Abstract
We describe gene targeting experiments involving a human cell line (RAN10) containing, in addition to its endogenous alleles, two ectopic alleles of the interferon-inducible gene 6–16. The frequency of gene targeting at one of the ectopic 6–16 alleles (H3.7) was 34-fold greater than the combined frequency of gene targeting involving endogenous 6-16 alleles in RAN10. Preference for H3.7 was maintained when the target loci in RAN10 were transcriptionally activated by interferon. Despite the 34-fold preference for H3.7, the absolute gene targeting efficiency in RAN10 was only 3-fold higher than in the parental HT1080 cell line. These data suggest that different alleles can compete with each other, and perhaps with non-homologous loci, in a step which is necessary, but not normally rate-limiting, for gene targeting. The efficiency of this step can therefore be more sensitive to chromosomal position effects than the rate-determining steps for gene targeting. The nature of the position effects involved remains unknown but does not correlate with transcription status, which in our system has a very modest influence on the frequency of gene targeting. In summary, our work unequivocally identifies a position effect on gene targeting in human cells.
INTRODUCTION
The relationship between the function of a segment of genomic DNA and its chromosomal position is an important focus of attention in molecular genetics. Most often this has been investigated by observing how gene expression is affected by changes in its chromosomal position or sequence context (1), but there are other aspects of DNA function, including recombination, whose sensitivity to chromosomal position may be informative. It is therefore interesting to know how gene targeting, a biologically and technically important form of DNA recombination in mitotic mammalian cells, is influenced by chromosomal position.
Gene targeting is the introduction of defined changes into the genome via homologous recombination (HR) between a chromosomal target locus and a transfected DNA molecule (2,3). HR is an important DNA repair pathway in mammalian cells (4,5), but it must be tightly regulated to avoid harmful chromosomal rearrangements, and gene targeting is generally very inefficient. Improvements in the efficiency of gene targeting would greatly aid its use as a tool in somatic cell genetics (6,7) and as a potential route to therapeutic gene repair (8). For these reasons, and also to improve our understanding of mitotic HR, a better definition of the factors that influence gene targeting efficiencies is desirable.
Gene targeting frequencies are very variable, for reasons that are not always clear. There are many variables known or suspected to influence gene targeting frequencies and pathways (8,9), including the amount (10,11) and quality (12,13) of the region of homology, the nature and positioning of selectable markers (14–17) and the method by which the targeting construct is delivered to cells (9,18–20). Never theless, even targeting vectors that have been similarly designed and delivered to the same cell type can have very different targeting frequencies, leading to the idea that the nature of the target locus is important.
It is likely that specific sequences in the target locus, either within or close to the region of homology with the targeting vector, promote HR. Sequence ‘hotspots’ that promote recombination have been studied in systems from bacteria, yeast and man (21). In eukaryotes, most attention has been paid to meiotic recombination hotspots (22). Evidence for mitotic recombination hotspots in yeast (23) and human cells (24,25) has also been reported, however. Microsatellite (26,27) and minisatellite (28) sequences have been shown to promote HR between extrachromosomal substrates in mammalian cells. Direct evidence for an effect of such sequences on gene targeting itself, however, is still lacking (29).
It is also possible that aspects of chromatin structure at the target locus that cannot be predicted from nucleotide sequence correlate with gene targeting frequencies, just as aspects of chromatin structure, reflected in the degree of histone acetylation, DNase hypersensitivity and DNA demethylation, are known to be associated with expressed genes (30,31). While there is evidence that transcription can stimulate intrachromosomal HR (32), extrachromosomal HR (33) and gene targeting (34), it is also known that untranscribed genes can be disrupted by gene targeting with appreciable efficiencies (35). Meanwhile, the relationship, if any, between chromatin structure and gene targeting efficiencies remains to be explored.
Prior to conducting such analyses it is of interest to establish unequivocally the importance of chromosomal location in gene targeting. To do this it is necessary to vary the chromosomal location of the target locus whilst holding constant the nucleotide sequence of the target. In yeast, the effect of chromosomal position on interallelic recombination (36) or gene targeting (37) was remarkably small. In many respects, however, including its high efficiency and sensitivity to target copy number (38,39), gene targeting in yeast is very different to that in mammalian cells. To our knowledge the only studies of this kind involving mammalian cells were by Lin et al. (40) and Thomas et al. (41), both in mouse L cells. While suggestive of a strong chromosomal position effect, the former study was complicated by the use of carrier DNA and the calcium phosphate delivery method. The latter study did not report major variations in targeting frequencies, but only three cell lines were compared, and where a cell line contained multiple copies of target genes, potential frequency variations between copies were not investigated.
To re-investigate this problem we chose as our target the human interferon (IFN)-inducible gene 6–16 (42), which has several attractions in this context. First, the 6-16 gene is small, making it relatively easy to introduce additional alleles at ectopic chromosomal sites. Secondly, in contrast to commonly used model target genes, such as that encoding neomycin phosphotransferase (neo), endogenous target alleles exist and these can serve as internal reference targets. Furthermore, the ability of mammalian cells to recognise and modify foreign DNA (43,44) is less of a complication for 6–16 gene targeting. Thirdly, because the 6–16 gene is transcriptionally inducible by IFN, transcription can be studied as a variable at the same time as chromosomal position. Lastly, by use of a promoter-trap enrichment procedure, the proportion of stably transfected clones that are targeted at the 6–16 locus is ∼1/3, making targeted clones easy to isolate (45).
Here we find that cell lines carrying one or more ectopic copies of 6–16 show only small variations in gene targeting frequency. In the line with the best targeting frequency (RAN10), however, we detected a strong preference for gene targeting at a particular ectopic 6–16 allele. The extent of this preference (34-fold) did not reflect a correspondingly elevated targeting efficiency at the ectopic allele because the absolute targeting frequency in RAN10 was only 3-fold higher than in parental HT1080 cells. We therefore suggest that different 6–16 alleles compete for interactions with the targeting construct or for an essential recombination factor, in a step that is not normally rate-limiting for gene targeting, and that this competition is subject to chromosomal position effects.
MATERIALS AND METHODS
Construction of plasmids
DNA cloning was carried out according to standard procedures. Plasmid p6–16neo contains the human 6–16 gene, including the promotor and all five exons, linked to a neo selection marker. An 8.5 kb XhoI fragment carrying the entire 6–16 gene and 2.3 kb of 5′ sequence and a 2.9 kb AccI–BamHI fragment containing the neo expression cassette of pSV2neo (46) were cloned into the EcoRI and XbaI sites, respectively, of pBSIIKS+ (Stratagene). The targeting construct p6gpt has been previously described (47). It contains a pBSIIKS+ backbone, 6.2 kb of 6–16 genomic sequence (the 3′ end of the genomic XhoI fragment, lacking the 6–16 promotor) and a promotorless gpt marker within 6–16 intron 1. Both p6–16neo and p6gpt were built with HT1080-isogenic DNA. pBSHPRTdSexhyg was constructed by cloning the hygro cassette from pSV2hyg into SexA1-cut pBSHPRT (47).
