Abstract
Overexpression of vascular endothelial growth factor (VEGF) is implicated in a number of diseases. It is therefore critical that mechanisms exist to strictly regulate VEGF expression. A hypoxia-responsive (HR) region of the VEGF promoter which binds the HIF-1 transcription factor is a target for many signals that up-regulate VEGF transcription. Repressors targeting the HIF-1 transcription factor have been identified but no repressors directly binding the HR promoter region had been reported. We now report a novel mechanism of repression of the VEGF HR region involving DNA binding. We find that single strand DNA-specific cold shock domain (CSD or Y-box) proteins repress the HR region via a binding site downstream of the HIF-1 site. The repressor site is functional in unstimulated, normoxic fibroblasts and represents a novel means to prevent expression of VEGF in the absence of appropriate stimuli. We characterized complexes forming on the VEGF repressor site and identified a previously unreported nuclear CSD protein complex containing dbpA. Nuclear dbpA appears to bind as a dimer and we determined a means by which nuclear CSD proteins may enter double strand DNA to bind to their single strand sites to bring about repression of the VEGF HR region.
INTRODUCTION
Vascular endothelial growth factor (VEGF) is a growth factor for vascular endothelial cells that induces proliferation and promotes cell migration (1–3). Disregulation of VEGF plays a key role in the development and maintenance of solid tumors by promoting tumor angiogenesis (4,5). The importance of VEGF in the pathogenesis of solid tumors is evidenced by the ability of agents inhibiting VEGF activity to inhibit tumor growth and to cause tumor regression (6). Disregulated VEGF expression also contributes to the progression of a number of other diseases characterized by abnormal angiogenesis (5). High levels of VEGF expression by tumor cells can be brought about by the action of activated oncogenes, the loss or mutation of tumor suppressors, the overexpression of growth factors and low oxygen levels (hypoxia) (3,4,7–10). Non-tumor cells, such as fibroblasts, surrounding tumor tissue are also sources of VEGF overexpression (11). Little is known about the regulation of VEGF expression in fibroblasts.
Transcriptional regulation plays a major role in both basal and inducible expression of the VEGF gene in both tumor and non-tumor cells (3,4). A key region of the VEGF promoter, located at approximately –930 from the transcription start site, is a target for a number of signals. This ∼50 bp region is responsive to hypoxia, oncoproteins and growth factor activation, primarily via activation of HIF-1 (hypoxia inducible factor 1), which binds to this region [hypoxia responsive (HR) region, Fig. 1A] (10,12–18). Expression from this region has been shown to be repressed under non-stimulated/normoxic (normal oxygen) conditions by the p53 and von Hippel Lindau (VHL) tumor suppressors which function by regulation of stability of the HIF-1α component of the HIF-1 transcription factor (9,10,12). No mechanisms of repression involving direct binding of a repressor to HR region DNA have been reported.
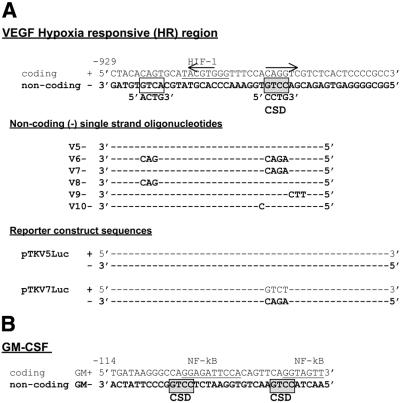
Figure 1.
Sequence of oligonucleotides for gel retardation assays and cloning. (A) The sequence of the mouse VEGF HR region coding (+) and non-coding (–) strands are shown (35). The binding site for the HIF-1 transcription factor (49,50) and the 5′-CCTG-3′ CSD protein-binding site identified here are indicated. An imperfect 5 bp inverted repeat (36,37) is also indicated by arrows. Non-coding (–) strand retardation oligonucleotide sequences (V5– to V10–) and sequences within reporter constructs (pTKV5Luc and pTKV7Luc) are given below with only those bases that vary from the wild-type indicated. (B) The sequence of coding (+) and non-coding (–) oligonucleotides containing a GM-CSF CSD protein-binding region are shown (29–31). CSD protein-binding repressor sites on the non-coding strand are indicated (30).
Cold shock domain (CSD) proteins (also called Y-box proteins) (19,20) have been shown to be potent repressors of a number of growth factor and stress response genes via both DNA binding and non-DNA binding mechanisms (21,22). There are two major CSD proteins in non-germ cells, dbpB (or YB-1) and dbpA (19–22), in addition to a splice variant of dbpA (23,24) and a small proteolytically processed form of dbpB (25). CSD proteins bind primarily to single strand DNA and RNA and can in some cases bind with low affinity to double strand DNA (21,22,26–28). CSD proteins were first identified as binding to an inverted 5′-CCAAT-3′ repeat but a general consensus binding site for CSD proteins has not been established and they are generally considered to bind to CT-rich sequences (21,22). We have, however, previously identified a specific pair of 5′-CCTG-3′ CSD protein-binding sites required for repression in the granulocyte macrophage colony stimulating factor (GM-CSF) gene and have identified a CSD protein nuclear complex, NF-GMb, containing dbpB/YB-1, that binds to these sequences (29–31). In general, there is little data on the nature of nuclear CSD protein complexes. Our data suggested that the CSD protein within NF-GMb binds as a monomer (29). CSD binding to the GM-CSF gene is single strand DNA specific and contacts sequences on the non-coding (–) strand of a TNFα-responsive region operational in fibroblasts (Fig. 1B).
We now report a single strand-specific CSD protein-binding sequence, on the non-coding (–) strand of the VEGF HR region, with repressor function in unstimulated/normoxic fibroblasts. The repressor sequence is located downstream of the HIF-1 site. In contrast to GM-CSF, the VEGF repressor site requires only a single 5′-CCTG-3′ element and binds a novel nuclear complex, NF-V1, containing dbpA, which appears to bind as a dimer. We demonstrate the ability of CSD proteins to control VEGF expression and determine a potential means by which single strand binding CSD proteins gain entry to the VEGF promoter within double strand DNA. This work reveals a novel means of preventing inappropriate expression of VEGF in unstimulated/normoxic cells.
MATERIALS AND METHODS
Plasmid constructs
The constructs pTKV5Luc and pTKV7Luc were constructed by cloning respective wild-type and mutant mouse VEGF promoter HR region oligonucleotide sequences (Fig. 1A) into pXPGTK229 (a gift from P. Cockerill, IMVS, Adelaide, Australia). Promoter fragments were inserted upstream of the thymidine kinase (TK) gene. The same oligonucleotide sequences were also cloned into pTK81Luc (32). pTK81Luc was used in overexpression experiments as pXPGTK229 was repressed slightly by dbpB/YB-1 and dbpA (10–20%). pTK81Luc was not repressed. The dbpB/YB-1 and dbpA CSD overexpression constructs pSGdbpB and pSGdbpA contained full-length human cDNA sequences cloned into pSG5 as previously described (30). Construction of expression vectors producing recombinant human GST–dbpA (pGEXA) and GST–dbpB (pGEXBT) was previously described (29,33).
