Abstract
In vitro compartmentalisation (IVC), a technique for selecting genes encoding enzymes based on compartmentalising gene translation and enzymatic reactions in emulsions, was used to investigate the interaction of the DNA cytosine-5 methyltransferase M.HhaI with its target DNA (5′-GCGC-3′). Crystallog raphy shows that the active site loop from the large domain of M.HhaI interacts with a flipped-out cytosine (the target for methylation) and two target recognition loops (loops I and II) from the small domain make almost all the other base-specific interactions. A library of M.HhaI genes was created by randomising all the loop II residues thought to make base-specific interactions and directly determine target specificity. The library was selected for 5′-GCGC-3′ methylation. Interestingly, in 11 selected active clones, 10 different sequences were found and none were wild-type. At two of the positions mutated (Ser252 and Tyr254) a number of different amino acids could be tolerated. At the third position, however, all active mutants had a glycine, as in wild-type M.HhaI, suggesting that Gly257 is crucial for DNA recognition and enzyme activity. Our results suggest that recognition of base pairs 3 and 4 of the target site either relies entirely on main chain interactions or that different residues from those identified in the crystal structure contribute to DNA recognition.
INTRODUCTION
Although structural studies of enzymes by X-ray crystallography and NMR can provide a great deal of insight into their mode of action, a more thorough understanding of the relationship between structure and function requires the use of other complementary techniques (1). The ability to construct proteins differing by only a single amino acid at a predetermined position has proven particularly powerful, enabling sophisticated questions to be asked about the role of any amino acid in a protein (1). However, the systematic substitution of every individual residue in a protein is a major undertaking; for a small, 100 amino acid protein this would require the creation and testing of 1900 variants. Alanine scanning (2), in which each residue is systematically changed to alanine, reduces the task to testing less than 100 variants (for a 100 amino acid protein) but does not provide information as to whether any residue other than alanine could be functional. In addition, neither approach provides information about the cooperative effect of simultaneous changes at more than one position, which can sometimes be significant (3).
If more than one position is to be mutated, the number of variants that need to be analysed increases dramatically (4). However, libraries of enzymes randomised simultaneously at several positions can be processed using high throughput screening (5) or selection strategies (6,7). Selection is particularly useful where only a small number of active variants are present in the background of numerous inactive ones. However, existing direct, in vitro selections for enzymatic activities are of rather limited scope (6,8). In this context, we have developed in vitro compartmentalisation (IVC) for the selection of genes encoding enzymes. This completely in vitro system is based on compartmentalising reactions in the aqueous droplets of a water-in-oil emulsion. The small size of the compartments (∼5 fl) means that IVC can select many genes (there are ∼1010 aqueous compartments in an emulsion made from 50 µl of reaction mix dispersed in 1 ml of oil), is highly sensitive and economical. We have previously demonstrated the fundamentals of IVC by selecting for the DNA cytosine-5 methyltransferase HaeIII (9) (Fig. 1) and a bacterial phosphotriesterase (A.D.Griffiths and D.S.Tawfik, manuscript in preparation). Compartmentalis ation in emulsions has also recently been used for the directed evolution of Taq DNA polymerase based on the polymerase replicating its own gene (10). In this study we have used IVC to investigate the function of residues in the DNA cytosine-5 methyltransferase HhaI (M.HhaI).
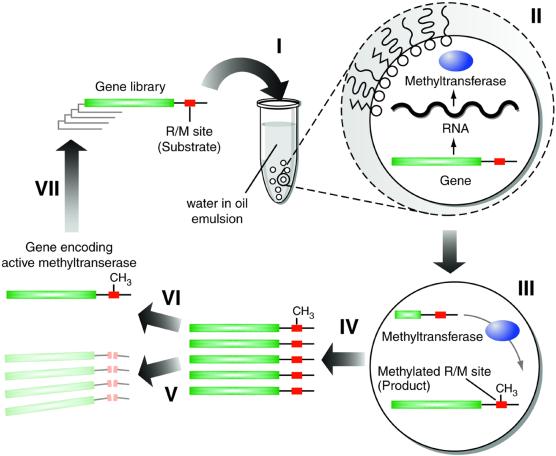
Figure 1.