Cell culture and transfection
Human fibrosarcoma HT1080 cells (48), or their RAN derivatives, were grown and electroporated as previously described (45). ‘Diploid’ cultures of HT0180 cells routinely contain a proportion (∼5%) of tetraploid cells even after recloning. RAN10 and other tetraploid clones behave similarly to ‘diploid’ cultures during all cellular manipulations we have used. DNA used for transfections was SalI-linearised p6–16neo (8 µg), SalI-linearised p6gpt (8 µg), an 8 kb SmaI–SalI fragment of p6gpt lacking the pBSIIKS+ backbone (4–6.5 µg) or SalI-linearised pBSHPRTdSexhyg (8 µg). Drug selection was started 48 h after electroporation and maintained throughout. The RAN cell lines were selected initially in 400 µg/ml G418, reducing the concentration to 200 µg/ml after ∼4 weeks in culture. Selection for targeted RAN clones was as follows. Cells (2.5 × 106/175 cm2 plate, 6 plates/sample) were selected in 200 µg/ml G418, 2.5–10 µg/ml mycophenolic acid (MPA), 100 µg/ml xanthine, in the presence or absence of 100 IU/ml IFN (Wellferon, a mixture of human type I IFNs; Glaxo-Wellcome). Between 14 and 19 days after electroporation the surviving colonies were either stained with crystal violet and scored, or picked and amplified for further analyses. Selection for targeting in parental HT1080 cells was as described for RAN cell lines but omitting G418. Selection of pBSHPRTdSexhyg-transfected cells was in hygromycin (100 µg/ml).
To test the effect of IFN or trichostatin A (TSA) pretreatment on 6–16 gene targeting in RAN10, cells were treated with or without 100 IU/ml IFN or 100 nM TSA, each in the presence of 200 µg/ml G418, for a 72 h period (from 24 h before electroporation until 48 h after electroporation) and then selected in 200 µg/ml G418, 10 µg/ml MPA, 100 µg/ml xanthine and 100 IU/ml IFN. Between 14 and 16 days after electroporation the surviving colonies were picked and expanded for further analyses. The effect of IFN pretreatment on 6–16 gene targeting in wild-type HT1080 cells was analysed likewise but omitting G418.
To test whether the MPA resistance of isolated cell clones was IFN-dependent we split cells grown in 200 µg/ml G418, 10 µg/ml MPA, 100 µg/ml xanthine and 100 IU/ml IFN into duplicate samples (1.5 × 104 cells/well, 6-well plate) and selected in the presence or absence of IFN. Differential growth was assessed after ∼6 days culture by visual inspection of both the colour of the medium and the cell density.
Flow cytometry
Ploidy was assessed by analyses of propidium iodide stained nuclei, as previously described (49).
Molecular analyses
PCR screening for 6–16 gene targeting was performed with oligonucleotides O1 and O2 as previously described (45). This reaction amplifies a 1 kb fragment specific to cells targeted at 6–16 by using a primer from the 6–16 promotor and another one from the gpt marker.
Southern blots for 6–16 analysis were done as previously described, using genomic DNA from amplified HT1080 clones and a 240 bp HaeIII–BamHI fragment from the 6–16 promotor as probe (45). Some blots were also hybridised using SmaI-digested pBSIIKS+ as probe. DNA size markers were a 1 kb ladder (M; Gibco BRL) or HindIII-digested bacteriophage λ DNA (λ).
Northern analysis of 6–16 expression was essentially as previously reported (45). Briefly, HT1080 cells were lysed with guanidine thiocyanate and RNA was purified by CsCl centrifugation. The blots were probed with a 0.6 kb 6–16 cDNA.
RESULTS
The 6–16 gene targeting system
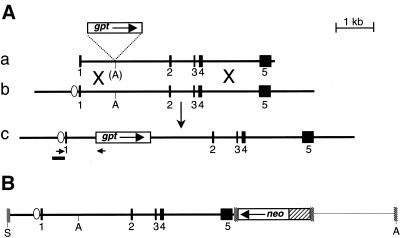
The 6–16 gene targeting construct (p6gpt) contains a promoterless gpt gene incorporated into a promoterless fragment of the 6–16 gene (Fig. 1A, a). When expressed, the bacterial gpt gene confers resistance to the purine synthesis inhibitor MPA on mammalian cells, if xanthine is also included in the growth medium. HR between p6gpt and a chromosomal 6–16 gene places the gpt gene under the control of the IFN-responsive 6–16 promoter (Fig. 1A). Targeted clones are therefore resistant to MPA and xanthine (MPA/X) if IFN is also present. Molecular analyses (PCR and Southern blots) are used to confirm that IFN-dependent MPA/X-resistant colonies are targeted. Most randomly integrated p6gpt molecules fail to express gpt and their host cells die in MPA/X, with or without IFN. A few random integration events, however, activate gpt by chance and give rise to colonies that grow in MPA/X. Unlike targeted clones, such random integrants do not depend on IFN for their resistance and are negative in molecular assays for targeted integration. In this system, therefore, the proportion of selected clones that are targeted (the relative targeting frequency, RTF) is conveniently high (∼1/3) (47), but the proportion of transfected cells that are targeted (the absolute targeting frequency, ATF) remains low (∼1/106).
Figure 1.
(A) The 6–16 gene targeting system. (a) Targeting construct p6gpt with vector sequences removed. (b) The 6–16 gene. (c) The product of gene targeting involving (a) and (b). (B) SalI-cut p6–16neo, used to generate ectopic 6–16 alleles. DNA is represented as follows: black boxes, 6–16 gene exons, numbered 1–5; white ellipse, IFN-responsive region of the 6–16 gene promoter; thick lines, other 6–16 gene DNA; stippled boxes, multiple cloning sites of pBSIIKS+; thin line, other pBluescript DNA; white boxes, drug resistance cassettes; hatched box, SV40 early promoter; black bar, target-specific probe in Southern analyses; short horizontal arrows, primers used in PCR assay for targeting. A, Asp718; S, SalI.
Generation and screening of cell lines with ectopic 6–16 alleles
To generate clones that carry one or more ectopic copies of the 6–16 gene integrated at random sites, a plasmid (p6–16neo, Fig. 1B) carrying the 6–16 gene linked to a neo expression cassette was electroporated into HT1080 cells and G418-resistant clones were isolated. Eleven such cell lines (RAN1, RAN2, etc.) were then transfected with the targeting construct p6gpt. After electroporation, duplicate samples were selected in G418/MPA/X, in the presence or absence of IFN, and colony numbers were scored. Previous experiments (47) showed that, for p6gpt-transfected HT1080 cells, approximately one-third of colonies selected in MPA/X/IFN are IFN-dependent, the remainder being random integrants. It was therefore expected that for HT1080 the number of colonies selected with IFN present would be ∼1.5-fold higher than the number selected without, and that in any clones with elevated targeting efficiencies this ratio would be higher. The observed ratios (Supplementary Material) were variable and surprisingly low, some even being <1. This probably indicates that the plating efficiencies are impaired by the presence of IFN. Nevertheless, two clones (RAN8 and RAN10) had elevated ratios (averages of 1.75 and 2.42, respectively, n = 2), and RAN10 was chosen for further analysis.
Absolute 6–16 gene targeting efficiency is elevated 3-fold in RAN10
A more detailed comparison of gene targeting in HT1080 and RAN10 was then made. Colonies selected in MPA/X/IFN after transfection of p6gpt were tested for gene targeting by PCR. Numbers of PCR-positive (T) and PCR-negative (R) colonies were used to estimate the relative targeting frequency. For most experiments these values were also used to determine the random integration frequency (RIF) and ATF. The data for each experiment are summarised in Table 1 and mean values are compared in Tables 2–4. Significant increases, of 2.2- and 2.9-fold, respectively, in the RTF and ATF for RAN10, compared to HT1080, were measured (Table 2). A 2.1-fold decrease in RIF was noted, but was outside the 95% confidence limits. In separate experiments, RAN10 and HT1080 formed hygromycin-resistant colonies with similar efficiencies after transfection with a plasmid (pBSHPRTdSexhyg-SalI) carrying a hygromycin resistance cassette but no 6–16 gene sequences (not shown). The ∼3-fold increase in ATF therefore does not need to be adjusted upward to compensate for any reduced transfectability of RAN10.