Oligonucleotides and probe preparation
Oligonucleotides for cloning or gel retardation assays were synthesized by Geneworks (Adelaide, Australia) and purified from non-denaturing polyacrylamide gels. Single strand DNA probes for gel retardation assays were prepared by end-labeling coding (+) or non-coding (–) single strand oligonucleotides with [γ-32P]ATP and T4 polynucleotide kinase followed by gel purification. Double strand DNA probes were prepared by annealing 32P-labeled coding (+) strand oligonucleotides with unlabeled non-coding (–) strand oligonucleotides followed by gel purification. VEGF and GM-CSF (29) gene oligonucleotide sequences are shown in Figure 1.
Preparation of recombinant and nuclear protein
The Escherichia coli strain MC1061 transformed with pGEXBT or pGEXA was induced with isopropyl-1-thio-β-d-galactopyranoside to produce recombinant GST–dbpB and GST–dbpA, respectively (29). The fusion proteins were purified on glutathione–Sepharose beads as described by the manufacturer (Promega). Crude nuclear extracts from mouse Balb/c 3T3 fibroblasts were prepared as previously described (29).
FPLC gel filtration of nuclear extracts
Crude nuclear extract was applied at a flow rate of 0.35 ml/min to a Superdex 200 column (10 mm diameter, 20 ml bed volume) pre-equilibrated with buffer containing 150 mM KCl, 20 mM Tris–HCl pH 7.6, 20% glycerol, 1.5 mM MgCl2, 2 mM DTT, 0.4 mM PMSF and 1 mM Na3VO4. The NF-V1 complex was eluted with the same buffer and 0.5 ml fractions collected. The molecular mass of the complex was estimated from this column by comparison with the elution volumes of γ-globulin, bovine serum albumin, ovalbumin, myoglobin and vitamin B12.
Antibodies
An anti-CSD peptide polyclonal antibody was raised by immunizing rabbits with the peptide IKKNNPRKYLRSVGD (human dbpB amino acids 89–103; peptide conserved in mouse and human dbpA and dbpB) conjugated to keyhole limpet hemocyanin (Imject conjugation kit; Pierce) as previously described (33). The anti-dbpA polyclonal antibody was a gift from Maria Balda (University College, London, UK).
Gel retardation analysis, competitions and antibody analysis
Gel retardation assays were performed using 0.25 ng single strand 32P-labeled oligonucleotide probe or 0.5 ng 32P-labeled double strand oligonucleotide [only coding (+) strand labeled], in a 10 µl reaction mix of 0.5× TM buffer (29) containing 200 mM KCl, 0.4 µg poly(dI·dC) and either 1 µg crude nuclear extract or 25 ng recombinant CSD fusion protein (GST–dbpB or GST–dbpA). Retardation assays using recombinant protein also contained 2 µg bovine serum albumin. Reactions were incubated at room temperature for 20 min and analyzed on 12 or 6% non-denaturing polyacrylamide gels for analysis of nuclear extracts and recombinant protein, respectively. Competition with unlabeled single strand oligonucleotides was performed by addition of protein and 5 ng unlabeled probe, followed by immediate addition of the 32P-labeled probe. Competition with digested or undigested double strand plasmid DNA (pTK81Luc or pTK81V5Luc) was performed by addition of protein and 1.2 µg plasmid DNA, which was incubated for 20 min at room temperature before addition of 32P-labeled probe. Competitions were performed in the presence or absence of 10 mM MgCl2 (27). Antibody blocking experiments were performed by adding protein and antibody and incubating for 5 min at room temperature before adding the 32P-labeled probe. The reaction was incubated for an additional 20 min at room temperature before being analyzed on polyacrylamide gels. Oligonucleotides used in retardation assays are shown in Figure 1.
UV crosslinking
Nuclear extracts were bound to 32P-labeled single strand DNA probes in a 25 µl gel retardation reaction and fractionated on a 12% polyacrylamide gel as described above. The gel was exposed to UV light (340 nm) for 15 min and retarded complexes were excised after exposure overnight to X-ray film. Protein in excised bands was analyzed on 12% SDS–polyacrylamide gels as previously descibed (29).
Cell culture and transfection
Balb/c 3T3 mouse fibroblasts were grown in Dulbecco’s modified Eagle’s medium and 10% fetal calf serum. Aliquots of 2 × 105 cells were transfected with 500 ng luciferase reporter constructs or co-transfected with 500 ng reporter constructs and 250 ng dbpB (pSGdbpB) and dbpA (pSGdbpA) overexpression constructs. Transfections were performed using Lipofectamine™ 2000 (Gibco BRL) according to the manufacturer’s directions. Twenty-four hours after transfection, cells were either placed in hypoxic conditions (<1% O2, 5–8% CO2) or left in normoxic conditions (20% O2) for 16 h. Hypoxic conditions were generated by ascorbic acid depletion of atmospheric oxygen in a sealed 2.5 l container (AnaeroGen AN25; Oxoid, UK) as previously described (34). After treatment of cells, luciferase activity was determined as described (33).
RESULTS
A single strand DNA-specific nuclear CSD complex binds to a 5′-CCTG-3′ site in the VEGF HR region
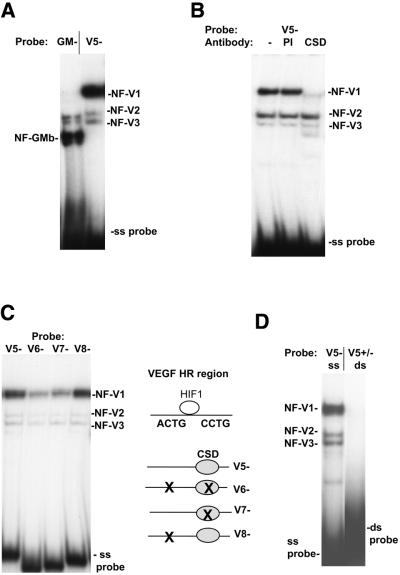
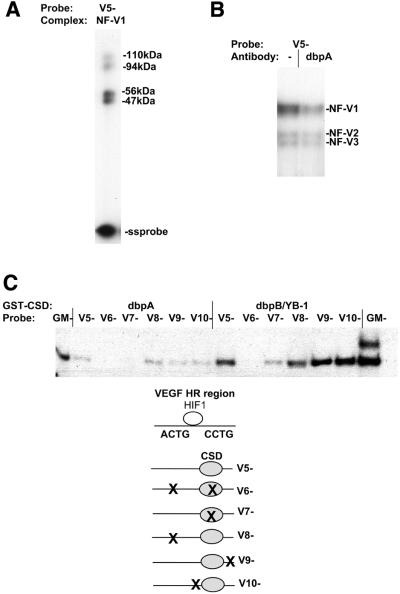
We have previously reported that a pair of 5′-CCTG-3′ sites, on the non-coding strand of a TNFα-responsive region in the GM-CSF gene, bind CSD proteins in a single strand-specific manner (29–31). A single 5′-CCTG-3′ sequence can also be observed on the non-coding (–) strand of the VEGF HR region, located downstream of the HIF-1-binding site (Fig. 1A). As no proteins, other than those binding the HIF-1 site, have been identified binding to and regulating the VEGF HR region, we investigated this region for nuclear CSD protein complex formation. From our previous analysis, we found that the GM-CSF CSD protein-binding sites bound a CSD protein nuclear complex called NF-GMb (29,31). Both CSD protein-binding sites were required for this binding. NF-GMb complex formation was detected in all cell types examined and contained a 42 kDa CSD protein, which is the reported size for dbpB/YB-1. The formation of this complex on the GM-CSF single strand oligonucleotide (GM–) using mouse Balb/c 3T3 fibroblast nuclear extracts in a gel retardation assay is shown in Figure 2A. The authenticity of the NF-GMb complex in Balb/c 3T3 fibroblasts was verified by antibody and UV crosslink analysis (data not shown). The single strand non-coding (–) VEGF HR region sequence (V5–, Fig. 1), however, did not form a complex co-migrating with the NF-GMb complex, but instead formed a major slower migrating complex (NF-V1) and two minor complexes (NF-V2 and NF-V3) (Fig. 2A). Preincubation of nuclear extracts with an anti-CSD antibody (CSD) blocked formation of the NF-V1 complex but not the NF-V2 or NF-V3 complexes (Fig. 2B). Pre-immune serum had no effect on complex formation. The VEGF HR region therefore binds a novel CSD protein nuclear complex, NF-V1, that is not detected on the GM-CSF sequence. The NF-V1 complex was detected in all cell lines tested (data not shown).