Selection of DNA methyltransferases by in vitro compartmentalisation. An in vitro transcription–translation reaction mixture containing a library of genes encoding mutant methyltransferases, each with restriction/methylation (R/M) sites appended to the gene, is dispersed to form a water-in-oil emulsion with typically one gene per aqueous compartment (I). The genes are transcribed and translated within their compartments (II). Proteins with methyltransferase activity methylate the R/M sites (III). Compartmentalisation prevents the methylation of genes in other compartments. The emulsion is broken, all reactions are stopped and the aqueous compartments combined (IV). Digestion with the cognate restriction enzyme results in the digestion of unmethylated genes (which do not encode active methyltransferases) (V) and the survival of methylated genes (which encode active methyltransferases) (VI). The surviving genes can be amplified using PCR and compartmentalised for further rounds of selection (VII).
DNA cytosine-5 methyltransferases catalyse the transfer of a methyl group from S-adenosylmethionine (AdoMet) to a DNA cytosine ring carbon (C5) in a sequence-specific manner. They are found in bacteria, archaea and eukaryotes. In bacteria, DNA methylation has roles in the control of DNA replication and in restriction–modification systems (11) wherein the host DNA is protected from cleavage by restriction endonucleases which recognise the same target site. In eukaryotes, cytosine-5 methyltransferases are responsible for CpG methylation, which is thought to be involved in regulation of chromatin structure and gene expression (12) and DNA methylation has been implicated in the development of cancer (13,14).
M.HhaI is a bacterial methyltransferase which methylates the internal cytosine of the tetranucleotide restriction/methylation (R/M) site 5′-GCGC-3′. Crystallographic studies reveal that M.HhaI folds into two domains, separated by a large cleft that holds the DNA (15,16), and that the DNA target cytosine is flipped out of the helix (16). The large domain contains the catalytic site and the cofactor (AdoMet) binding site. Its structure is very similar to the large domain of other cytosine-C5, adenine-N6 and cytosine-N4 DNA methyltransferases, and indeed all but two of the 25 crystallographically determined AdoMet-dependent methyltransferases share this common structural core (17,18). The large domains of DNA cytosine-5 methyltransferases also contain a series of conserved sequence motifs involved in catalysis and cofactor binding (19). The small domains of DNA cytosine-5 methyltransferases are responsible for target recognition and their sequences as well as structures vary greatly from one enzyme to another. This renders sequence comparison of little use in comprehending differences in DNA recognition and sequence specificity. Crystallographic studies show that the small domain of M.HhaI contains two glycine-rich target recognition loops (loops I and II) which contact the target DNA sequence (16) (Fig. 2). These two loops are thought to be responsible for almost all the base-specific interactions with the 5′-GCGC-3′ target site; the only other amino acid in the enzyme which also interacts with the bases is Ile86, which interacts with G1. This general arrangement, with two loops from the small domain interacting with the R/M site, is also seen in the crystal structure of DNA cytosine-5 methyltransferase HaeIII (M.HaeIII) (20).
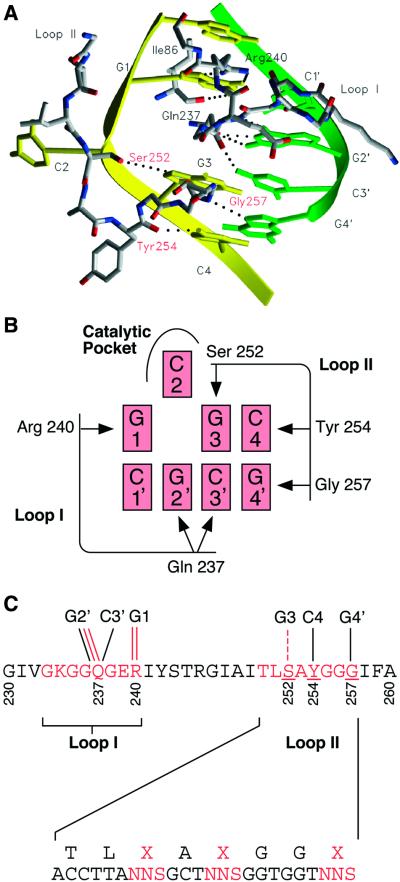
Figure 2.
Residues in the small target recognition domain of methyltransferase HhaI (M.HhaI) thought to interact with bases in the target sequence (16) and design of the loop II library. (A) Model of the interaction between bases and the two target recognition loops (loops I and II) in the small domain (16). The two DNA strands are in yellow and green. Loops I and II and Ile86 (the one residue not in the loops thought to make a base-specific interaction) are shown with atoms coloured: grey, carbon; red, oxygen; blue, nitrogen. Putative hydrogen bonding interactions are indicated by dotted lines. Interactions with the flipped out base, C2, are omitted. (B) Schematic diagram of the proposed interactions between bases and the two target recognition loops (loops I and II) in the small domain. (C) Sequence of the region of the small domain including target recognition loops I and II and the diversity introduced into the loop II library. Direct interactions with bases in the target sequence are shown by solid lines and water-mediated interactions by dashed lines. Red lines indicate interactions involving amino acid side chains and black lines indicate main chain interactions.