Table 1. Summary of gene targeting experiments in HT1080 and RAN10.
| Exp. | Cells | DNA | Seln | N | C | T + R | T | R | RTF | RIF | ATF |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HT1080 | 8, p | 2.5 | 27 | 10 | 17 | 0.37 | ||||
| 2.1 | HT1080 | 6.5, i | 2.5 | 13.5 | 25 | 15 | 4 | 11 | 0.27 | 1.36 | 0.49 |
| 2.2 | HT1080 | 6.5, i | 10 | 16.2 | 29 | 17 | 7 | 10 | 0.41 | 1.05 | 0.74 |
| 2.3 | HT1080 | 4, i | 10 | 16.2 | 39 | 23 | 9 | 14 | 0.39 | 1.47 | 0.94 |
| 3.1 | HT1080 | 8, p | 10 | 12.5 | 16 | 15 | 8 | 7 | 0.53 | 0.6 | 0.68 |
| 3.2 | HT1080 | 8, p | 10 | 12.5 | 20 | 18 | 9 | 9 | 0.50 | 0.8 | 0.8 |
| 3.3 | HT1080 | 8, p | 10, I | 15 | 14 | 13 | 10 | 3 | 0.77 | 0.22 | 0.72 |
| 3.4 | HT1080 | 8, p | 10, I | 12.5 | 23 | 20 | 15 | 5 | 0.75 | 0.46 | 1.38 |
| 4 | RAN10 | 4, i | 10 | 15 | 26 | 18 | 17 | 1 | 0.94 | 0.10 | 1.64 |
| 5.1 | RAN10 | 4, i | 10 | 10 | 28 | 26 | 18 | 8 | 0.69 | 0.86 | 1.94 |
| 5.2 | RAN10 | 4, i | 10 | 15 | 52 | 28 | 21 | 7 | 0.75 | 0.87 | 2.6 |
| 5.3 | RAN10 | 4, i | 10, I | 7.5 | 33 | 30 | 28 | 2 | 0.93 | 0.29 | 4.11 |
| 5.4 | RAN10 | 4, i | 10, I | 15 | 71 | 23 | 20 | 3 | 0.87 | 0.62 | 4.12 |
| 6.1 | RAN10 | 4, i | 10 | 3 | 3 | 3 | 0 | 1.0 | |||
| 6.2 | RAN10 | 4, i | 10, T | 5 | 4 | 4 | 0 | 1.0 |
Exp., experiments with the same whole number (e.g. 2.1, 2.2) were done in parallel, on the same day. DNA, DNA electroporated; numbers indicate µg used; p, SalI-linearised p6gpt; i, purified SmaI–SalI insert from p6gpt. Seln, selection conditions; numbers indicate concentration (µg/ml) of MPA; I, IFN-pretreated; T, TSA-pretreated. N, number of cells electroporated (×10–6). C, total number of colonies selected in MPA/X/IFN. T + R, number of colonies tested in PCR assay for gene targeting. T, number of targeted integrants as detected by PCR. R, number of random integrants as detected by PCR. RTF, relative targeting frequency = T/(T + R). RIF, random integration frequency (×106) = RC/N(T + R). ATF, absolute targeting frequency (×106) = TC/N(T + R).
Table 2. Targeted and random integration frequencies: RAN10 versus HT1080.
| HT1080 | Experiments | RAN10 | Experiments | Fold difference | P | |
|---|---|---|---|---|---|---|
| ATF | 0.72 ± 0.22 | 2.1, 2.2, 2.3 | 2.06 ± 0.49 | 4, 5.1, 5.2 | +2.85 | 0.01 |
| RIF | 1.29 ± 0.21 | 2.1, 2.2, 2.3 | 0.61 ± 0.44 | 4, 5.1, 5.2 | –2.12 | 0.07 |
| RTF | 0.36 ± 0.07 | 1, 2.1, 2.2, 2.3 | 0.80 ± 0.13 | 4, 5.1, 5.2 | +2.21 | 0.002 |
Table 4. Targeted and random integration frequencies: effect of IFN-pretreatment in RAN10.
| –IFN | Experiments | +IFN | Experiments | Fold difference | P | |
|---|---|---|---|---|---|---|
| ATF | 2.27 ± 0.47 | 5.1, 5.2 | 4.11 ± 0.007 | 5.3, 5.4 | +1.81 | 0.03 |
| RIF | 0.86 ± 0.004 | 5.1, 5.2 | 0.46 ± 0.23 | 5.3, 5.4 | –1.90 | 0.13 |
| RTF | 0.72 ± 0.04 | 5.1, 5.2 | 0.90 ± 0.05 | 5.3, 5.4 | +1.25 | 0.052 |
See legend to Table 2.
RAN10 is tetraploid with two ectopic 6–16 alleles
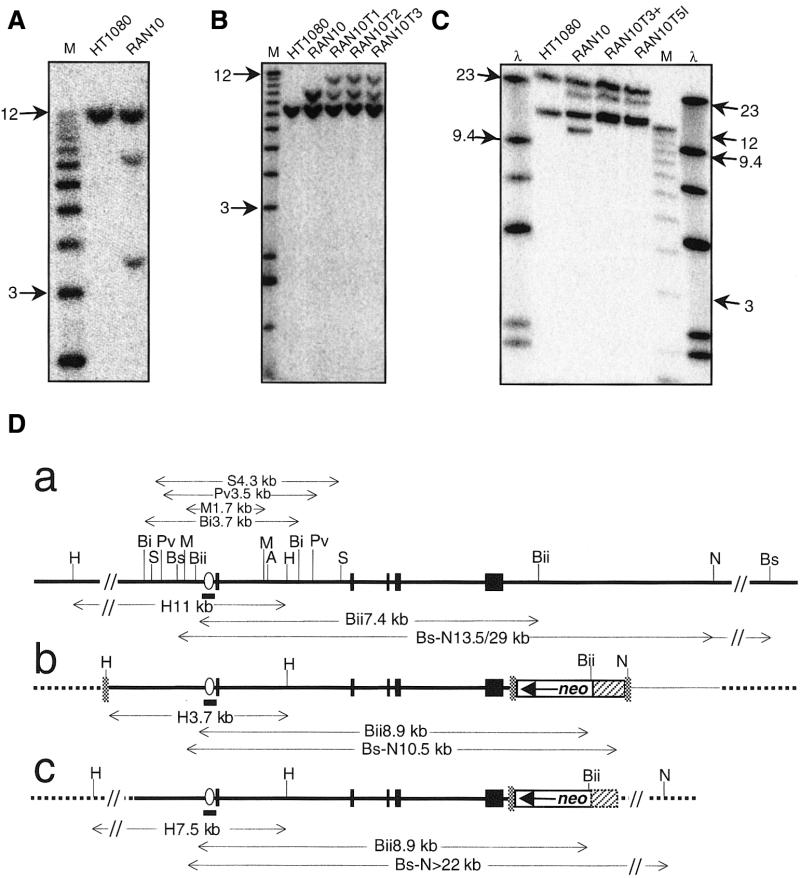
Flow cytometry showed that RAN10 is tetraploid (data not shown). In Southern blots, a probe spanning the 6–16 gene promotor detected HindIII fragments of 11, 7.5 and 3.7 kb (Fig. 2A). The 11 kb band was expected for the endogenous 6–16 alleles and was 4-fold more intense than either the 7.5 or 3.7 kb bands. The latter two bands represent distinct single copy 6–16 alleles integrated at ectopic chromosomal sites, and we will refer to these as the H7.5 and H3.7 alleles, respectively. The 3.7 kb fragment is exactly as expected for random integration of intact SalI-cut p6–16neo. The 7.5 kb band, on the other hand, is likely to derive from a p6–16neo molecule that has suffered loss of some 5′ sequences, including its HindIII site. The same 6–16 promoter probe detected only single bands in blots of RAN10 genomic DNA digested with BglI, PvuII, MfeI or SacI (not shown). The maximum loss of 6–16 DNA from the 5′ end of the H7.5 allele is therefore 419 bp (the distance between the BglI and SalI sites in p6–16neo). Both ectopic 6–16 alleles therefore retain >1.8 kb of 6–16 sequences 5′ of exon 1, far more than is sufficient for IFN-inducible transcription. The two ectopic alleles also differ at their 3′ ends: while they both retain the neo-derived BglII site (Fig. 2B), the H7.5 allele has lost the pBSIIKS+-derived NotI site (Fig. 2C). Finally, SmaI and HindIII digests of RAN10, hybridised with either the 6–16 promotor probe or a pBSIIKS+ probe, showed that the two chromosomal copies of p6–16neo are not arranged in tandem and must have separate integration sites (not shown). Thus, despite their differences (summarised in Fig. 2D), both ectopic 6–16 alleles appear to have intact 6–16 genes fully capable of undergoing HR with p6gpt to generate MPAr clones.
