic rate constant for the association process.
In the second regime, association reactions were measured in the presence of a large molar excess of dI. Under these conditions, the accumulation of deoxyribozyme–dI complex with time would follow a monoexponential function. Experiments were performed with various concentrations of dI to obtain several different pseudo-first order rate constants; these were then plotted against [dI] to yield the second-order association rate constant.
Discontinuous assays based on radioactive substrates
In assays employing radioactive substrates, cleavage was measured under single-turnover conditions (DNAzyme 0.5–2 µM; internally labeled substrate ∼1 nM), to ensure that the product dissociation step did not affect the observed kinetics (9,31). Substrate and catalyst were separately heated at 95°C for 2 min to disrupt potential aggregates, spun briefly in a microcentrifuge and equilibrated for 10 min at the reaction temperature in a thermostated water bath. After adding MgCl2, CaCl2 or MnCl2 to both tubes, reactions were initiated by mixing the deoxyribozyme (or ribozyme) with its substrate (total volume of the reaction mixture, 40 µl). Time-points were collected at appropriate intervals and further reaction was quenched by adding formamide and excess EDTA. Products and substrates were separated on 7 M urea/20% polyacrylamide gels and their ratio was quantitated using a Molecular Imager (Bio-Rad). Rate constants for cleavage were obtained by non-linear least-squares fitting the reaction time-courses using Sigma Plot (SPSS Inc.).
RESULTS
Background and design of the ethidium assay
Ethidium is a well-known nucleic acid intercalator, characterized by a substantially higher affinity for double-stranded nucleic acids as compared with single-stranded (32–36). Intercalation of an ethidium molecule into a duplex covers at least two adjacent base pairs and can be considered, to a very first approximation, sequence-independent (32–35). It is also well known that the fluorescence emission of ethidium increases dramatically (∼20-fold) once the dye is intercalated (34). Thanks to these properties, ethidium is often used as an exogenous fluorophoric probe in nucleic acids studies for measuring structural transitions that involve the formation or the disruption of double-helical segments. For example, kinetic assays have been reported that exploit ethidium for measuring the annealing of complementary nucleic acids (37) or the unwinding of double-stranded DNA operated by helicases (38).
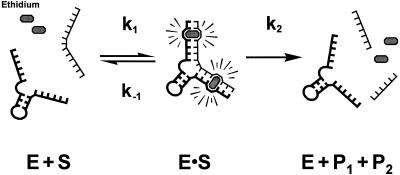
We aimed at establishing a continuous method for studying the kinetics of the small RNA-cleaving 8-17 deoxyribozyme (Fig. 1), originally isolated by Santoro and Joyce through in vitro selection (7). In the reaction carried out by 8-17, the DNA enzyme binds to its substrate through the formation of new helical segments (Fig. 2, left). Subsequently, in the presence of divalent metal ions, the substrate is cleaved and the products are released, with loss of helical structure (Fig. 2, right). Thus, in principle, the signal arising from binding and dissociation of ethidium can be used to monitor, in single turnover reactions, the formation of the deoxyribozyme–substrate complex and the release of the reaction products. Under many conditions, the latter process is very fast compared with the actual transphosphorylation reaction, so that the rate of dissociation of the products reflects, in fact, the rate of substrate cleavage.
Figure 2.
Schematic representation of the reaction carried out by the 8-17 deoxyribozyme. Binding of the catalytic DNA (E, thick line) to its substrate (S, thin line) occurs through base pairing; the formation of new helical segments allows the intercalation of ethidium ions. Subsequently, the substrate is cleaved and the products, P1 and P2, dissociate (the estimated thermodynamic dissociation constants for P1 and P2 are both >0.5 µM; 48,50). Upon product dissociation, the number of available intercalation sites drops and ethidium is released.
In order for ethidium to serve as a convenient probe for measuring the kinetic processes depicted in Figure 2, several criteria need to be met. First, the presence of ethidium must not affect the steps that are being monitored, e.g. substrate cleavage or product dissociation. Establishing this requires control experiments. However, except for cases in which very tight ethidium binding sites exist, any inhibitory effect of the dye should be negligible when ethidium is present in largely substoichiometric amounts, i.e., when only a small fraction of the intercalation sites are occupied at any given time.
Second, to ensure a good signal-to-background ratio, the amount of bound ethidium should vary substantially with variations in the amount of double strand. This requirement is best satisfied at concentrations of intercalation sites ≤Kd, where Kd is the dissociation constant of ethidium for a single intercalation site. The average Kd value, at room temperature and moderate ionic strength, can be taken as ∼4 µM (39–41). Since the 8-17 deoxyribozyme–substrate complex contains up to eight potential intercalation sites (Fig. 1; at most, each helical segment can bind one ethidium ion every 2 bp), the sensitivity of the assay should be maximal at concentrations of deoxyribozyme–substrate complex ∼0.5 µM.
Third, the rates of ethidium association–dissociation must be much faster than the rates of nucleic acids annealing– unwinding so as not to limit the observed kinetics. It is known that, at room temperature and moderate ionic strength, the rate of ethidium dissociation from an intercalation site in a DNA duplex is ∼600 min–1 (40,41). Moreover, since the bimolecular rate constant of ethidium association is ∼109 M–1 min–1 (40,41), binding at micromolar concentrations of sites would occur with a pseudo first-order rate constant >1000 min–1. This means that any annealing–unwinding transition occurring at rates of 100 min–1, or slower, should be measurable using ethidium.