Figure 2.
A single strand-specific nuclear CSD protein complex, NF-V1, binds to a 5′-CCCTG-3′ sequence in the VEGF HR region. (A) Balb/c 3T3 mouse fibroblast nuclear extracts were bound to labeled non-coding (–) single strand mouse VEGF HR region (V5–) or GM-CSF (GM–) oligonucleotides (Fig. 1) in a gel retardation assay. VEGF (NF-V1 to NF-V3) and GM-CSF (NF-GMb) nuclear complexes and free single strand oligonucleotides (ss probe) are indicated. (B) Nuclear extract for binding to the VEGF probe (V5–) was pretreated with a CSD protein-specific polyclonal antibody (CSD), pre-immune serum (PI) or left untreated (–) and assayed by gel retardation. (C) Nuclear extract was bound to wild-type (V5–) and mutant (V6–, V7– and V8–) VEGF HR region single strand oligonucleotides in a gel retardation assay. (D) Nuclear extract was bound to the single strand non-coding (–) VEGF HR region oligonucleotide (V5–) or to a double strand HR region probe (V5+/–) where only the coding (+) strand was labeled and assayed by gel retardation. Free double strand and single strand probe (ds and ss probe) is indicated.
To determine the sequence requirements of the NF-V1 complex, crude Balb/c 3T3 nuclear extracts were bound to wild-type and mutant VEGF HR region non-coding (–) strand sequences in a gel retardation assay (Fig. 2C). Mutation of the 5′-CCTG-3′ site (V7–) to 5′-AGAC-3′ resulted in reduction of NF-V1 complex formation, whereas mutation of an imperfect 5′-ACTG-3′ site (V8–) upstream of the HIF-1 site had no effect on binding. A slight further reduction in binding was seen when both sites were mutated (V6–). Hence the primary site for nuclear CSD protein complex formation on the VEGF HR region is the single 5′-CCTG-3′ sequence, on the non-coding strand, downstream of the HIF-1 site.
CSD protein binding to many genes is single strand specific, as reviewed (21,22,30,31). CSD proteins have also been reported to bind to double strand DNA in some genes with low affinity (26–28). To determine the single strand versus double strand DNA specificity of the NF-V1 complex, binding to double strand VEGF HR region DNA (V5+/–) was tested (Fig. 2D). NF-V1 complex formation was undetectable on double strand DNA, under the conditions used in the gel retardation assay, and hence is specific to the non-coding single strand.
The 5′-CCTG-3′ CSD protein-binding sequence is a repressor site in normoxic fibroblasts
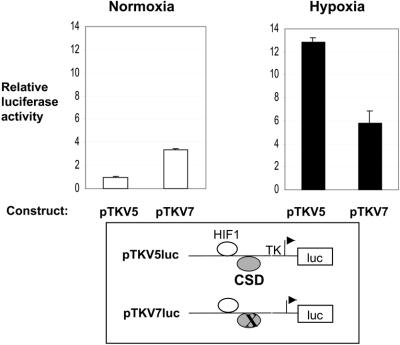
CSD proteins have been shown to function as repressors of expression from a number of growth factor and stress response genes, as reviewed (21,22). Repression required the presence of both 5′-CCTG-3′ CSD protein-binding sites in the GM-CSF gene (30,31). We and others proposed that CSD proteins maintain promoters in a single strand structure preventing the binding of activators that require double strand DNA. Activation by CSD proteins has also been reported, but most commonly in viral rather than cellular genes (21,22). To test the function of the single VEGF HR region 5′-CCTG-3′ CSD protein-binding site, expression from the 47 bp HR region, on a heterologous TK promoter in a luciferase reporter vector, was tested in transient transfection assays in mouse Balb/c 3T3 fibroblasts (Fig. 3). In unstimulated cells (normoxic, normal oxygen conditions) mutation of the 5′-CCTG-3′ site (pTKV7Luc) resulted in a 3.4-fold increase in expression relative to the wild-type construct (pTKV5Luc), hence revealing this site as a repressor of normoxic expression from the VEGF HR region. The CSD site therefore plays a role in preventing inappropriate expression in unstimulated cells. A repressor site within the VEGF HR region has not previously been reported.
Figure 3.
The 5′-CCCTG-3′ CSD protein-binding sequence in the VEGF HR region is a repressor site in normoxic fibroblasts. Wild-type and mutant mouse VEGF HR region reporter constructs (pTKV5Luc and pTKV7Luc) were transfected into mouse Balb/c 3T3 fibroblasts and grown under normoxic (unstimulated) or hypoxic (activated) conditions and the cells assayed for luciferase activity. Data is pooled from four experiments, each assayed in triplicate, and is presented as luciferase activity for each construct relative to that for the backbone vector under normoxic conditions. Backbone vector activity was not affected by hypoxia.
In hypoxic (low oxygen) conditions the wild-type HR region construct (pTKV5Luc) is efficiently activated (12.8-fold above normoxic expression). This is the first demonstration of hypoxic response of the HR region in fibroblasts. This is consistent with our previous demonstration of hypoxic induction of VEGF mRNA levels (34) and activation of the whole VEGF promoter (–1217 to +30) (35) in transient transfections in Balb/c 3T3 fibroblasts (data not shown). In contrast to the situation in normoxic cells, mutation of the 5′-CCTG-3′ CSD protein-binding site (pTKV7Luc) resulted in a decrease (55%) rather than an increase in hypoxic expression. This suggests that either (i) single strand binding CSD repressor proteins are replaced by an activator during hypoxic induction or (ii) that CSD proteins themselves are involved in activation. The former seemed most likely as work by us and others suggests that CSD proteins are removed/inhibited from binding single strand DNA upon activation, allowing the binding of double strand DNA binding activator proteins, as reviewed (22,29,33).
Overexpression of the dbpA and dbpB/YB-1 CSD proteins represses VEGF HR region expression
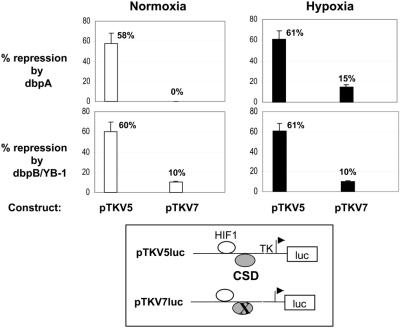
To address the role of CSD proteins in both unstimulated (normoxic) cells and activated (hypoxic) cells, VEGF HR region luciferase reporter constructs were co-transfected with CSD protein overexpression constructs in Balb/c 3T3 fibroblasts (Fig 4). In both normoxic and hypoxic cells, overexpression of dbpA (pSGdbpA) and dbpB (pSGdbpB) CSD proteins resulted in efficient repression (∼60%) from the wild-type HR region construct (pTKV5Luc), confirming a repressor rather than an activator function for CSD proteins on the HR region. In both cases the degree of repression was drastically reduced by mutation of the CSD protein-binding site (pTKV7Luc), confirming that this site is the target for repression by CSD proteins. This was most evident with overexpression of dbpA in normoxic cells, where no repression was observed when the CSD protein-binding site was mutated (pTKV7Luc).