Figure 2.
Structure of 6–16 alleles in RAN10. Genomic DNA was digested with HindIII (A), BglII (B) or BstZ17I and NotI (C), blotted and probed with the target-specific probe shown in Figure 1. Genomic DNA was from HT1080, RAN10 or targeted RAN10 clones (RAN10T1, etc.) whose MPA resistance was partially IFN-dependent. Size markers (M or λ) are described in Materials and Methods and sizes (kb) of arrowed markers are indicated. (D) Schematic representation of the endogenous (a) and ectopic copies (b and c) of 6–16 in RAN10. DNA is represented as in Figure 1. Relevant sites are shown for Asp718 (A), BglI (Bi), BglII (Bii), BstZ17I (Bs), HindIII (H), PvuII (Pv), MfeI (M), NotI (N) and SacI (S).
Some 6–16 gene expression in RAN10 is IFN-independent
In parental HT1080 cells the ∼1 kb 6–16 transcript is barely detectable prior to IFN treatment and is strongly up-regulated by IFN (Fig. 3A) (50). In RAN10, however, two transcripts of 1 and 1.3 kb were detectable in the absence of IFN and both were strongly up-regulated by IFN. A degree of inappropriate ‘background’ expression is typical for transgenes that lack their usual sequence context and has been observed before for 6–16 (42). Other RAN clones tested had both transcripts and similar but variable levels of uninduced expression (Fig. 3). The presence of the 1.3 kb band in other RAN clones suggests that some aspect of p6–16neo structure causes spurious transcriptional initiation or termination, or aberrant RNA splicing.
Figure 3.
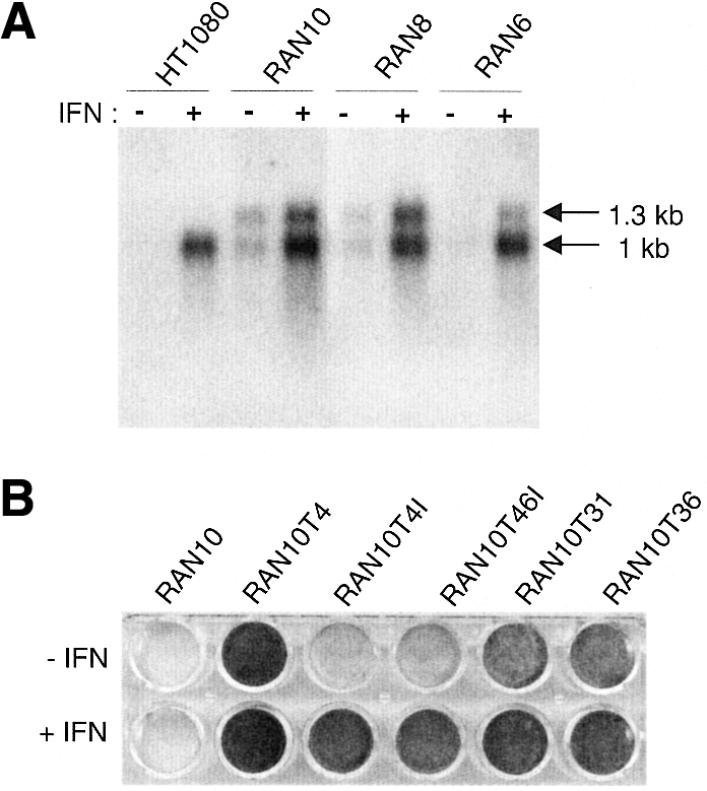
Evidence for constitutive 6–16 transcription of the H3.7 allele in RAN cell lines. (A) Northern blot of RNA isolated from cell lines grown with or without IFN treatment, as indicated. (B) Tests for IFN dependence of resistance to MPA. Cells were seeded at low density and grown in selective medium with or without IFN (Materials and Methods). Results for RAN10 and five p6gpt-transfected, MPAr derivatives are shown. RAN10T4 was not targeted at a 6–16 allele, as judged by PCR and Southern analyses. The remaining clones were targeted at an endogenous allele (RAN10T4I and RAN10T46I) or at the H3.7 allele (RAN10T31 and RAN10T36), as judged by PCR and Southern analyses.
6–16 gene targeting in RAN10 is strongly biased toward the H3.7 allele
Southern blots were used to confirm that PCR-positive clones were targeted at a 6–16 locus and to analyse the relative frequency of gene targeting for the six 6–16 alleles present in RAN10 (Fig. 4). Gene targeting with p6gpt increases the size of 6–16 HindIII fragments by 1.8 kb, i.e. from 11 to 12.8 kb for the endogenous alleles (Fig. 4, lanes 2 and 3) and, for the ectopic alleles, from 3.7 to 5.5 kb (Fig. 4, lane 4) or from 7.5 to 9.3 kb (Fig. 4, lane 5). Remarkably, PCR-positive RAN10 subclones showed a near complete bias for targeting at the H3.7 allele: of the 56 PCR-positive clones generated in experiments 4, 5.1 and 5.2 (Table 1), 48 were clearly targeted at the H3.7 allele, and none was targeted at an endogenous 6–16 allele (Fig. 4). Furthermore, six of the remaining eight clones appeared to have undergone unusual targeting events involving the H3.7 allele. Thus three clones (see, for example, Fig. 4, lane 10) had gained a 5.5 kb HindIII band without losing the 3.7 kb band [these probably represent ‘pick-up’ events (51,52)] and another three clones (see, for example, Fig. 4, lanes 8, 9 and 11) had lost the 3.7 kb band but had gained a band that was not the predicted size of 5.5 kb. One of the latter clones (Fig. 4, lane 11) had also undergone an unpredicted change at the H7.5 allele. The remaining two clones had the same HindIII pattern as RAN10 (lane 2 in Fig. 4), even though they were targeted at 6–16 according to the PCR screening (repeated for confirmation). In summary, 96% (54/56) of clones had undergone a gene targeting or gene targeting-related event involving the H3.7 allele. Of equal note to this bias was the apparent lowering of the ATF for the endogenous alleles to an undetectable level. Thus, for experiments 4, 5.1 and 5.2 combined, the expected number of clones with targeted endogenous 6–16 alleles is 20 [using the ATF for HT1080 of 0.72 ± 0.22 × 10–6 and the formula ATF × N(T + R)/C], but no such clones were detected.