Binding of ethidium to the free 8-17 deoxyribozyme and to the deoxyribozyme–substrate and deoxyribozyme–inhibitor complexes
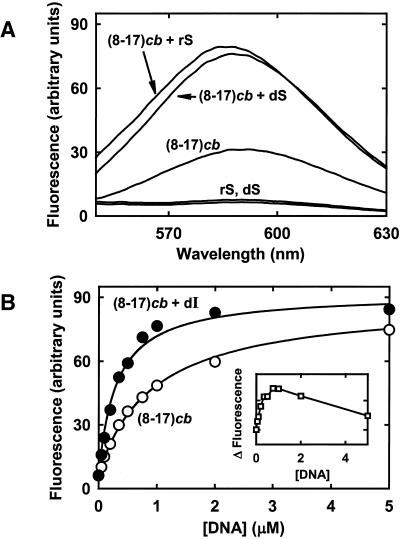
We collected fluorescence emission spectra of ethidium in the presence of the free (8-17)cb deoxyribozyme and of its complexes with the two substrates rS and dS (see above). Under the adopted experimental conditions, which had been chosen based on the arguments above, a substantial fluorescence increase was observed upon binding of the substrates to the 8-17 deoxyribozyme (Fig. 3A). This also implicated that a substantial fluorescence decrease would accompany dissociation of the products from the deoxyribozyme arms.
Figure 3.
(A) Fluorescence emission spectra of ethidium bromide (0.5 µM) in the presence of either 0.5 µM substrate (dS or rS), or 0.5 µM deoxyribozyme or 0.5 µM deoxyribozyme–substrate complex. The spectra were collected in PIPES–NaOH pH 7.4 at 25°C; the excitation wavelength was 520 nm. (B) Fluorescence emission of ethidium (0.5 µM) as a function of the concentration of (8-17)cb deoxyribozyme (open circles) or of the deoxyribozyme–dI complex (closed circles) in the presence of 3 mM CaCl2 [other conditions were as in (A); the emission wavelength was 590 nm]. The solid lines through the data points represent tentative fittings to hyperbolic functions, yielding K0.5 values of 0.9 µM for the deoxyribozyme and 0.3 µM for deoxyribozyme–dI complex, respectively. Inset, the difference between the two fluorescence datasets.
The spectra in Figure 3A were collected in the absence of divalent metal ions, to prevent substrate cleavage; however, the presence of modest concentrations of divalent metal ions should not affect substantially the overall emission or the extent of fluorescence change upon substrate binding. Figure 3B shows an experiment conducted in the presence of 3 mM Ca2+, using the non-cleavable substrate analog, dI. Emission was measured upon titrating ethidium bromide with either the free (8-17)cb construct or the deoxyribozyme–dI complex. The difference between the two curves confirms that working at nucleic acid concentrations ∼0.5 µM ensures the largest variation in fluorescence between the free catalyst and the deoxyribozyme–substrate complex.
The data in Figure 3B were also used to estimate K0.5 (the concentration of DNA giving 50% of the maximum fluorescence enhancement), which can be taken as an average dissociation constant of the ethidium–DNA complex. The calculated K0.5 values were 0.9 µM for (8-17)cb alone and 0.3 µM for the (8-17)cb–dI complex. Essentially identical K0.5 values were obtained in the presence of 3 mM Mg2+ or in the absence of divalent metal ions (not shown).
A 3-fold difference in K0.5 is consistent with the higher number of intercalation sites in the deoxyribozyme–inhibitor complex as compared with the free catalyst. The pre dicted secondary structure of free (8-17)cb (42; and J.SantaLucia,Jr, M.Zuker, A.Bommarito and R.J.Irani, unpublished results; DNA folding calculations were carried out at the website http://www.bioinfo.rpi.edu/applications/mfold/old/dna/form1.cgi) includes one potential intercalation site within the 3 bp core stem and another site formed by partial self-annealing of the 3′ arm of the DNAzyme. On the other hand, the (8-17)cb–dI complex could comprise up to eight intercalation sites.
Substrate cleavage by (8-17)cb probed by ethidium bromide fluorescence
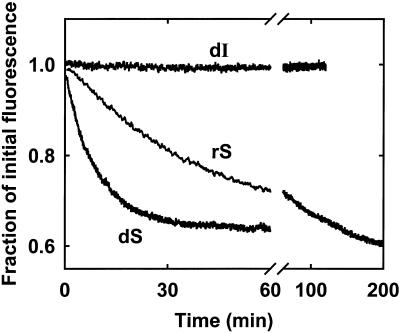
Figure 4 shows the decays in fluorescence emission occurring after reacting the deoxyribozyme–rS complex or deoxy ribozyme–dS complex with 3 mM Ca2+, in the presence of ethidium. The observed decays could only be attributed to release of the products subsequent to cleavage, as the fluorescence emission of the deoxyribozyme–dI complex, under the same experimental conditions, remained constant for hours. Furthermore, product release appeared to be nearly complete, as the final fluorescence value was very close to the emission expected for ethidium bound to free (8-17)cb.
Figure 4.