Figure 4.
Overexpression of dbpA and dbpB/YB-1 CSD proteins represses expression from the VEGF HR region. VEGF HR region reporter constructs (pTKV5Luc and pTKV7Luc) were co-transfected with dbpA (pSGdbpA) or dbpB/YB-1 (pSGdbpB) overexpression vectors or the pSG5 overexpression backbone vector, into Balb/c 3T3 fibroblasts. Cells were grown under normoxic (unstimulated) or hypoxic (activated) conditions and assayed for luciferase activity. Percent repression represents the degree to which luciferase activity of each HR region reporter construct is reduced when co-transfected with the pSGdbpA (dbpA) or pSGdbpB (dbpB/YB-1) overexpression vector, relative to luciferase activity from reporter constructs co-transfected with the backbone overexpression vector (pSG5) alone. Data is pooled from four experiments. Luciferase activity from the backbone reporter vector was not affected by dbpA or dbpB overexpresssion.
Taken together, the transfection and binding data (Figs 2–4) demonstrate, first, that the single strand-specific CSD proteins are repressors of VEGF HR region expression during normoxia via the 5′-CCTG-3′ CSD protein-binding site and, hence, play an important role in preventing unwanted expression from the VEGF promoter in the absence of activation signals. Secondly, it shows that CSD proteins do not function as activators in hypoxic cells, suggesting that repressive single strand binding CSD proteins are replaced by an activator during hypoxia. A hypoxic activator site has been reported that requires sequences overlapping the HR region CSD site, in addition to sequences immediately downstream of the CSD protein-binding site (36,37). Nuclear CSD protein binding does not require these downstream sequences (data not shown). This is also observed for recombinant CSD protein binding (see Fig. 5C). The nature of the activator proteins binding to these sequences has not been determined, but we have identified a nuclear complex that binds to the activator sequences that is dependent on double strand DNA for binding (P.Diamond, L.S.Coles and G.J.Goodall, unpublished data). The ability of CSD protein overexpression to repress HR region expression during hypoxia suggests that overexpressed CSD proteins may in turn be able to replace/displace the double strand activator.
Figure 5.
The NF-V1 complex contains the dbpA CSD protein. (A) Balb/c 3T3 nuclear extract was bound to the non-coding single strand VEGF HR region oligonucleotide (V5–) and nuclear complexes were separated by gel retardation assay and exposed to UV light to crosslink proteins to DNA. Crosslinked proteins in the NF-V1 complex were then analyzed by SDS–PAGE. The sizes of crosslinked proteins in kDa and free single strand probe (ss probe) are indicated. Crosslinked protein sizes were determined by subtraction of the molecular weight of the single strand oligonucleotide (V5–). (B) Nuclear extract was pretreated with a dbpA polyclonal antibody (dbpA) or left untreated (–) and then bound to the single strand VEGF oligonucleotide (V5–) in a gel retardation assay. Nuclear complexes are indicated (NF-V1, NF-V2 and NF-V3). (C) Recombinant GST–dbpA and GST–dbpB/YB-1 was bound to wild-type (V5–) and mutant (V6– to V10–) single strand VEGF and GM-CSF (GM–) oligonucleotides in a gel retardation assay.
The VEGF HR region NF-V1 nuclear complex contains dbpA
Somatic cells contain two types of CSD protein, dbpA and dbpB. We have previously reported that a CSD protein nuclear complex, NF-GMb, binding to the GM-CSF CSD sites as shown in Figure 2A, contained a 42 kDa CSD protein of the correct size for dbpB/YB-1 (30,31). Data suggested that this protein bound as a monomer (29). The difference in mobility of the NF-V1 complex on the VEGF HR region CSD protein-binding site, compared to the mobility of the NF-GMb complex (Fig. 2A) suggested that the NF-V1 complex had a different protein composition to NF-GMb.
To analyze the proteins within NF-V1, UV crosslink analysis was performed (Fig. 5A). Nuclear extract was bound to the single strand VEGF HR sequence (V5–) and complexes were separated on a retardation gel. The gel was exposed to UV light to crosslink proteins to DNA, the NF-V1 complex was excised from the gel and crosslinked proteins were sized by SDS–PAGE. In contrast to NF-GMb, the NF-V1 complex contained crosslinked proteins of 56, 47, 110 and 94 kDa. The sizes of crosslinked proteins are determined by subtraction of the size of the single strand DNA probe. As 56 and 47 kDa are the appropriate size for dbpA and a dbpA splice variant (23,24,38), nuclear extracts were treated with a dbpA-specific polyclonal antibody and tested for binding to the VEGF sequence (V5–) in a gel retardation assay (Fig. 5B). As expected, formation of the CSD protein-containing NF-V1 complex was reduced by the dbpA antibody. NF-V2 and NF-V3 complex formation was not affected. NF-V1, therefore, appears to contain dbpA. Little is known about the composition of CSD protein nuclear complexes and a nuclear complex containing dbpA has not previously been reported. The lack of NF-GMb complexes binding to the VEGF HR region suggests that the VEGF HR region exclusively binds nuclear dbpA. Consistent with this, the VEGF HR region sequence (V5–) does not compete for NF-GMb complex formation on the GM-CSF (GM–) sequence (data not shown). This was the same for all cell types tested (data not shown). This is not due to the lack of dbpB/YB-1 in nuclear extracts, as the NF-GMb complex on the GM-CSF sequence is readily formed in all cell types, including Balb/c 3T3 fibroblasts (Fig. 2A).
The binding of dbpA to the VEGF HR sequence (V5–) was confirmed using recombinant dbpA in a gel retardation assay (Fig. 5C). As for nuclear NF-V1 complex formation this binding was dependent on the 5′-CCTG-3′ site (mutated in V7–). Mutations immediately upstream (V10–) or downstream of this site (V9–) had no effect on binding. The sequences mutated in V9– are part of a hypoxic activator sequence (36,37).
To determine if the preference of the VEGF sequence for nuclear dbpA complex formation was simply due to increased affinity for dbpA relative to dbpB, recombinant dbpB was also bound to the VEGF (V5–) and GM-CSF (GM–) sequences (Fig. 5C). As expected, the GM-CSF sequence binds dbpB more strongly than dbpA (previously reported) (29,30) given the selective binding of the nuclear NF-GMb complex to this sequence. Surprisingly, however, the VEGF sequence (V5–) also bound dbpB more strongly than dbpA. This is reflected in the ability of both overexpressed dbpA and dbpB to repress the VEGF HR region (Fig. 4). As formation of the NF-GMb complex, on the GM-CSF sequence, suggests that there is available dbpB/YB-1 in Balb/c 3T3 nuclear extracts, CSD protein complex formation must be altered in nuclear extracts to allow preferential binding of nuclear dbpA to the VEGF sequence. One possibility was that the additional crosslinked proteins (94 and 110 kDa) in the NF-V1 complex enabled preferential binding of dbpA.