Here we have explored the role of the residues in target recognition loop II thought to interact with bases in the target sequence (Fig. 2) (16). Libraries of mutant M.HhaI genes with the codons for these residues randomised to give all 20 possible amino acids were selected and the active mutants isolated and characterised.
MATERIALS AND METHODS
Oligonucleotides
HhaIBc, 5′-GCATCTGACAAGCTTAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGATTGAAATAAAAGATAAACAGCTCACAGG-3′; HhaIFoNc, 5′-CGAGC TAGAGGTACCTTATTAATATGGTTTGAAATTTAATGATGAACCAATG-3′; pBR-ClaIba, 5′-GCATCTGACATCGATTAATACGACTCACTATAGGG-3′; pBRHha232-EcoRIfo, 5′-GCATCTGACGAATTCGGTCTCCTACAATACCAAGTCGAACTG-3′; pGEMHha258-Hind3Ba, 5′-GCATCTGACAAGCTTATTTTCGCTAAGACAGGGGG-3′; pGEM-EcoRIfo, 5′-GCATCTGACGAATTCTTATTAATATGGTTTGAAAT-3′; tet1, 5′-CAGGACGGGTGTGGTCGC-3′; Hha245-Bsafo2, 5′-GTCCTCGCAACTGGTCT CCTCTTGTGCTATAAATTCGTTCTCCTTGCCCGCCTTTTCCTACAATACCAAGTCGAACTG-3′; LMB2-1Bio, 5′-biotin-CAGGCGCCATTCGCCATT-3′; pGEMHha246- BsaIba, 5′-GTCCTCGCAACTGGTCTCCAAGAGGCATTGCAATTACCTTANNSGCTNNSGGTGGTNNSATTTTCGCTAAGACAGGGGG-3′; tet6, 5′-CAGTGCTCCGAGAA CGGG-3′; McrLMB2-2Bio, 5′-biotin-AAGTTCAGCCTG-GTTAAGGCCGGCCGCGCGCGCAGCTTGGATCTTAGTCGCTTGCACTGCTATCGGGCCGGCCGCGCGCGCCA GGCTGCGCAACTGTTG-3′; tet7, 5′-TGCGCATAGAAATTGCAT-3′; McrLMB2-3Bio, 5′-biotin-AAGTTCAGCCTGGTTAAGGCCGGCCGCGCGCGCAGCTTGGATCTTAGTCGCTTGCACTGCTATCGGGCCGGCCGCGCGCGCGG AAGGGCGATCGGTGCG-3′; tet8, 5′-CAACGCATATAGCGCTAG-3′; McrLMB2-4Bio, 5′-biotin-AAGTTCAGCCTGGTTAAGGCCGGCCGCGCGCGCAGCTTGGATCTTA GTCGCTTGCACTGCTATCGGGCCGGCCGCGCGCGCGGCCTCTTCGCTATTACG-3′; tet10, 5′-GGCGATGCTGTCGGAATG-3′; LMB2, 5′-GTAAAACGACGGCCAGT-3′; pCR2.1ba, 5′-ACGGCCGCCAGTGTGCTGGAA-3′; pCR2.1fo, 5′-AGTGTGATGGATATCTGCAG-3′; HindIIIPCR, 5′-AGGATGACGATGAGCGCATTG-3′.