Figure 4.
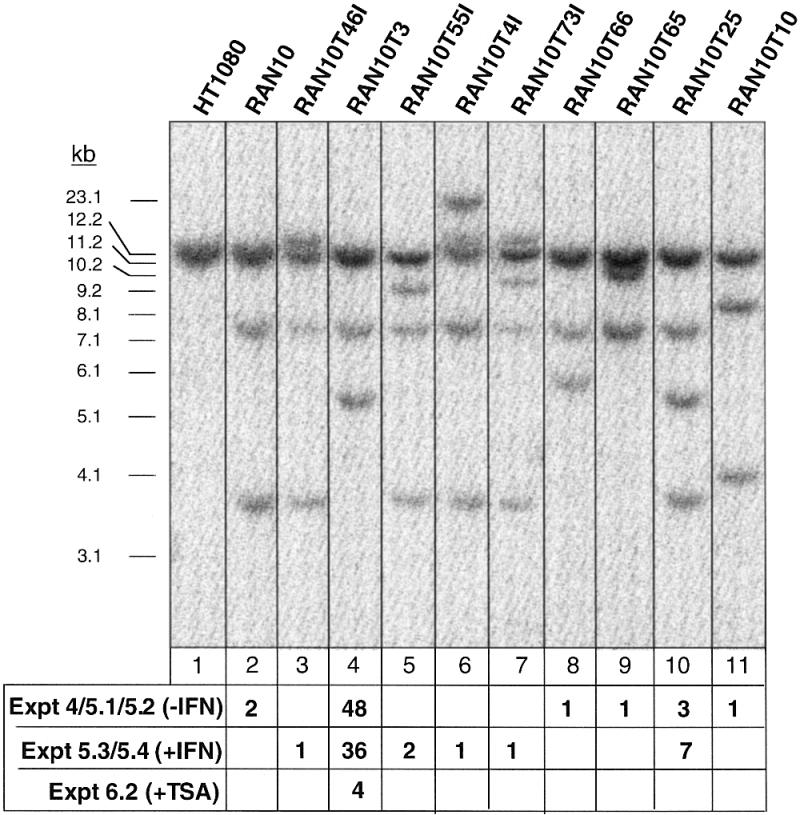
Southern blot analyses of HindIII-digested genomic DNA from HT1080, RAN10 and nine representative p6gpt-transfected RAN10 derivatives. In total, 108 MPAr, PCR-positive clones, derived from experiments 4–6 (Table 1), were analysed in this way and each generated one of the 10 representative hybridisation patterns shown in lanes 2–11, as indicated below the autoradiograph. The probe was as shown in Figure 1A.
Clones with targeted H3.7 alleles are only partially IFN-dependent
As described (45), targeted modification of an endogenous 6–16 allele with p6gpt generates HT1080 cells whose resistance to MPA is IFN-dependent. In initial tests for such IFN dependence, none of the 56 PCR-positive clones from experiments 4, 5.1 and 5.2 was found to be IFN-dependent. In more detailed analyses, however, involving selection at lower cell densities (Materials and Methods), it was clear that most clones (37/39) grew less well in the absence of IFN than in its presence, the remaining two being completely IFN-independent (Fig. 3B). We conclude from this that the H3.7 allele is at least partly responsible for the IFN-independent 6–16 transcription detected in RAN10 (Fig. 3A) and that this is sufficient to confer, after targeting with p6gpt, partial resistance to MPA. The two clones whose growth was not improved by IFN corresponded to clones that had atypical Southern patterns (Fig. 4, lanes 2 and 8), suggesting that they have undergone an unusual event in which a 6–16 allele has become linked to a constitutively active promoter.
An improved ability of targeted RAN clones to grow without IFN may help to explain some of the surprisingly low +IFN:–IFN ratios seen during the screening of RAN clones.
Targeting bias in RAN10 persists when all 6–16 alleles are transcribed
It was possible that preferential targeting at the H3.7 allele in RAN10 was a consequence of the appreciable transcription of this allele seen in the absence of IFN. To test this possibility we performed a 6–16 gene targeting experiment in RAN10 cells that had been pretreated with IFN (Table 1, experiments 5.3 and 5.4). In this case, all four endogenous 6–16 alleles (and possibly the H7.5 allele), as well as the H3.7 allele, are actively transcribed at the time of transfection with p6gpt. Thus, if the targeting bias in RAN10 observed without IFN pretreatment is due to differential transcription, we would expect the bias to disappear in pretreated cells. After selection in G418/IFN/MPA/X we recovered 53 clones, 48 of which were targeted at 6–16 according to the PCR screening. Southern blot analyses of the 48 PCR-positive clones showed that 36/48 (43/48 if pick-up events are included) were targeted at the H3.7 allele, 2/48 at the H7.5 allele (both of them pick-up events; Fig. 4, lane 5), 2/48 at an endogenous allele (Fig. 4, lanes 3 and 6) and 1/48 at both the H7.5 allele and an endogenous allele (Fig. 4, lane 7). Thus, IFN pretreatment had little effect on the bias towards targeting of the H3.7 allele and we conclude that preferential targeting at H3.7 is not primarily the result of IFN-independent transcription of this allele. In both HT1080 and RAN10 cells we detected small increases (1.4- and 1.8-fold, respectively) in the ATF as a result of IFN pretreatment, although only the latter was significant (Tables 3 and 4). While suggesting that our IFN pretreatment was effective, these small or insignificant increases contrast with the 3- to 20-fold stimulation reported for another targeting system (34).
Table 3. Targeted and random integration frequencies: effect of IFN-pretreatment in HT1080.
| –IFN | Experiments | +IFN | Experiments | Fold difference | P | |
|---|---|---|---|---|---|---|
| ATF | 0.74 ± 0.08 | 3.1, 3.2 | 1.05 ± 0.47 | 3.3, 3.4 | +1.41 | 0.46 |
| RIF | 0.70 ± 0.14 | 3.1, 3.2 | 0.34 ± 0.17 | 3.3, 3.4 | –2.07 | 0.15 |
| RTF | 0.52 ± 0.02 | 3.1, 3.2 | 0.76 ± 0.01 | 3.3, 3.4 | +1.47 | 0.006 |
See legend to Table 2.
Although we did detect three clones with a targeted endogenous 6–16 allele, this was again fewer than the eight clones predicted [using the ATF for HT1080 of 0.72 ± 0.22 × 10–6 and the formula ATF × N(T + R)/C].