Nuclear dbpA may bind as a dimer to the VEGF HR region
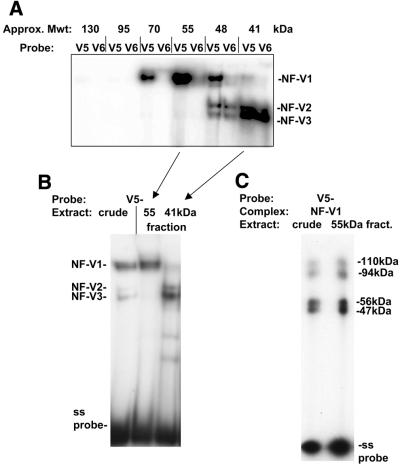
It was apparent, however, that the additional sized proteins of 110 and 94 kDa detected by UV crosslink analysis of the NF-V1 complex were approximately double the size of the 56 and 47 kDa dbpA proteins. It was possible, therefore, that these proteins represented crosslinking of a dimer of dbpA proteins rather than being additional proteins. To investigate this, crude nuclear extracts were fractionated by FPLC gel filtration and fractions tested, by gel retardation assay, for NF-V1 complex formation on the non-coding single strand VEGF HR region sequence (V5–). Fractions were also bound to the mutant HR sequence (V6–, Fig. 1A). NF-V1 complex activity peaked in a fraction of ∼55 kDa and was clearly separated from NF-V2/3 complex activity (Fig. 6A and B). The nature of this complex was verified by UV crosslinking and was found to contain the 56, 47, 110 and 94 kDa proteins observed with the NF-V1 complex from crude nuclear extracts (Fig. 6C). As a 55 kDa fraction (monomer size of dbpA) is sufficient to form the NF-V1 complex, this strongly suggests that the larger 110 and 94 kDa proteins are in fact two molecules (dimers) of the smaller 56 and 47 kDa proteins binding to DNA. The decreased mobility of the NF-V1 complex (Fig. 2A) compared to NF-GMb (monomer of dbpB) also supports this. The ability of nuclear proteins to bind to DNA as a dimer may play a role in the preferential binding of nuclear dbpA to the VEGF HR region.
Figure 6.
Nuclear dbpA may bind to the VEGF HR region as a dimer. (A) Balb/c 3T3 crude nuclear extract was fractionated by FPLC gel filtration and protein in fractions assayed for binding to the VEGF HR region single strand oligonucleotide (V5–) or to the CSD protein-binding site mutant (V6–) in a gel retardation assay. Consecutive 0.5 ml fractions containing NF-V1-binding activity are shown. NF-V1 to NF-V3 complexes are indicated. The approximate sizes of protein fractions, determined by comparison to elution profiles of protein standards, is given in kDa. (B) Balb/c 3T3 crude nuclear extract and gel filtration fractions containing the peak of NF-V1 activity (55 kDa) and NF-V2/3 activity (41 kDa) are shown bound to the VEGF HR region single strand oligonucleotide (V5–) in a gel retardation assay. Nuclear complexes (NF-V1, NF-V2 and NF-V3) and free single strand probe (ss probe) are indicated. (C) NF-V1 complexes from crude nuclear and fractionated extract were UV crosslinked to the VEGF HR region oligonucleotide (V5–) and crosslinked proteins analyzed by SDS–PAGE. The sizes of crosslinked proteins are indicated and were calculated by subtraction of the molecular weight of the V5– probe.
The NF-V1 nuclear CSD protein complex recognizes supercoiling-induced structures within VEGF HR region DNA
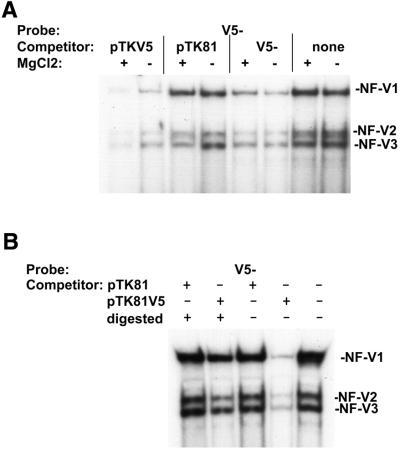
Given the single strand DNA specificity of the NF-V1 CSD protein complex for the VEGF HR region (Fig. 2D) and the proposed mechanisms of repression by CSD proteins via single strand DNA binding, it was important to determine whether/how nuclear CSD proteins gain access to their single strand binding sites within double strand DNA. It was possible that nuclear CSD proteins bind to structures induced by supercoiling, as such structures contain single strand DNA (27,39). To investigate this, NF-V1 complex formation on the non-coding single strand HR sequence (V5–) was competed with supercoiled plasmid DNA in the presence or absence of Mg2+ (Fig. 7A). Mg2+ stabilizes single strand-containing structures within supercoiled double strand DNA (27,39). The NF-V1 complex was readily competed by a plasmid containing the VEGF HR region sequence (pTKV5Luc) but was not competed by the backbone vector (pTK81Luc) and this competition was enhanced in the presence of MgCl2. NF-V1 complex formation alone was not affected by MgCl2. Competition with single strand competitor (V5–) was also not affected. This suggested that competition with the supercoiled HR region plasmid was due to the presence of the HR region sequences and that this region contains single strand DNA, as competition is enhanced by MgCl2.
Figure 7.
Nuclear CSD proteins bind to a supercoiling-induced single strand structure within the VEGF HR region. (A) Balb/c 3T3 fibroblast nuclear extract was preincubated with supercoiled plasmid DNA containing the VEGF HR region (pTKV5Luc), with the supercoiled backbone vector (pTK81Luc), with single strand self competitor (V5–) or left untreated (none). Extracts were then bound to labeled VEGF HR region single strand oligonucleotide (V5–) in the presence (+) or absence (–) of MgCl2 in a gel retardation assay. MgCl2 stabilizes supercoiling-induced single strand structures (27,39). Nuclear complexes (NF-V1, NF-V2 and NF-V3) are indicated. (B) Nuclear extracts were preincubated with restriction enzyme-digested (+) (relieves supercoiling and supercoiling-induced single strand structures) or undigested (–) (supercoiled) pTKV5Luc and pTK81Luc plasmids and bound to the labeled VEGF oligonucleotide (V5–) in a gel retardation assay.
To further investigate this, competitions were performed with supercoiled (non-digested) and non-supercoiled (restriction enzyme digested) plasmids (Fig. 7B). Supercoiling is required to generate structures in double strand DNA that contain single strand DNA. Restriction enzyme digestion will relieve supercoiling and hence supercoiling-induced structures. Competition with the non-supercoiled (digested +) HR region-containing pTKV5Luc plasmid was much less efficient than with the supercoiled (digested –) plasmid. The NF-V1 complex therefore binds with much greater efficiency to the supercoiled plasmid than to the non-supercoiled linear plasmid. There was no change in competition with the backbone pTK81Luc plasmid. Taken together (Figs 2D and 7), these data suggest that the NF-V1 complex can recognize supercoiling-induced structures (single strand DNA containing) within VEGF HR region DNA, suggesting a mechanism for entry of nuclear CSD proteins, in particular dbpA, to their target binding sites in normoxic cells.