Real-time monitoring of the (8-17)cb reaction using the ethidium assay. Changes in the fluorescence emission of ethidium bromide (0.5 µM) were followed after reacting either 0.5 µM (8-17)cb–rS complex, or 0.5 µM (8-17)cb-dS, or 0.5 µM (8-17)cb–dI complex with 3 mM Ca2+. Other conditions: 50 mM PIPES–NaOH buffer pH 7.4 at 25°C. The excitation wavelength was 520 nm; the emission wavelength was 590 nm.
It should be noted that the time-courses measured by following product release through the ethidium assay are not expected to strictly obey an exponential equation, mainly because the correlation between the amount of bound ethidium and the amount of double strand present is only roughly linear under the conditions of the assay. Nevertheless, both the fluorescence decays presented in Figure 4 (as well as those collected in the presence of other metals; data not shown) fitted reasonably well to single exponential functions, yielding kobs values of 0.016 and 0.16 min–1, respectively.
Note also that, if the product release step were rate-limiting under the adopted conditions, the kobs values measured using ethidium would be substantially slower than the rates measured using radioactive substrates (radiolabeled substrates allow monitoring the cleavage irrespective of whether the products are released or not). However, in the (8-17)cb system, the rate of cleavage within the ES complex is generally much slower than the release of products (unpublished results); therefore, the kinetics observed through the ethidium assay are expected to reflect the rate of chemical cleavage. This was confirmed by the systematic near-identity between the kobs values obtained using the ethidium assay and the k2 values measured employing radioactive substrates, under single-turnover conditions (Table 1).
Table 1. Comparison between the reaction rates of (8-17)cb measured using a radioactive substrate or through the ethidium assay.
| Metal ion cofactora | Substrate | k2, 32P-substrate assayb (min–1) | k2, ethidium assayc (min–1) |
|---|---|---|---|
| Mg2+ | rS | 0.010 | 0.012 |
| dS | 0.060 | 0.080 | |
| Ca2+ | rS | 0.018 | 0.016 |
| dS | 0.08 | 0.13 | |
| Mn2+ | rS | 0.4 | 0.5 |
| dS | 1.5 | 1.8d | |
| dS, pH 8e | n.d. | 4.9d |
aAll reactions were carried out at 25°C, in the presence of 3 mM metal ion cofactor.
bIn assays employing a radioactive substrate, a large deoxyribozyme excess (0.5–2 µM versus ∼1 nM substrate) was used, ensuring that product release was not rate-limiting.
cIn ethidium assays, equimolar concentrations of deoxyribozyme and substrate were used (0.5 µM). Ethidium bromide was also typically 0.5 µM. In some control experiments in which the intercalator concentration was varied, the k2 values obtained at ethidium concentrations between 0.2 and 1 µM were identical within error (±25%). Substantial (>2-fold) inhibition was observed only at ethidium concentrations >3 µM.
dThese reactions were measured through the ethidium assay employing a stopped-flow apparatus, as described in the text.
eThe reaction was carried out in 50 mM HEPES–NaOH buffer pH 8.0, in the presence of 3 mM MnCl2. Under these conditions, substrate cleavage was too fast to be reliably measured using the radioactive substrate assay.
Stopped-flow measurements of rapid deoxyribozyme kinetics
Manganese ions are very effective in activating the 8-17 cleavage reaction (9,10). For this reason, the reaction of (8-17)cb with dS in the presence of Mn2+ could be measured accurately only by resorting to stopped-flow instrumentation. When the deoxyribozyme and the substrate were reacted together in the presence of ethidium and Mn2+, the time-course showed an initial rapid rise in fluorescence, followed by a slower decay (with an apparent rate constant of 1.8 min–1) corresponding to the release of the reaction products (Fig. 5).
Figure 5.
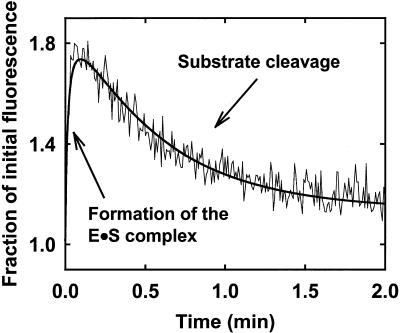
Stopped-flow time-course for the reaction of (8-17)cb with its substrate dS in the presence of Mn2+. The deoxyribozyme (0.5 µM final) was mixed with a stoichiometric amount of dS in the presence of 0.5 µM ethidium and 3 mM manganese chloride (PIPES–NaOH pH 7.4 at 25°C). The excitation wavelength was 520 nm; emission was collected at wavelengths >530 nm (selected using a Corning 3-69 cut-off filter). The thin line shows the experimental time-course, whereas the thick line represents the best fit of the data points to the sum of two functions. The initial rise in fluorescence was analyzed according to equation 1, yielding a bimolecular association rate constant of ∼108 M–1 min–1. The second phase was fit to an exponential decay, occurring with a rate constant of 1.8 min–1.
The initial increase in fluorescence emission was not limited by ethidium association. In a control experiment in which ethidium (0.5 µM) was allowed to bind to the preformed deoxyribozyme–substrate complex (0.5 µM), association of the dye was completed within 15 ms (not shown), as expected based on the published rate constants for ethidium intercalation (40,41). This suggested that the initial phase of the kinetic time-course reflected formation of the deoxyribozyme–substrate complex, occurring with an approximate second-order rate constant of 108 M–1 min–1. This value approaches the rate constant measured for association of another deoxyribozyme, the 10-23 construct, to its substrate (43).