The 48 PCR-positive clones were tested for the IFN dependence of their resistance to MPA. Again, the majority (46/48) showed a partial dependence on IFN. The remaining two clones were fully IFN-dependent (Fig. 3B) and corresponded to those that had been targeted at an endogenous 6–16 allele (lanes 3 and 6 in Fig. 4). The lower than expected number of clones targeted at an endogenous 6–16 allele therefore does not appear to be the result of any impaired transcription of endogenous 6–16 alleles in RAN10.
TSA treatment does not alter the gene targeting bias in RAN10
To test whether the 6–16 gene targeting bias in RAN10 was determined by allelic differences in histone acetylation, we carried out experiments in cells pretreated with TSA, a known inhibitor of histone deacetylation. Pilot experiments were conducted to titrate TSA in HT1080 cells and concentrations were chosen that allowed cell growth and normal survival after electroporation, even though they caused morphological changes. We electroporated 6–16gpt into RAN10 cells pretreated with or without TSA and selected in G418/IFN/MPA/X (Table 1, experiment 6). Five colonies were recovered from TSA-treated cells, four of which were expanded and analysed by Southern blot for 6–16 gene targeting (Fig. 4). All four of them were targeted at the H3.7 allele.
DISCUSSION
The key observation in this study is the pronounced preference for gene targeting to one particular 6–16 allele (H3.7) in a cell line (RAN10) that contains five other 6–16 alleles. Thus, of 108 candidate clones analysed (Fig. 4), 101 involved the H3.7 allele and only three involved an endogenous 6–16 allele. Targeting of the 6–16 gene in RAN10 is therefore 34-fold more likely to occur at the H3.7 allele than at any of the endogenous 6–16 alleles.
The cloned 6–16 DNA used to make the targeting construct and the two ectopic target loci was derived from the HT1080 cell line used for these experiments. The H3.7 allele is therefore identical in its nucleotide sequence to three of the five other 6–16 alleles in RAN10; the remaining two (endogenous) copies may or may not be identical depending on whether polymorphisms exist at this locus in HT1080. Some feature of the chromatin or DNA sequence context of the H3.7 allele must therefore be responsible for its preferability. At present we do not know what this feature is. A preliminary experiment with TSA suggests that the bias does not depend on histone deacetylation, but more detailed analyses of this kind would be of interest. Characterisation of the H3.7 integration site will also be informative as this may reveal genomic sequences (e.g. tandem or inverted repeats) as candidates responsible for the position effect. Such sequences might be naturally occurring or result from rearrangements generated during random integration of the H3.7 allele. Another possibility is that the SV40 ori/enhancer in the neo cassette close to the preferred target gene is responsible, because this may have been lost during integration of the other ectopic allele (H7.5). The SV40 ori/enhancer has been detected as a site of chromatin remodeling in SV40 (53) and Drosophila (54) chromosomes and has been associated with genomic instability in mouse cell lines (55). If the SV40 enhancer is responsible it seems unlikely that its effect can be explained simply in terms of its potential to stimulate transcription because we were able to show that the bias was not altered when transcription of all six loci was induced by IFN. While the SV40 ori/enhancer is not a normal constituent of the mammalian genome, any sequence capable of influencing gene targeting is of interest and could provide clues as to the nature of analogous mammalian genomic sequences.
Regardless of the underlying cause, our results clearly show that sequence and/or chromosome context can have a profound influence on gene targeting. Given the known existence of recombination hotspots in a variety of recombination systems, and that chromosomal context is well known to have important influences on gene expression, an effect of chromosomal context on gene targeting is not unexpected. Nevertheless, unequivocal data in support of a chromosomal position effect on gene targeting in mammalian cells have not previously been reported. Our data not only provide such support but also, as outlined below, suggest ways in which the effect might be acting.
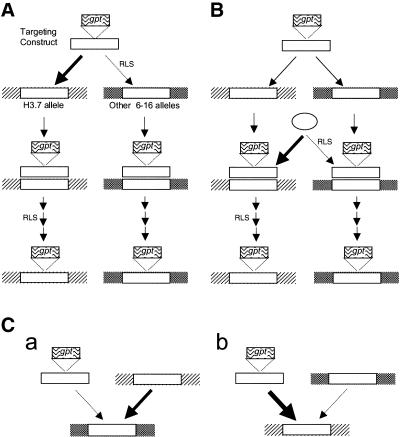
The effect of chromatin and/or sequence context on gene targeting cannot simply be to stimulate the overall efficiency of gene targeting at the H3.7 allele. If that were true, the 34-fold preference for targeting at the H3.7 allele would be mirrored by a similar increase in the ATF for RAN10. In fact, the ATF in RAN10 was only 3-fold higher than in HT1080. Any model to explain preferential targeting of the H3.7 allele must not only account for this relatively mild stimulation of the ATF in RAN10, but also for the significantly lower than expected frequency of targeting events involving 6–16 alleles other than H3.7 in RAN10. In Figure 5 we outline three distinct models that satisfy these requirements. Variations on these models can be envisaged and further work will be required to determine which, if any, is correct. All three models depend on an enhanced ability of the H3.7 allele, compared to other 6–16 sequences, to interact with a key, sometimes limiting, component in the gene targeting pathway.
Figure 5.
Three models to explain preferential targeting to H3.7 without increased ATF. Regions of homology are shown as white boxes. Hatched and stippled regions represent chromosomal flanking sequences at H3.7 and other 6–16 alleles, respectively. RLS, rate-limiting step. (A) Competition between H3.7 and other alleles for interaction with limiting amounts of targeting construct. (B) Competition between H3.7 and other 6–16 alleles for recruitment of a limiting factor (ellipse) required for HR. (C) Interallelic interference. (a) The H3.7 allele physically interacts with endogenous 6–16 alleles, preventing access by the targeting construct. (b) Endogenous 6–16 alleles do not impair access to the H3.7 allele by the targeting construct. See text for further details.
In the first model (Fig. 5A), the H3.7 allele interacts preferentially with the targeting construct during the initial homology search stage of gene targeting. Several studies (39,41,56,57) indicate that the homology search is not rate-limiting for gene targeting, and this would help to explain the limited increase in ATF in RAN10. To explain the much lower than expected frequency of targeting events involving 6–16 alleles other than H3.7, it is suggested that the targeting construct is limiting so that its preferential recruitment to the H3.7 allele makes it unavailable for targeting at other 6–16 alleles. We do not know how many DNA molecules reach the nucleus in our electroporation protocol, but it is known that electroporation can be particularly efficient in generating transfectants with single copy integrations (58), and it is therefore conceivable that intact targeting constructs are limiting. Furthermore, of those targeting constructs that do reach the nucleus intact, the majority are likely to be channeled into non-homologous end-joining pathways, leading to integration at random sites, and are therefore unavailable for gene targeting. Finally, it is possible that the H3.7 allele is capable of interacting with multiple targeting constructs at once, rather than just one, further limiting the availability of targeting constructs at other 6–16 loci. Nevertheless, the model predicts that, for conditions where targeting construct availability is not limited (e.g. high concentrations of targeting construct), preferential targeting at H3.7 will no longer be observed.
In the second model (Fig. 5B), all 6–16 alleles interact with the targeting construct with similar efficiencies during the homology search, but the H3.7 allele preferentially recruits a factor that is essential for a later step in the gene targeting pathway. Preferential recruitment of this factor to the H3.7 allele does not greatly stimulate the targeting efficiency, suggesting that recruitment of the factor is not normally rate-limiting for gene targeting. Preferential recruitment to the H3.7 allele does, however, reduce the concentration of factor available for recruitment to other 6–16 alleles, to the extent that recruitment becomes rate-limiting for gene targeting at these alleles. In this model, preferential targeting to the H3.7 allele is predicted to be independent of the concentration of targeting construct. Another testable prediction is that targeting events involving the H3.7 allele would suppress HR pathways that do not involve 6–16 sequences.