DISCUSSION
A novel repressor site in the VEGF HR region
Inappropriate expression of VEGF by both cancer cells and surrounding cells, such as fibroblasts, is involved in solid tumor formation (4–6,11). It is therefore very important that the VEGF gene be maintained in a repressed state in the absence of appropriate stimuli in normoxic cells. Tumor suppressors such as p53 and VHL have been shown to repress VEGF expression by regulating HIF-1α protein stability (9,10,12), but no DNA-binding repressors had been identified.
We now report a novel mechanism of repression of the VEGF gene involving a DNA-binding repressor. We identify a repressor site binding CSD proteins, downstream of the HIF-1-binding site, that is involved in maintaining the VEGF HR region in a repressed state under normoxic conditions in fibroblasts. The repressor function of CSD proteins, through the identified HR region CSD protein-binding site, was verified by overexpression of CSD proteins. Proteins binding to sites flanking the HIF-1 site have not previously been identified. The single strand DNA specific CSD protein-binding site (5′-CCTG-3′) on the non-coding strand of the HR region sequence is identical to the sequence of a pair of CSD protein-binding sites in the GM-CSF gene. Even though there is no universal consensus for CSD protein-binding sites (22,30), the identification of 5′-CCTG-3′ sites in both the GM-CSF and VEGF genes verifies this sequence as an important target for CSD protein binding. The VEGF site, however, has quite different characteristics to the GM-CSF sites. The VEGF 5′-CCTG-3′ site can function as a single element to bring about repression, whereas the GM-CSF 5′-CCTG-3′ sites do not act individually, with both sites being required for CSD protein binding and repression. The VEGF repressor site also binds a novel CSD protein nuclear complex (NF-V1) containing dbpA, while we have previously reported that the GM-CSF sequences bind a complex, NF-GMb, containing dbpB/YB-1 (29,30). Most reported studies on CSD protein action have investigated dbpB/YB-1 (21,22). There are a few reports, however, demonstrating a repressor function of dbpA (23,24,38,40). The exact binding sites for dbpA have not been determined in these genes, but the binding region within the nicotinic acetylcholine receptor delta subunit gene contains a single sequence, 5′-TCCTGA-3′, identical to the VEGF CSD protein-binding site (23). A similar nuclear complex to that observed here (NF-V1) may also regulate this gene.
Mechanisms of repression by CSD proteins in normoxic cells
The identification of a CSD protein-binding repressor site in the VEGF gene, in normoxic cells, further supports a role for CSD proteins as key regulators of growth factor and stress response genes. As for the VEGF gene, in most cases, the genes regulated by CSD proteins require strict regulation and inappropriate expression leads to disease (22,29). It has been observed in a number of cases that CSD proteins bind to DNA in a single strand-specific manner to the repressed genes (22). We have also observed DNA binding to the VEGF HR region to be single strand specific with no binding to double strand DNA. We and others have hypothesized, for genes repressed by CSD proteins, that the binding of CSD proteins to single strand DNA in unstimulated cells stabilizes a single strand structure and inhibits the binding of double strand DNA-dependent activators (25,27,29–31,41). Single strand DNA binding CSD proteins, in particular dbpA, could similarly inhibit activators binding to the HR region.
As DNA in the cell is in double strand form we determined a means by which nuclear CSD proteins, within the NF-V1 complex, could target their single strand binding sites in double strand VEGF promoter DNA. Such studies have not previously been performed with nuclear proteins and there are no reports on the mechanisms of entry of dbpA into double strand DNA. We show here that CSD proteins can target supercoiling-induced structures that produce single strand DNA, within the VEGF HR region. Regions of DNA with a propensity to form single strand sites often contain inverted repeats (39). An imperfect 5 bp inverted repeat is found overlapping the HR region CSD protein-binding repressor site (36,37) (Fig. 1). Such a repeat could play a role in single strand DNA formation. Interestingly, 5 bp inverted repeats are also found overlapping the binding sites for dbpB/YB-1 in the collagen α 1(I) and gelatinase A genes. dbpB/YB-1 has been shown to bind to single strand DNA within double strand DNA regions of these genes in vitro (41–43). dbpA and dbpB/YB-1 may therefore have similar mechanisms of entry to promoter sequences. Consistent with the identification of single strand regions within CSD protein-binding sites in vitro is the identification of such single strand regions in CSD protein-binding genes in vivo (26,27). Taken together, this suggests that single strand sites are in fact targets for CSD proteins in endogenous genes supporting a role for single strand binding in bringing about repression.
CSD protein derepression and hypoxic activation of the VEGF HR region
Activation of CSD protein-repressed genes appears to require derepression of CSD protein action. Proposed mechanisms of derepression involve interaction of CSD proteins with induced factors to remove/inhibit CSD protein binding to single strand DNA, allowing the binding of activators dependent on double strand DNA, such as the HR HIF-1 complex (22,29,33). Derepression is most clearly demonstrated for the ErbB2 gene, whereby ZO-1 sequesters dbpA, preventing its repressive action (38). Consistently, the VEGF HR region CSD protein-binding site did not act as a repressor in hypoxic (activated) cells. Activator function was instead revealed by mutation of the CSD protein-binding site. A hypoxic activator site overlapping the CSD protein-binding repressor site has been reported, but the protein binding to this site has not been identified (36,37). As CSD protein overexpression under hypoxic conditions resulted in repression rather than activation of the HR region, it is not likely that CSD proteins are involved in hypoxic activation. In addition, the reported activator site (36,37) also includes bases immediately downstream of the CSD repressor site and we have shown that these bases are not involved in CSD protein binding (Fig. 5, V9–). We have, however, identified a double strand DNA-binding nuclear complex that binds to the activator sequences (unpublished results). It is therefore likely that repressive single strand-binding CSD proteins are replaced by a double strand DNA-binding activator in hypoxic cells. Return of the HR region to double strand form would also allow binding of the double strand DNA-dependent HIF-1 complex. Forced overexpression of CSD proteins in hypoxic cells appears to be able to reverse this process.
Taken together, our data suggest a model whereby in normoxic cells, CSD proteins bind to single strand DNA-containing structures, induced by torsional stress in the VEGF HR region. This could stabilize the HR region in single strand form and prevent the binding of double strand DNA-dependent activators, present at low levels in normoxic cells. In combination with the p53 and VHL tumor suppressors this would provide strict regulation of the HR region during normoxia. In hypoxic cells the increased levels of double strand DNA-binding activators, such as those binding to sites directly overlapping the CSD protein-binding site, probably results in removal/displacement of CSD proteins allowing binding of the HIF-1 complex.
Role of nuclear dbpA in VEGF HR region regulation
It is of interest that the nuclear NF-V1 complex detected on the VEGF HR region appears to only contain dbpA, even though the HR region can bind both recombinant dbpA and dbpB. This apparent lack of nuclear dbpB complex formation is not due to the lack of dbpB/YB-1, as the GM-CSF NF-GMb complex (dbpB/YB-1) forms efficiently in the fibroblast extracts. Little is known about the way nuclear CSD proteins bind DNA and there are no reports of nuclear complexes containing dbpA. Our work suggests, however, that nuclear dbpA may bind as a dimer in the presence of DNA. In contrast, studies of the GM-CSF gene suggest that nuclear dbpB binds as a monomer (29–31). Recombinant dbpB/YB-1 has, however, been shown to dimerize, but this dimerization occurred in solution (28). The differences between recombinant and nuclear CSD protein binding to the VEGF HR region may be due to modifications of CSD nuclear proteins that enhance dimer formation/binding or be due to the presence of additional proteins in the NF-V1 complex that were not crosslinked to the HR region DNA. The latter possibility is probably ruled out as we found that a gel filtration fraction of the size of dbpA could reform the NF-V1 complex. Modification of CSD proteins in vitro has been shown to affect binding (20) and we have observed that dephosphorylation of nuclear protein alters the formation of the NF-V1 complex (unpublished results). The selective binding of dbpA (NF-V1) and dbpB/YB-1 (NF-GMb) nuclear complexes to the VEGF and GM-CSF genes, respectively, suggests different functional roles for these proteins in regulation of the two genes. One possibility is that nuclear dbpA and dbpB/YB-1 proteins binding to these genes are interacting with different transcription factors as part of the process of repression or derepression. dbpA and dbpB have, for example, been shown to interact with different NF-Y subtypes in regulation of a MHC class II gene (24).