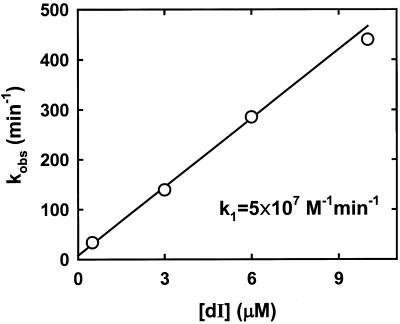
We used the stopped-flow technique to study in more detail the association of (8-17)cb with the substrate analog, dI, in the presence of ethidium. Conceptually similar approaches to measure hybridization rates have been used before by others (37). In our experiments, the deoxyribozyme was reacted with an excess of the substrate analog (at various concentrations) in the presence of ethidium and 3 mM Mn2+, and the reaction time-courses showed only an increase in fluorescence, since dI was not cleaved by the deoxyribozyme. The measured pseudo-first order rate constants were directly proportional to the concentration of DNAzyme, allowing determination of a second-order rate constant of 0.5 × 108 M–1 min–1 (Fig. 6). Very similar second-order rate constants for association of (8-17)cb with dI were measured in the absence of divalent metal ions or in the presence of 3 mM Mg2+ or 3 mM Ca2+ (not shown).
Figure 6.
Exploiting ethidium to determine the kinetic association constant of dI to (8-17)cb. Reaction of deoxyribozyme (0.5 µM) with various amounts of dI was monitored by stopped-flow (other experimental conditions and instrumental setups were as in Fig. 5). The observed increases in fluorescence were fit to monoexponential functions, and the kobs values were plotted versus the deoxyribozyme concentration. The solid line represents the linear least-squares fit of the data, and its slope provides the second-order rate constant for association of the substrate analog to the deoxyribozyme. The intercept on the y-axis was 7 ± 9 min–1.
Application of the ethidium assay for monitoring the 10-23 deoxyribozyme kinetics
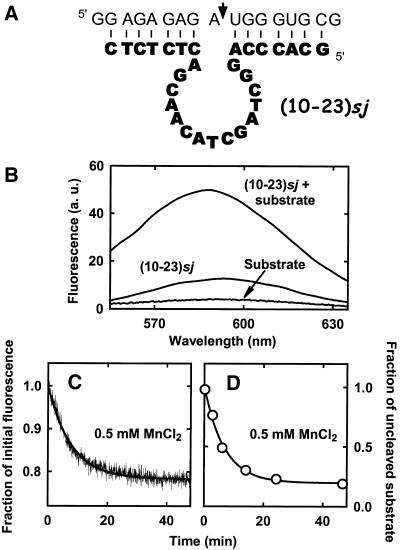
The 10-23 DNA enzyme (7) is another versatile catalyst, widely employed for cleaving RNA both in vitro and in vivo (15–17,21). Since the 10-23 deoxyribozyme shows a size and a topology very similar to that of 8-17, we tested the possibility of monitoring the 10-23 reaction through the use ethidium. To this end, we employed a 10-23 construct previously studied by Santoro and Joyce (43) (Fig. 7A) and we adopted the same experimental conditions (buffer, pH, temperature, nucleic acid concentration) of the previously described 8-17 experiments.
Figure 7.
(A) Structure of the 10-23 deoxyribozyme used in this study. The DNA enzyme is shown in bold letters and the RNA substrate is shown in non-bold letters. RNA cleavage occurs at the site indicated by the arrow. After cleavage, at least one of the two products would completely dissociate under the adopted reaction conditions (48,50). (B) Fluorescence emission spectra of ethidium bromide (1 µM) in the presence of 0.5 µM 10-23 deoxyribozyme alone or of 0.5 µM deoxyribozyme–substrate complex. PIPES–NaOH buffer pH 7.4 at 25°C; the excitation wavelength was 520 nm. (C) The 10-23 deoxyribozyme reaction monitored using the ethidium assay. The decrease in fluorescence of ethidium bromide (1 µM) was followed upon reacting the deoxyribozyme–substrate complex (0.5 µM) with 0.5 mM Mn2+. Buffer, pH and temperature were as in (B); the excitation wavelength was 520 nm and the emission wavelength was 590 nm. The experimental time-course is shown by the thin line, whereas the thick line represents the best fit of the data to a single exponential decay, yielding kobs = 0.13 min–1. (D) Time-course of the 10-23 cleavage reaction, monitored using a radioactive substrate. The deoxyribozyme (8 µM) was in large excess with respect to its substrate (∼1 nM); the other conditions were as in (C) (ethidium was omitted). The thick line through the data points represents the best fit to a monoexponential function, yielding kobs = 0.15 min–1.
In analogy with the results obtained using the 8-17 deoxyribozyme, the free 10-23 catalyst interacted weakly with ethidium, as judged by the low fluorescence intensity, but a strong enhancement of emission was observed upon substrate binding (Fig. 7B). Addition of 0.5 mM Mn2+ brought about a significant fluorescence decrease, which occurred at a rate of 0.13 min–1, essentially identical to the rate of the cleavage step (Fig. 7C and D). Other experiments, carried out in the presence of different metal ions, confirmed the correspondence between the rate constants obtained through the ethidium assay and the k2 values measured employing radioactive substrates: in the presence of 3 mM Ca2+, the cleavage rate measured fluorometrically was 0.014 min–1, whereas k2 measured using radioactive RNA was 0.013 min–1; in the presence of 3 mM Mg2+, the observed rate constants were 0.009 and 0.007 min–1, respectively (data not shown).