In the third model (Fig. 5C) the H3.7 allele competes with the targeting construct for interaction with other 6–16 alleles. Thus the H3.7 allele either prevents the targeting construct from interacting with other 6–16 alleles or disrupts any established interaction. This effect is non-reciprocal in the sense that access to the H3.7 allele by the targeting construct is relatively unaffected by the other 6–16 alleles. The model is based on a study (59) in which repair by HR of a damaged repetitive LINE element in mouse cells used other genomic LINE elements as recombination partners. Choice of the donor LINE elements appeared to depend more on its availability than on its degree of homology with the damaged element. We can thus imagine that, on the rare occasions when a 6–16 allele is competent to undergo HR, the H3.7 allele is in a chromosomal context that makes it more available as an HR partner than either the targeting construct or the other 6–16 alleles (Fig. 5C, a). Conversely, when the H3.7 is competent to undergo HR, other 6–16 alleles are not so favourably positioned to compete with the targeting construct (Fig. 5C, b). This model does not necessarily require any component to be present in limiting amounts and could accommodate an independence of preferential targeting on targeting construct concentration. The model also predicts that HR pathways in RAN10 that do not involve 6–16 DNA will not be affected by targeting at the H3.7 allele.
The chromosomal position effect we have observed has its main influence on target locus preference with a relatively small but significant effect on the overall efficiency of gene targeting. It might therefore appear that this effect is of minor importance in typical gene targeting situations, where there are usually only two target loci at equivalent chromosomal locations on homologous chromosomes, and is therefore unable to explain wide variations in gene targeting efficiencies for different target genes. It is interesting to consider the possibility, however, that non-homologous sequences also compete with the target loci during the initial genome scan. If this were true, the chromosomal position effect we have observed might have an important effect on the choice between random and targeted integration pathways. This may explain the fact that our estimates of RIF for the 6–16 gene targeting construct, but not for a simple expression cassette, were lower in RAN10 than in HT1080 cells. If this is correct, differences between the different target loci in their ability to compete with the rest of the genome for interaction with the targeting construct might account for at least some of the reported variation in gene targeting efficiencies.
In conclusion, our data provide clear evidence for a strong influence of chromosomal position on gene targeting in human cells. This influence appears to be on a step that is not rate-determining, such as the homology search, or on a later step for which an essential factor is limiting. The position effect thus has its main influence on the choice between identical targets at different loci in the same cell, with relatively little effect on the overall frequency of gene targeting.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Clare Huxley and Karen Brown for critical reading of the manuscript.
REFERENCES
- 1.Festenstein R. and Kioussis,D. (2000) Locus control regions and epigenetic chromatin modifiers. Curr. Opin. Genet. Dev., 10, 199–203. [DOI] [PubMed] [Google Scholar]
- 2.Cappechi M.R. (1989) Altering the genome by homologous recombination. Science, 244, 1288–1292. [DOI] [PubMed] [Google Scholar]
- 3.Smithies O., Koralewski,M.A., Song,K.Y. and Kucherlapati,R.S. (1984) Homologous recombination with DNA introduced into mammalian cells. Cold Spring Harbor Symp. Quant. Biol., 49, 161–170. [DOI] [PubMed] [Google Scholar]
- 4.Haber J.E. (2000) Partners and pathways repairing a double-strand break. Trends Genet., 16, 259–264. [DOI] [PubMed] [Google Scholar]
- 5.van Gent D.C., Hoeijmakers,J.H. and Kanaar,R. (2001) Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet., 2, 196–206. [DOI] [PubMed] [Google Scholar]
- 6.Porter A. (1998) Controlling your losses: conditional gene silencing in mammals. Trends Genet., 14, 73–79. [DOI] [PubMed] [Google Scholar]
- 7.Sedivy J.M. and Dutriaux,A. (1999) Gene targeting and somatic cell genetics—a rebirth or a coming of age? Trends Genet., 15, 88–90. [DOI] [PubMed] [Google Scholar]
- 8.Yáñez R.J. and Porter,A.C.G. (1998) Therapeutic gene targeting. Gene Ther., 5, 149–159. [DOI] [PubMed] [Google Scholar]
- 9.Vasquez K.M., Marburger,K., Intody,Z. and Wilson,J.H. (2001) Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA, 98, 8403–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng C. and Capecchi,M.R. (1992) Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol., 12, 3365–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith A.J. and Kalogerakis,B. (1990) Replacement recombinant events targeted at immunoglobulin heavy chain DNA sequences in mouse myeloma cells. J. Mol. Biol., 213, 415–435. [DOI] [PubMed] [Google Scholar]
- 12.te Riele H., Maandag,E.R. and Berns,A. (1992) Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl Acad. Sci. USA, 89, 5128–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Deursen J. and Wieringa,B. (1992) Targeting of the creatine kinase M gene in embryonic stem cells using isogenic and nonisogenic vectors. Nucleic Acids Res., 20, 3815–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasty P., Crist,M., Grompe,M. and Bradley,A. (1994) Efficiency of insertion versus replacement vector targeting varies at different chromosomal loci. Mol. Cell. Biol., 14, 8385–8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmer A. and Reynolds,K. (1994) Gene targeting constructs: effects of vector topology on co-expression efficiency of positive and negative selectable marker genes. Biochem. Biophys. Res. Commun., 201, 943–949. [DOI] [PubMed] [Google Scholar]
- 16.Nairn R.S., Adair,G.M., Porter,T., Pennington,S.L., Smith,D.G., Wilson,J.H. and Seidman,M.M. (1993) Targeting vector configuration and method of gene transfer influence targeted correction of the APRT gene in Chinese hamster ovary cells. Somat. Cell Mol. Genet., 19, 363–375. [DOI] [PubMed] [Google Scholar]
- 17.Ward M.A., Abramow Newerly,W. and Roder,J.C. (1993) Effect of vector topology on homologous recombination at the CHO aprt locus. Somat. Cell Mol. Genet., 19, 257–264. [DOI] [PubMed] [Google Scholar]
- 18.Russell D.W. and Hirata,R.K. (1998) Human gene targeting by viral vectors. Nature Genet., 18, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yáñez R.J. and Porter,A.C.G. (1999) Influence of DNA delivery method on gene targeting frequencies in human cells. Somat. Cell Mol. Genet., 25, 27–31. [DOI] [PubMed] [Google Scholar]
- 20.Zimmer A. and Gruss,P. (1989) Production of chimaeric mice containing embryonic stem (ES) cells carrying a homoeobox Hox 1.1 allele mutated by homologous recombination. Nature, 338, 150–153. [DOI] [PubMed] [Google Scholar]
- 21.Smith G.R. (1994) Hotspots of homologous recombination. Experientia, 50, 234–241. [DOI] [PubMed] [Google Scholar]
- 22.Jeffreys A.J., Barber,R., Bois,P., Buard,J., Dubrova,Y.E., Grant,G., Hollies,C.R., May,C.A., Neumann,R., Panayi,M. et al. (1999) Human minisatellites, repeat DNA instability and meiotic recombination. Electrophoresis, 20, 1665–1675. [DOI] [PubMed] [Google Scholar]