A broader role for CSD proteins in VEGF expression
CSD proteins may play a broader role in VEGF regulation than just being repressors of the VEGF gene directly, as CSD proteins have been shown to regulate genes and proteins involved in VEGF regulation. These include the ErbB2, EGFR and c-myc genes and the p53 tumor suppressor (26,38,44,45). It is also evident that disregulation of CSD protein expression could be involved in inappropriate expression of VEGF in cancer cells, as is observed when p53 and VHL tumor suppressor expression is altered. CSD proteins have in fact been implicated in a number of cancers, with cancer cells showing both quantitative and qualitative changes in CSD protein expression (46–48). The data presented here also demonstrate that overexpression of CSD proteins may be useful as a tool in controlling inappropriate VEGF expression from cells that overexpress VEGF.
In summary, we have identified CSD proteins as repressors of the VEGF HR region under normoxic conditions in fibroblasts and have identified a novel mechanism of repression of this region. The primary nuclear CSD protein involved in repression appears to be dbpA and the nuclear CSD protein complex is single strand specific and enters the HR region via intrinsic single strand structure within this region. CSD protein overexpression can regulate both normoxic and hypoxic HR region expression. This work demonstrates the growing importance of CSD proteins as key regulators of growth factor and stress response genes and the mechanisms by which nuclear CSD proteins, in particular dbpA, bind to single strand DNA and bring about repression.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Maria Balda (University College, London) for her generous gift of dbpA antibody. This work was supported by a National Health and Medical Research Council Program Grant, a Cancer Council (South Australia) project grant and an IMVS/RAH project grant.
REFERENCES
- 1.Yancopoulos G.D., Davis,S., Gale,N.W., Rudge,J.S., Wiegand,S.J. and Holash,J. (2000) Vascular-specific growth factors and blood vessel formation. Nature, 407, 242–248. [DOI] [PubMed] [Google Scholar]
- 2.Neufeld G., Cohen,T., Gengrinovitch,S. and Poltorak,Z. (1999) Vascular endothelial growth factor (VEGF) and its receptors. FASEB J., 13, 9–22. [PubMed] [Google Scholar]
- 3.Ferrara N. and Davis-Smyth,T. (1997) The biology of vascular endothelial growth factor. Endocr. Rev., 18, 4–25. [DOI] [PubMed] [Google Scholar]
- 4.Siemeister G., Martiny-Baron,G. and Marme,D. (1998) The pivotal role of VEGF in tumor angiogenesis: molecular facts and therapeutic opportunities. Cancer Metastasis Rev., 17, 241–248. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet P. and Jain,R.K. (2000) Angiogenesis in cancer and other diseases. Nature, 407, 249–257. [DOI] [PubMed] [Google Scholar]
- 6.Kim K.J., Li,B., Winer,J., Armanini,M., Gillett,N., Phillips,H.S. and Ferrara,N. (1993) Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature, 362, 841–844. [DOI] [PubMed] [Google Scholar]
- 7.Semenza G.L. (2000) HIF-1 and human disease: one highly involved factor. Genes Dev., 14, 1983–1991. [PubMed] [Google Scholar]
- 8.Semenza G.L. (2000) Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem. Pharmacol., 59, 47–53. [DOI] [PubMed] [Google Scholar]
- 9.Richard D.E., Berra,E. and Pouyssegur,J. (1999) Angiogenesis: how a tumor adapts to hypoxia. Biochem. Biophys. Res. Commun., 266, 718–722. [DOI] [PubMed] [Google Scholar]
- 10.Semenza G.L. (1999) Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell. Dev. Biol., 15, 551–578. [DOI] [PubMed] [Google Scholar]
- 11.Fukumura D., Xavier,R., Sugiura,T., Chen,Y., Park,E.C., Lu,N., Selig,M., Nielsen,G., Taksir,T., Jain,R.K. and Seed,B. (1998) Tumor induction of VEGF promoter activity in stromal cells. Cell, 94, 715–725. [DOI] [PubMed] [Google Scholar]
- 12.Milanini J., Vinals,F., Pouyssegur,J. and Pages,G. (1998) p42/p44 MAP kinase module plays a key role in the transcriptional regulation of the vascular endothelial growth factor gene in fibroblasts. J. Biol. Chem., 273, 18165–18172. [DOI] [PubMed] [Google Scholar]
- 13.Pages G., Milanini,J., Richard,D.E., Berra,E., Gothie,E., Vinals,F. and Pouyssegur,J. (2000) Signaling angiogenesis via p42/p44 MAP kinase cascade. Ann. N. Y. Acad. Sci., 902, 187–200. [DOI] [PubMed] [Google Scholar]
- 14.Berra E., Pages,G. and Pouyssegur,J. (2000) MAP kinases and hypoxia in the control of VEGF expression. Cancer Metastasis Rev., 19, 139–145. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Ocejo O., Viloria-Petit,A., Bequet-Romero,M., Mukhopadhyay,D., Rak,J. and Kerbel,R.S. (2000) Oncogenes and tumor angiogenesis: the HPV-16 E6 oncoprotein activates the vascular endothelial growth factor (VEGF) gene promoter in a p53 independent manner. Oncogene, 19, 4611–4620. [DOI] [PubMed] [Google Scholar]
- 16.Sodhi A., Montaner,S., Patel,V., Zohar,M., Bais,C., Mesri,E.A. and Gutkind,J.S. (2000) The Kaposi’s sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res., 60, 4873–4880. [PubMed] [Google Scholar]
- 17.Laughner E., Taghavi,P., Chiles,K., Mahon,P.C. and Semenza,G.L. (2001) HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell. Biol., 21, 3995–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zelzer E., Levy,Y., Kahana,C., Shilo,B.Z., Rubinstein,M. and Cohen,B. (1998) Insulin induces transcription of target genes through the hypoxia-inducible factor HIF-1alpha/ARNT. EMBO J., 17, 5085–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graumann P.L. and Marahiel,M.A. (1998) A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci., 23, 286–290. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto K. and Wolffe,A.P. (1998) Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol., 8, 318–323. [DOI] [PubMed] [Google Scholar]
- 21.Shannon M.F., Coles,L.S., Vadas,M.A. and Cockerill,P.N. (1997) Signals for activation of the GM-CSF promoter and enhancer in T cells. Crit. Rev. Immunol., 17, 301–323. [DOI] [PubMed] [Google Scholar]
- 22.Shannon M.F., Coles,L.S., Attema,J. and Diamond,P. (2001) The role of architectural transcription factors in cytokine gene transcription. J. Leukoc. Biol., 69, 21–32. [PubMed] [Google Scholar]