These results indicate that the ethidium assay can be easily employed to monitor the 10-23 kinetics. The results also suggest that the assay should be useful in the study of other small RNA-cleaving deoxyribozymes such as the ML-6 construct, originally described by Sugimoto et al. (12), which is in fact a minimized version of the 10-23 motif.
Hammerhead ribozyme: a high-affinity ethidium binding site interferes with application of the ethidium assay
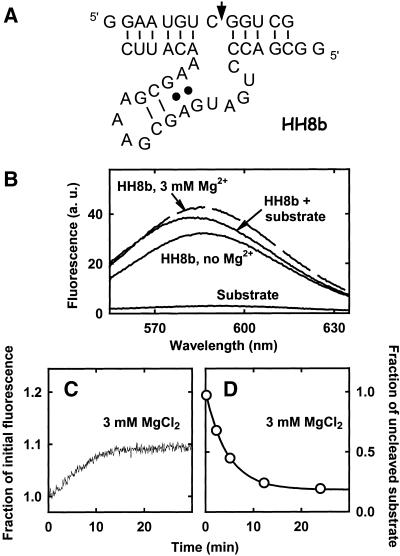
Application of the ethidium assay is based on the assumption that the free catalyst (and free substrate) contains little or no secondary structure. In fact, if the free nucleic acid enzyme contained a substantial amount of duplex and, hence, intercalation sites, the change in fluorescence arising from substrate association or product dissociation would be minimal. In the ribozyme field, most of the naturally occurring RNA-cleaving catalysts are large and structured, which makes them not amenable to be studied through the ethidium assay. An obvious exception is the hammerhead ribozyme (Fig. 8A; reviewed in 44), which shows a small size and a topology analogous to those of the 8-17 and 10-23 deoxyribozymes. We therefore tested whether ethidium could also be employed to monitor the reaction carried out by this ribozyme.
Figure 8.
(A) Structure of the hammerhead ribozyme used in this study; the black dots indicate two non-standard G·A pairs that are present in the ribozyme structure; the arrow indicates the site of substrate cleavage. HH8b is derived from the kinetically well-behaved HH8 ribozyme (51). With respect to the original HH8, one extra C has been added at the 3′ end to stabilize substrate binding (the dissociation constant of the HH8–substrate complex was 49 nM in 10 mM MgCl2; 51) while stem II in the ribozyme core has been shortened from 4 to 2 bp. After cleavage, both reaction products are predicted to completely dissociate from the ribozyme arms (49). (B) Fluorescence emission spectra of complexes of the hammerhead ribozyme with ethidium. The solid lines represent spectra of ethidium bromide (0.5 µM) in the presence of 0.5 µM ribozyme substrate, or 0.5 µM hammerhead ribozyme or of 0.5 µM ribozyme + 0.5 µM substrate, in the absence of metal ions. The dashed line represents the spectrum of ethidium bromide (0.5 µM) plus 0.5 µM ribozyme in the presence of 3 mM MgCl2. PIPES–NaOH buffer pH 7.4 at 25°C; the excitation wavelength was 520 nm. (C) Monitoring the HH8b ribozyme reaction in the presence of ethidium. The change in fluorescence of ethidium bromide (0.5 µM) was followed upon reacting the deoxyribozyme–substrate complex (0.5 µM) with 3 mM Mg2+. Buffer, pH and temperature as in (B); the excitation wavelength was 520 nm and the emission wavelength was 590 nm. A tentative fitting of the experimental time-course to a single exponential increase yielded kobs = 0.12 min–1 (not shown). (D) Time-course of the HH8b cleavage reaction, monitored using a radioactive substrate. The deoxyribozyme (0.6 µM) was in large excess with respect to its substrate (∼1 nM); the other conditions were as in (C) (ethidium was omitted). The thick line through the data points represents the best fit to a monoexponential function, yielding kobs = 0.23 min–1.
We found that the free hammerhead ribozyme could bind ethidium with rather high affinity, so that formation of the ribozyme–substrate complex brought about only a relatively small increase in fluorescence (Fig. 8B). In addition, whereas for the free 8-17 and 10-23 deoxyribozymes addition of modest concentrations of metal ions did not significantly change the ethidium emission (data not shown), fluorescence of the free hammerhead ribozyme increased by over 30% upon addition of 3 mM Mg2+ (Fig. 8B) or 3 mM Mn2+ (not shown).
These properties of the hammerhead system resulted in the observation of peculiar kinetics with the ethidium assay. When the ribozyme–substrate complex was reacted with divalent metal ions, the change in emission signal was small, and in the presence of 3 mM Mg2+ the fluorescence increased slightly, instead of decreasing (Fig. 8C). In fact, under these conditions the substrate was cleaved (Fig. 8D) and the products released, regenerating the free ribozyme.
The data above imply that ethidium binds strongly not only to the hammerhead–substrate complex (which includes a substantial amount of duplex) but also to the free ribozyme, and that binding of the dye is further stabilized by divalent metal ions. This behavior is not unique of the HH8b construct employed in this study: we tested the ethidium-binding properties of another hammerhead construct, for which the three-dimensional structure has been solved (45), obtaining essentially the same results. The existence of at least one high-affinity ethidium binding site within the hammerhead structure is consistent with the results of kinetic experiments (carried out employing a radioactive HH8b substrate), which showed that sub-micromolar concentrations of ethidium inhibited catalysis (data not shown).