Creation of libraries of mutated M.HhaI genes
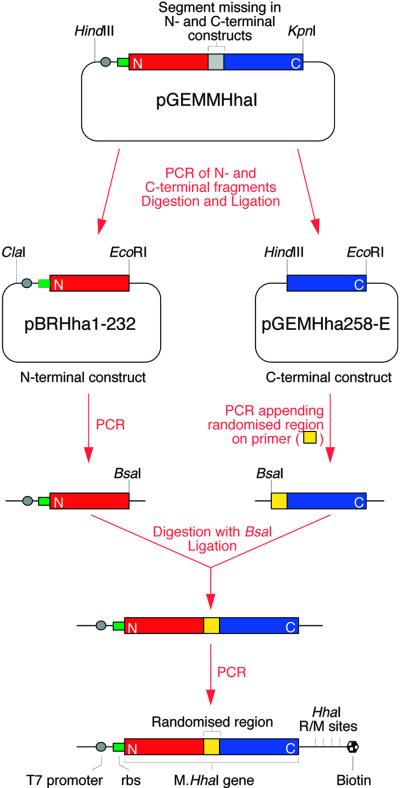
The procedure used to create libraries of mutated M.HhaI genes is outlined schematically in Figure 3. The M.HhaI (21) gene was first amplified from Haemophilus parahaemolyticus (ATCC 10014) using oligonucleotides HhaIBc and HhaIFoNc and cloned into HindIII and KpnI cut pGEM-4Z (Promega) to create pGEMMHhaI. To create a library of M.HhaI genes mutated in target recognition loop II, a fragment comprising the T7 promoter, T7 gene 10 ribosome binding site, and the N-terminal part of the M.HhaI gene (up to residue 232) was then re-amplified from pGEMMHhaI by PCR (with Pfu polymerase) using the primers pBR-ClaIba and pBRHha232-EcoRIfo, digested with ClaI and EcoRI, and cloned into pBR322 cut with the same enzymes to create pBRHha1-232. A fragment comprising the C-terminal part of the M.HhaI gene (from residue 258) was also amplified from pGEMMHhaI using the primers pGEMHha258-Hind3Ba and pGEM-EcoRIfo, digested with HindIII and EcoRI and cloned into pGEM-4Z digested with the same enzymes to create pGEMHha258-E. pBRHha1-232 was amplified by PCR using the primers tet1 (5′-end-labelled with 32P) and Hha245-Bsafo2. pGEMHha258-E was amplified with LMB2-1Bio (5′-end-labelled with biotin) and pGEMHha246-BsaIba. The fragments were digested with BsaI, purified using a Wizard PCR Preps DNA Purification System (Promega), quantitated by measuring the OD260 nm and each fragment added to a 35 µl ligation reaction to give a concentration of 0.095 µM (2 × 1012 molecules of each fragment). Dynabeads M-280 Streptavidin (Dynal) (2 mg) were added to the ligation mix, allowed to bind and the supernatant removed. The beads were washed three times with W+B buffer (2 M NaCl, 10 mM Tris–HCl, 1 mM EDTA pH 7.4), twice with PCR buffer (minus Mg2+) and resuspended in 50 µl of PCR buffer. The ligation efficiency was estimated to be >10% from scintillation counting 10% of the beads and the supernatants. Loop II library DNA, ready for selection, was created by taking 5 µl of resuspended beads (carrying an estimated >2 × 1010 ligated DNA molecules) and PCR amplifying the genes in a 200 µl reaction with primers tet6 and McrLMB2-2Bio (5′-biotinylated and with four HhaI R/M sites).
Figure 3.
Schematic diagram of the procedure for creating the library of mutated M.HhaI genes. See text for details.
Selection for DNA methylation by IVC
Selections for catalysis were performed with DNA at 0.5 nM for 2 h at 25°C as described (9), but digesting with the restriction endonuclease HhaI (as opposed to HaeIII) (Fig. 1). After the first round of selection, the surviving DNA was amplified with primers tet7 and McrLMB2-3Bio (which nest inside primers tet6 and McrLMB2-2Bio), analysed on a gel and quantitated by measuring the absorbance at 260 nm. A second round of selection was performed using this DNA exactly as above and the surviving DNA was PCR amplified with primers tet8 and McrLMB2-4Bio (which nest inside primers tet7 and McrLMB2-3Bio). The unselected DNA and DNA amplified after the first or second rounds of selection were added (to a final concentration of 5 nM) to 10 µl of in vitro transcription–translation reactions (Promega Escherichia coli S30 extract system for linear DNA), supplemented with 2000 U T7 RNA polymerase and 80 µM AdoMet. After 1 h at 25°C, the substrate DNA DIG-D1.3-biotin (a 942 bp DNA fragment PCR amplified with a biotinylated forward primer and a digoxigenin-labelled back primer, containing one HhaI R/M site and four HaeIII R/M sites) was added to give a concentration of 10 nM and incubated for 15 min at 25°C. The reaction was stopped and methylation of DIG-D1.3-biotin assayed by measuring protection from cleavage by the cognate restriction endonuclease R.HhaI using DIG-biotin ELISA (Roche) as described (9). Samples were also digested with the non-cognate restriction endonuclease R.HaeIII and left undigested. The percentage undigested DNA was calculated from the ratio of the ELISA signal observed with or without restriction enzyme digestion (with the background signal subtracted—where background is the signal, after digestion with R.HhaI, of a sample with no methyltransferase activity).