- 23.Li W.W., Hsiung,Y., Wong,V., Galvin,K., Zhou,Y., Shi,Y. and Lee,A.S. (1997) Suppression of grp78 core promoter element-mediated stress induction by the dbpA and dbpB (YB-1) cold shock domain proteins. Mol. Cell. Biol., 17, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lloberas J., Maki,R.A. and Celada,A. (1995) Repression of major histocompatibility complex I-A beta gene expression by dbpA and dbpB (mYB-1) proteins. Mol. Cell. Biol., 15, 5092–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald G.H., Itoh-Lindstrom,Y. and Ting,J.P. (1995) The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J. Biol. Chem., 270, 3527–3533. [DOI] [PubMed] [Google Scholar]
- 26.Kolluri R., Torrey,T.A. and Kinniburgh,A.J. (1992) A CT promoter element binding protein: definition of a double-strand and a novel single-strand DNA binding motif. Nucleic Acids Res., 20, 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horwitz E.M., Maloney,K.A. and Ley,T.J. (1994) A human protein containing a “cold shock” domain binds specifically to H-DNA upstream from the human gamma-globin genes. J. Biol. Chem., 269, 14130–14139. [PubMed] [Google Scholar]
- 28.Izumi H., Imamura,T., Nagatani,G., Ise,T., Murakami,T., Uramoto,H., Torigoe,T., Ishiguchi,H., Yoshida,Y., Nomoto,M. et al. (2001) Y box-binding protein-1 binds preferentially to single-stranded nucleic acids and exhibits 3′→5′ exonuclease activity. Nucleic Acids Res., 29, 1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coles L.S., Diamond,P., Occhiodoro,F., Vadas,M.A. and Shannon,M.F. (2000) An ordered array of cold shock domain repressor elements across tumor necrosis factor-responsive elements of the granulocyte-macrophage colony-stimulating factor promoter. J. Biol. Chem., 275, 14482–14493. [DOI] [PubMed] [Google Scholar]
- 30.Coles L.S., Diamond,P., Occhiodoro,F., Vadas,M.A. and Shannon,M.F. (1996) Cold shock domain proteins repress transcription from the GM-CSF promoter. Nucleic Acids Res., 24, 2311–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coles L.S., Occhiodoro,F., Vadas,M.A. and Shannon,M.F. (1994) A sequence-specific single-strand DNA binding protein that contacts repressor sequences in the human GM-CSF promoter. Nucleic Acids Res., 22, 4276–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordeen S.K. (1988) Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques, 6, 454–458. [PubMed] [Google Scholar]
- 33.Diamond P., Shannon,M.F., Vadas,M.A. and Coles,L.S. (2001) Cold shock domain factors activate the granulocyte-macrophage colony-stimulating factor promoter in stimulated Jurkat T cells. J. Biol. Chem., 276, 7943–7951. [DOI] [PubMed] [Google Scholar]
- 34.Dibbens J.A., Miller,D.L., Damert,A., Risau,W., Vadas,M.A. and Goodall,G.J. (1999) Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol. Biol. Cell, 10, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shima D.T., Kuroki,M., Deutsch,U., Ng,Y.S., Adamis,A.P. and D’Amore,P.A. (1996) The mouse gene for vascular endothelial growth factor. Genomic structure, definition of the transcriptional unit and characterization of transcriptional and post-transcriptional regulatory sequences. J. Biol. Chem., 271, 3877–3883. [DOI] [PubMed] [Google Scholar]
- 36.Kimura H., Weisz,A., Kurashima,Y., Hashimoto,K., Ogura,T., D’Acquisto,F., Addeo,R., Makuuchi,M. and Esumi,H. (2000) Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood, 95, 189–197. [PubMed] [Google Scholar]
- 37.Kimura H., Weisz,A., Ogura,T., Hitomi,Y., Kurashima,Y., Hashimoto,K., D’Acquisto,F., Makuuchi,M. and Esumi,H. (2001) Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J. Biol. Chem., 276, 2292–2298. [DOI] [PubMed] [Google Scholar]
- 38.Balda M.S. and Matter,K. (2000) The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J., 19, 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank-Kamenetskii M.D. and Mirkin,S.M. (1995) Triplex DNA structures. Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 40.Sapru M.K., Gao,J.P., Walke,W., Burmeister,M. and Goldman,D. (1996) Cloning and characterization of a novel transcriptional repressor of the nicotinic acetylcholine receptor delta-subunit gene. J. Biol. Chem., 271, 7203–7211. [DOI] [PubMed] [Google Scholar]
- 41.Norman J.T., Lindahl,G.E., Shakib,K., En-Nia,A., Yilmaz,E. and Mertens,P.R. (2001) The Y-box binding protein YB-1 suppresses collagen alpha 1(I) gene transcription via an evolutionarily conserved regulatory element in the proximal promoter. J. Biol. Chem., 276, 29880–29890. [DOI] [PubMed] [Google Scholar]
- 42.Mertens P.R., Alfonso-Jaume,M.A., Steinmann,K. and Lovett,D.H. (1998) A synergistic interaction of transcription factors AP2 and YB-1 regulates gelatinase A enhancer-dependent transcription. J. Biol. Chem., 273, 32957–32965. [DOI] [PubMed] [Google Scholar]
- 43.Mertens P.R., Alfonso-Jaume,M.A., Steinmann,K. and Lovett,D.H. (1999) YB-1 regulation of the human and rat gelatinase A genes via similar enhancer elements. J. Am. Soc. Nephrol., 10, 2480–2487. [DOI] [PubMed] [Google Scholar]
- 44.Chernukhin I.V., Shamsuddin,S., Robinson,A.F., Carne,A.F., Paul,A., El Kady,A.I., Lobanenkov,V.V. and Klenova,E.M. (2000) Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1 and the multivalent zinc finger factor, CTCF. J. Biol. Chem., 275, 29915–29921. [DOI] [PubMed] [Google Scholar]
- 45.Okamoto T., Izumi,H., Imamura,T., Takano,H., Ise,T., Uchiumi,T., Kuwano,M. and Kohno,K. (2000) Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene, 19, 6194–6202. [DOI] [PubMed] [Google Scholar]
- 46.Bargou R.C., Jurchott,K., Wagener,C., Bergmann,S., Metzner,S., Bommert,K., Mapara,M.Y., Winzer,K.J., Dietel,M., Dorken,B. and Royer,H.D. (1997) Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nature Med., 3, 447–450. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka T., Kondo,S., Iwasa,Y., Hiai,H. and Toyokuni,S (2000) Expression of stress-response and cell proliferation genes in renal cell carcinoma induced by oxidative stress. Am. J. Pathol., 156, 2149–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nambiar A., Kandala,J.C., Svoboda,J. and Guntaka,R.V. (1998) Cloning of a novel Y-box homology protein (chkYB-1HP) cDNA lacking the cold-shock domain. Biochim. Biophys. Acta, 1395, 1–6. [DOI] [PubMed] [Google Scholar]
- 49.Levy A.P., Levy,N.S., Wegner,S. and Goldberg,M.A. (1995) Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J. Biol. Chem., 270, 13333–13340. [DOI] [PubMed] [Google Scholar]
- 50.Forsythe J.A., Jiang,B.H., Iyer,N.V., Agani,F., Leung,S.W., Koos,R.D. and Semenza,G.L. (1996) Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol., 16, 4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]