We do not know where the strong ethidium binding site is located. A plausible possibility is that ethidium binds at or near the tandem G·A pairs in the ribozyme core, since these pairs interact through extensive stacking, and since they can form in the free ribozyme and their conformation is modulated by divalent metal ions (45–47). We are currently trying to test this hypothesis. In any case, with regard to the subject of this paper, we must conclude that the existence of a high-affinity ethidium binding site within the hammerhead structure impedes any straightforward application of the ethidium assay in this system.
DISCUSSION
We have devised a continuous assay for measuring in real time the RNA cleavage reactions catalyzed by different small catalytic DNAs. The assay is based on the presence, in the reaction mixture, of substoichiometric amounts of the familiar intercalating dye ethidium bromide. The technique is simple, fast and inexpensive, as it does not require the preparation of specially labeled substrates. Moreover, the technique should be easily adaptable to multi-well plates assays, allowing its use for the large-scale kinetic analysis of deoxyribozyme constructs.
The ethidium assay can also be employed to study the kinetics of deoxyribozymes by stopped-flow, permitting the measurement of processes occurring with rates >100 min–1. Thus far, stopped-flow approaches in the field of catalytic nucleic acids have been limited due to the lack of intrinsic ‘reporter’ groups in RNA and DNA. We have shown that the combination of an exogenous fluorophoric ligand and the stopped-flow technique makes it possible to measure, in a single experiment, both the rate of annealing of the catalyst to its substrate and the rate of chemical cleavage.
It must be noted that the cleavage assay described in this work requires the catalyst to be present at a concentration equal to (or even slightly larger than) its substrate: in other words, the assay does not function under multiple-turnover but rather under single-turnover conditions. Single-turnover kinetic measurements are commonplace in the field of deoxyribozymes and of catalytic nucleic acids in general (9,12,13,31,43) and provide direct information about the catalytic efficiency of the deoxyribozyme or ribozyme being tested. Moreover, it is predicted that the kobs obtained under single-turnover using the ethidium assay should match closely the kcat measured under multiple turnover conditions, even if kcat is limited by product release (a possible exception to this generalization is represented by cases in which the presence of excess substrate accelerates product release, e.g. through strand-displacement mechanisms).
Caveats and perspectives for the application of ‘exogenous fluorophore’ assays
In the ethidium assay, observation of a signal after substrate cleavage is subordinate to the release of the products from the deoxyribozyme arms. To achieve this, the concentration of catalyst in the assay should be lower than the thermodynamic dissociation constants for the DNAzyme–product complexes. A reasonable estimate of these dissociation constants can be obtained a priori, through the use of published nearest-neighbor stability parameters (42,48,49), and the experimental conditions can be adjusted so as to ensure complete release of at least one (and preferably both) of the products. In principle, three parameters can be changed to achieve this goal: the catalyst’s recognition arms may be shortened, the temperature of the assay may be raised, or the concentration of the catalyst (and, concomitantly, of the substrate) may be lowered.
Raising the temperature would reduce the amplitude of the spectroscopic signal, given the sensitivity of fluorescence emission to thermal quenching. By using the ethidium assay, we could collect kinetic data on the (8-17)cb at temperatures ranging from 15 to 40°C, obtaining in all cases rate constants very similar to those obtained through the use of radiolabeled substrates (data not shown). However, the signal-to-noise ratio in the fluorescence time-courses worsened significantly at the higher temperatures, due to the decrease in ethidium emission and possibly to reduced binding of the dye.
As for lowering the concentration of nucleic acids, it would also affect sensitivity, since at low catalyst concentrations less ethidium would bind (especially if the overall dye concentration is decreased, to maintain substoichiometric conditions) and the emission signal would be correspondingly diminished. Adopting stronger excitation sources might not be a practicable solution and might damage the sample. Alternatively, the assay described in this paper could be adapted to employ other double-strand binding fluorophores, possessing a higher quantum yield and/or a greater binding affinity compared with ethidium bromide.
Indeed, while in this study we have focused on ethidium because of its general availability and because so much is known about its mode of interaction with nucleic acids, preliminary tests indicate that other fluorescent dyes could also be effectively used in ‘exogenous fluorophore’ assays. For example, we found that DAPI (a minor-groove binder which, as compared with ethidium, shows a greater fluorescence enhancement upon binding to DNA) could be employed at concentrations of just 100 nM to monitor the (8-17)cb-catalyzed cleavage of dS (unpublished results). However, the same dye was not useful for monitoring the reaction of the 8-17 deoxyribozyme with its all-RNA substrate, rS, due to the rather low affinity of DAPI for RNA–DNA hybrid helices (36).
CONCLUSIONS
Assays based on exogenous fluorophores can represent a rapid and convenient way for monitoring the activity of small RNA-cleaving DNAzymes. In particular, application of such assays may result advantageous in preliminary characterization works, in large-scale, high-throughput kinetic analyses and in studies where fast reaction processes have to be measured. In fact, at variance with methods based on radioactive oligonucleotides, approaches like the ‘ethidium assay’ described in this work are continuous and allow the measurement of substantially faster time-courses. Furthermore, at variance with most conventional methods, these assays do not require specially labeled oligonucleotides and can be employed with both in vitro transcribed and chemically synthesized substrates. Among the limits of the exogenous fluorophore assays are the need of choosing experimental conditions under which the reaction products dissociate after cleavage and the difficulty of applying the method to catalysts that are large and structured or that contain unusually strong dye-binding sites.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Andrea Mozzarelli for use of his stopped flow apparatus and to Yi Lu for the kind gift of one of the hammerhead constructs used. We also thank Yi Lu, Gerald Joyce, Daniel Lafontaine and Barbara Campanini for comments on the manuscript. This work was supported by grants from the EMBO Young Investigator Programme and from the Italian National Research Council to A.P., and by a grant from the University of Parma to D.F.