- 23.Farah J.A., Hartsuiker,E., Mizuno,K., Ohta,K. and Smith,G.R. (2002) A 160-bp palindrome is a Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics, 161, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buzina A. and Shulman,M.J. (1996) An element in the endogenous IgH locus stimulates gene targeting in hybridoma cells. Nucleic Acids Res., 24, 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raynard S.J., Read,L.R. and Baker,M.D. (2002) Evidence for the murine IgH mu locus acting as a hot spot for intrachromosomal homologous recombination. J Immunol., 168, 2332–2339. [DOI] [PubMed] [Google Scholar]
- 26.Wahls W.P. and Moore,P.D. (1990) Homologous recombination enhancement conferred by the Z-DNA motif d(TG)30 is abrogated by simian virus 40 T antigen binding to adjacent DNA sequences. Mol. Cell. Biol., 10, 794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahls W.P., Wallace,L.J. and Moore,P.D. (1990) The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol. Cell. Biol., 10, 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahls W.P., Wallace,L.J. and Moore,P.D. (1990) Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell, 60, 95–103. [DOI] [PubMed] [Google Scholar]
- 29.Sargent R.G., Merrihew,R.V., Nairn,R., Adair,G., Meuth,M. and Wilson,J.H. (1996) The influence of a (GT)29 microsatellite sequence on homologous recombination in the hamster adenine phosphoribosyltransferase gene. Nucleic Acids Res., 24, 746–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards E.J. and Elgin,S.C. (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell, 108, 489–500. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 32.Nickoloff J.A. (1992) Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol., 12, 5311–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nickoloff J.A. and Reynolds,R.J. (1990) Transcription stimulates homologous recombination in mammalian cells. Mol. Cell. Biol., 10, 4837–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thyagarajan B., Johnson,B.L. and Campbell,C. (1995) The effect of target site transcription on gene targeting in human cells in vitro. Nucleic Acids Res., 23, 2784–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson R.S., Sheng,M., Greenberg,M.E., Kolodner,R.D., Papaioannou,V.E. and Spiegelman,B.M. (1989) Targeting of nonexpressed genes in embryonic stem cells via homologous recombination. Science, 245, 1234–1236. [DOI] [PubMed] [Google Scholar]
- 36.Lichten M. and Haber,J.E. (1989) Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics, 123, 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray M. and Honigberg,S.M. (2001) Effect of chromosomal locus, GC content and length of homology on PCR-mediated targeted gene replacement in Saccharomyces. Nucleic Acids Res., 29, 5156–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson J.H., Leung,W.Y., Bosco,G., Dieu,D. and Haber,J.E. (1994) The frequency of gene targeting in yeast depends on the number of target copies. Proc. Natl Acad. Sci. USA, 91, 177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng H. and Wilson,J.H. (1990) Gene targeting in normal and amplified cell lines. Nature, 344, 170–173. [DOI] [PubMed] [Google Scholar]
- 40.Lin F.L., Sperle,K. and Sternberg,N. (1985) Recombination in mouse L cells between DNA introduced into cells and homologous chromosomal sequences. Proc. Natl Acad. Sci. USA, 82, 1391–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas K.R., Folger,K.R. and Capecchi,M.R. (1986) High frequency targeting of genes to specific sites in the mammalian genome. Cell, 44, 419–428. [DOI] [PubMed] [Google Scholar]
- 42.Porter A.C.G., Chernajovsky,Y., Dale,T.C., Gilbert,C.S., Stark,G.R. and Kerr,I.M. (1988) Interferon response element of the human gene 6–16. EMBO J., 7, 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller K., Heller,H. and Doerfler,W. (2001) Foreign DNA integration. Genome-wide perturbations of methylation and transcription in the recipient genomes. J. Biol. Chem., 276, 14271–14278. [DOI] [PubMed] [Google Scholar]
- 44.Scrable H. and Stambrook,P.J. (1999) A genetic program for deletion of foreign DNA from the mammalian genome. Mutat. Res., 429, 225–237. [DOI] [PubMed] [Google Scholar]
- 45.Porter A.C.G. and Itzhaki,J.E. (1993) Gene targeting in human somatic cells. Complete inactivation of an interferon-inducible gene. Eur. J. Biochem., 218, 273–281. [DOI] [PubMed] [Google Scholar]
- 46.Southern P.J. and Berg,P. (1982) Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet., 1, 327–341. [PubMed] [Google Scholar]
- 47.Yáñez R.J. and Porter,A.C.G. (1999) Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther., 6, 1282–1290. [DOI] [PubMed] [Google Scholar]
- 48.Rasheed S., Nelson-Rees,W.A., Toth,E.M., Arnstein,P. and Gardner,M.B. (1974) Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer, 33, 1027–1033. [DOI] [PubMed] [Google Scholar]
- 49.Itzhaki J.E., Gilbert,C.S. and Porter,A.C.G. (1997) Construction by gene targeting in human cells of a “conditional” CDC2 mutant that rereplicates its DNA. Nature Genet., 15, 258–265. [DOI] [PubMed] [Google Scholar]
- 50.Kelly J.M., Porter,A.C.G., Chernajovsky,Y., Gilbert,C.S., Stark,G.R. and Kerr,I.M. (1986) Characterization of a human gene inducible by alpha- and beta-interferons and its expression in mouse cells. EMBO J., 5, 1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adair G.M., Nairn,R.S., Wilson,J.H., Seidman,M.M., Brotherman,K.A., MacKinnon,C. and Scheerer,J.B. (1989) Targeted homologous recombination at the endogenous adenine phosphoribosyltransferase locus in Chinese hamster cells. Proc. Natl Acad. Sci. USA, 86, 4574–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itzhaki J.E. and Porter,A.C.G. (1991) Targeted disruption of a human interferon-inducible gene detected by secretion of human growth hormone. Nucleic Acids Res., 19, 3835–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milavetz B.I. (2002) SP1 and AP-1 elements direct chromatin remodeling in SV40 chromosomes during the first 6 hours of infection. Virology, 294, 170–179. [DOI] [PubMed] [Google Scholar]
- 54.Alexiadis V., Varga-Weisz,P.D., Bonte,E., Becker,P.B. and Gruss,C. (1998) In vitro chromatin remodelling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. EMBO J., 17, 3428–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter D.J. and Gurney,E.G. (1994) The genomic instability associated with integrated simian virus 40 DNA is dependent on the origin of replication and early control region. J. Virol., 68, 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukacsovich T., Waldman,B.C. and Waldman,A.S. (2001) Efficient recruitment of transfected DNA to a homologous chromosomal target in mammalian cells. Biochim. Biophys. Acta, 1521, 89–96. [DOI] [PubMed] [Google Scholar]
- 57.Rommerskirch W., Graeber,I., Grassmann,M. and Grassmann,A. (1988) Homologous recombination of SV40 DNA in COS7 cells occurs with high frequency in a gene dose independent fashion. Nucleic Acids Res., 16, 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boggs S.S., Gregg,R.G., Borenstein,N. and Smithies,O. (1986) Efficient transformation and frequent single-site, single-copy insertion of DNA can be obtained in mouse erythroleukemia cells transformed by electroporation. Exp. Hematol., 14, 988–994. [PubMed] [Google Scholar]
- 59.Tremblay A., Jasin,M. and Chartrand,P. (2000) A double-strand break in a chromosomal LINE element can be repaired by gene conversion with various endogenous LINE elements in mouse cells. Mol. Cell. Biol., 20, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.