REFERENCES
- 1.Breaker R.R. (1997) DNA enzymes. Nat. Biotechnol., 15, 427–431. [DOI] [PubMed] [Google Scholar]
- 2.Sen D. and Geyer,C.R. (1998) DNA enzymes. Curr. Opin. Chem. Biol., 2, 680–687. [DOI] [PubMed] [Google Scholar]
- 3.Li Y. and Breaker,R.R. (1999) Deoxyribozymes: new players in the ancient game of biocatalysis. Curr. Opin. Struct. Biol., 9, 315–323. [DOI] [PubMed] [Google Scholar]
- 4.Emilsson G.M. and Breaker,R.R. (2002) Deoxyribozymes: new activities and new applications. Cell. Mol. Life Sci., 59, 596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breaker R.R. and Joyce,G.F. (1994) A DNA enzyme that cleaves RNA. Chem. Biol., 1, 223–229. [DOI] [PubMed] [Google Scholar]
- 6.Breaker R.R. and Joyce,G.F. (1995) A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem. Biol., 2, 655–660. [DOI] [PubMed] [Google Scholar]
- 7.Santoro S.W. and Joyce,G.F. (1997) A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faulhammer D. and Famulok,M. (1996) The Ca2+ ion as a cofactor for a novel RNA-cleaving deoxyribozyme. Angew. Chem. Int. Ed. Engl., 35, 2837–2841. [Google Scholar]
- 9.Peracchi A. (2000) Preferential activation of the 8-17 deoxyribozyme by Ca2+ ions. Evidence for the identity of 8-17 with the catalytic domain of the Mg5 deoxyribozyme. J. Biol. Chem., 275, 11693–11697. [DOI] [PubMed] [Google Scholar]
- 10.Li J., Zheng,W., Kwon,A.H. and Lu,Y. (2000) In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res., 28, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geyer C.R. and Sen,D. (1997) Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol., 4, 579–593. [DOI] [PubMed] [Google Scholar]
- 12.Sugimoto N., Okumoto,Y. and Ohmichi,T. (1999) Effect of metal ions and sequence of deoxyribozymes on their RNA cleavage activity. J. Chem. Soc. Perkin Trans. 2, 1381–1386. [Google Scholar]
- 13.Feldman A.R. and Sen,D. (2001) A new and efficient DNA enzyme for the sequence-specific cleavage of RNA. J. Mol. Biol., 313, 283–294. [DOI] [PubMed] [Google Scholar]
- 14.Roth A. and Breaker,R.R. (1998) An amino acid as a cofactor for a catalytic polynucleotide. Proc. Natl Acad. Sci. USA, 95, 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sioud M. and Leirdal,M. (2000) Therapeutic RNA and DNA enzymes. Biochem. Pharmacol., 60, 1023–1026. [DOI] [PubMed] [Google Scholar]
- 16.Khachigian L.M. (2000) Catalytic DNAs as potential therapeutic agents and sequence-specific molecular tools to dissect biological function. J. Clin. Invest., 106, 1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cairns M.J., Saravolac,E.G. and Sun,L.Q. (2002) Catalytic DNA: a novel tool for gene suppression. Curr. Drug Targets, 3, 269–279. [DOI] [PubMed] [Google Scholar]
- 18.Stojanovic M.N., Mitchell,T.E. and Stefanovic,D. (2002) Deoxyribozyme-based logic gates. J. Am. Chem. Soc., 124, 3555–3561. [DOI] [PubMed] [Google Scholar]
- 19.Li J. and Lu,Y. (2000) A highly sensitive and selective catalytic DNA biosensor for lead ions. J. Am. Chem. Soc., 122, 10466–10467. [Google Scholar]
- 20.Stojanovic M.N., de Prada,P. and Landry,D.W. (2000) Homogeneous assays based on deoxyribozyme catalysis. Nucleic Acids Res., 28, 2915–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pyle A.M., Chu,V.T., Jankowsky,E. and Boudvillain,M. (2000) Using DNAzymes to cut, process, and map RNA molecules for structural studies or modification. Methods Enzymol., 317, 140–146. [DOI] [PubMed] [Google Scholar]
- 22.Okumoto Y., Ohmichi,T. and Sugimoto,N. (2002) Immobilized small deoxyribozyme to distinguish RNA secondary structures. Biochemistry, 41, 2769–2773. [DOI] [PubMed] [Google Scholar]
- 23.Jenne A., Gmelin,W., Raffler,N. and Famulok,M. (1999) Real-time characterization of ribozymes by fluorescence resonance energy transfer (FRET). Angew. Chem. Int. Ed. Engl., 38, 1300–1303. [DOI] [PubMed] [Google Scholar]
- 24.Singh K.K., Parwaresch,R. and Krupp,G. (1999) Rapid kinetic characterization of hammerhead ribozymes by real-time monitoring of fluorescence resonance energy transfer (FRET). RNA, 5, 1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt C., Welz,R. and Muller,S. (2000) RNA double cleavage by a hairpin-derived twin ribozyme. Nucleic Acids Res., 28, 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenne A., Hartig,J.S., Piganeau,N., Tauer,A., Samarsky,D.A., Green,M.R., Davies,J. and Famulok,M. (2001) Rapid identification and characterization of hammerhead-ribozyme inhibitors using fluorescence-based technology. Nat. Biotechnol., 19, 56–61. [DOI] [PubMed] [Google Scholar]
- 27.Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 28.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 29.Bresloff J.L. and Crothers,D.M. (1975) DNA-ethidium reaction kinetics: demonstration of direct ligand transfer between DNA binding sites. J. Mol. Biol., 95, 103–123. [DOI] [PubMed] [Google Scholar]
- 30.Capellos C. and Bielski,B.H.J. (1972) Kinetic Systems: Mathematical Description of Chemical Kinetics in Solution, Wiley-Interscience. New York.
- 31.Stage-Zimmermann T.K. and Uhlenbeck,O.C. (1998) Hammerhead ribozyme kinetics. RNA, 4, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waring M.J. (1965) Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol., 13, 269–282. [DOI] [PubMed] [Google Scholar]
- 33.Waring M.J. (1966) Structural requirements for the binding of ethidium to nucleic acids. Biochim. Biophys. Acta, 114, 234–244. [DOI] [PubMed] [Google Scholar]
- 34.LePecq J.B. and Paoletti,C. (1967) A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J. Mol. Biol., 27, 87–106. [DOI] [PubMed] [Google Scholar]
- 35.Ren J. and Chaires,J.B. (1999) Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry, 38, 16067–16075. [DOI] [PubMed] [Google Scholar]
- 36.Ren J., Qu,X., Dattagupta,N. and Chaires,J.B. (2001) Molecular recognition of a RNA:DNA hybrid structure. J. Am. Chem. Soc., 123, 6742–6743. [DOI] [PubMed] [Google Scholar]
- 37.Yguerabide J. and Ceballos,A. (1995) Quantitative fluorescence method for continuous measurement of DNA hybridization kinetics using a fluorescent intercalator. Anal. Biochem., 228, 208–220. [DOI] [PubMed] [Google Scholar]
- 38.Eggleston A.K., Rahim,N.A. and Kowalczykowski,S.C. (1996) A helicase assay based on the displacement of fluorescent, nucleic acid-binding ligands. Nucleic Acids Res., 24, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson W.D., Krishnamoorthy,C.R., Wang,Y.H. and Smith,J.C. (1985) Mechanism of intercalation: ion effects on the equilibrium and kinetic constants for the interaction of propidium and ethidium with DNA. Biopolymers, 24, 1941–1961. [DOI] [PubMed] [Google Scholar]
- 40.Macgregor R.B. Jr, Clegg,R.M. and Jovin,T.M. (1985) Pressure-jump study of the kinetics of ethidium bromide binding to DNA. Biochemistry, 24, 5503–5510. [DOI] [PubMed] [Google Scholar]
- 41.Meyer-Almes F.J. and Porschke,D. (1993) Mechanism of intercalation into the DNA double helix by ethidium. Biochemistry, 32, 4246–4253. [DOI] [PubMed] [Google Scholar]
- 42.SantaLucia ,J. Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santoro S.W. and Joyce,G.F. (1998) Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry, 37, 13330–13342. [DOI] [PubMed] [Google Scholar]
- 44.Eckstein F. and Bramlage,B. (1999) The hammerhead ribozyme. Biopolymers, 52, 147–154. [DOI] [PubMed] [Google Scholar]
- 45.Pley H.W., Flaherty,K.M. and McKay,D.B. (1994) Three-dimensional structure of a hammerhead ribozyme. Nature, 372, 68–74. [DOI] [PubMed] [Google Scholar]
- 46.Orita M., Vinayak,R., Andrus,A., Warashina,M., Chiba,A., Kaniwa,H., Nishikawa,F., Nishikawa,S. and Taira,K. (1996) Magnesium-mediated conversion of an inactive form of a hammerhead ribozyme to an active complex with its substrate. An investigation by NMR spectroscopy. J. Biol. Chem., 271, 9447–9454. [DOI] [PubMed] [Google Scholar]
- 47.Bassi G.S., Murchie,A.I.H., Walter,F., Clegg,R.M. and Lilley,D.M.J. (1997) Ion-induced folding of the hammerhead ribozyme: a fluorescence resonance energy transfer study. EMBO J., 16, 7481–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugimoto N., Nakano,S., Katoh,M., Matsumura,A., Nakamuta,H., Ohmichi,T., Yoneyama,M. and Sasaki,M. (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- 49.Freier S.M., Kierzek,R., Jaeger,J.A., Sugimoto,N., Caruthers,M.H., Neilson,T. and Turner,D.H. (1986) Improved free-energy parameters for predictions of RNA stability. Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano S., Fujimoto,M., Hara,H. and Sugimoto,N. (1999) Nucleic acid duplex stability: influence of base composition on cation effects. Nucleic Acids Res., 27, 2957–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fedor M.J. and Uhlenbeck,O.C. (1992) Kinetics of intermolecular cleavage by hammerhead ribozymes. Biochemistry, 31, 12042–12054. [DOI] [PubMed] [Google Scholar]