Cloning and characterisation of the selected M.HhaI library
Aliquots of 1 µl of loop II library DNA, unselected and after one and two rounds of selection, were amplified using Taq polymerase and primers tet10 and LMB2 in a 50 µl hot start PCR (25 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 2 min, then 72°C for 10 min). These PCRs were then cloned into pCR2.1-TOPO using TOPO TA cloning (Invitrogen). Colonies were PCR screened for inserts using primers pCR2.1ba and pCR2.1fo (94°C for 10 min, then 25 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 2 min, then 68°C for 7 min). Those with inserts of the correct size were checked for the presence of methyltransferase activity by in vitro translation of the PCR fragment and DIG-biotin ELISA as above. Clones from the unselected library and both active and inactive clones from the selected libraries were sequenced using primers HindIIIPCR and LMB2. Rates (relative to wild-type M.HhaI) were also determined using a DIG-biotin ELISA as above, except that the template DNA was at 8 nM and after translation the reactions were diluted 10-fold with water and 20 µl methylation reactions were set up with 2 µl of diluted translation, 80 µM AdoMet and 10 nM DIG-D1.3-biotin substrate DNA in HhaI methyltransferase buffer (50 mM Tris–HCl, pH 7.5, 10 mM EDTA, 5 mM 2-mercaptoethanol). The reactions were incubated at 25°C for 10 min, at which point the rate of methylation by wild-type M.HhaI (and hence also all mutant clones) was still at or close to initial velocity, then stopped by diluting 60 times in 2 M NaCl, 10 mM Tris–HCl, 1 mM EDTA pH 7.4, 25 µg/ml yeast RNA, 4 µM S-adenosylhomocysteine and methylation assayed by DIG-biotin ELISA as above. All assays were performed in duplicate. The absorbance (450 nm minus 650 nm) of the DIG-biotin ELISA was directly proportional to the percentage of methylated (i.e. uncut) DNA from 0 to 100%. The amount of each mutant produced by in vitro transcription–translation was quantitated by translation in the presence of [35S]methionine, SDS–PAGE and storage phosphor autoradiography using a Typhoon 8600 Imager (Amersham Biosciences). Yields of full-length protein varied from 41 to 220% of wild-type and rates were normalised for differences in expression levels.
RESULTS
Creation of a library of mutant M.HhaI genes by randomising codons in target recognition loop II
A library of ∼3.3 × 104 mutant M.HhaI genes was created using PCR. Three codons in target recognition loop II, encoding Ser252, Tyr254 and Gly257 in wild-type M.HhaI, were randomised (Fig. 2C). These are the only three residues in loop II thought to interact with bases. The side chain hydroxyl of Ser252 (Oγ) is thought to form a hydrogen bond with N7 of G3 through a water molecule, and Tyr254 and Gly257 are thought to have direct contacts through main chain atoms (O of Tyr and N of Gly) with C4 (N4) and G4′ (N7), respectively (16) (Fig. 2).
As described in Figure 3, the library was created by first splitting the M.HhaI gene in the region encoding target recognition loop II and subcloning each non-overlapping fragment into a different vector to ensure that the wild-type M.HhaI gene would not be present in the library. These two vectors were then amplified using PCR. The backward primer for the ‘C-terminal’ fragment of the M.HhaI gene contained the region encoding the three randomised codons. This primer and the forward primer for the ‘N-terminal’ fragment both contained sites for the restriction endonuclease BsaI: a Type IIs restriction enzyme that cuts to one side of the recognition sequence enabling the cohesive ends to match the sequence of the gene and the recognition sequence to be removed. The BsaI-digested ‘N-terminal’ and ‘C-terminal’ fragments were ligated to give intact genes. This allowed mutations to be introduced at any chosen site. The ligated gene libraries were amplified by PCR to give double-stranded linear DNA with a single biotin at one end (appended on a PCR primer). Each gene contained a T7 promoter and ribosome-binding site (rbs) for efficient expression in vitro. Each gene also had four HhaI R/M sites (5′-GCGC-3′) appended to the gene by the biotinylated primer. A sample from the library was cloned and 20 clones were sequenced. Each sequence was unique, with a random distribution of amino acids at the three mutated residues, and none had the wild-type amino acid sequence.
Selection by IVC
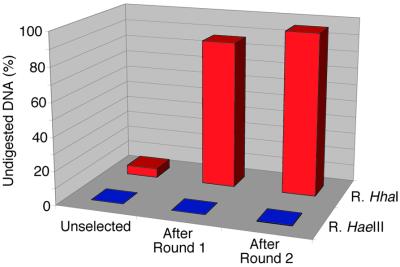
The library was subjected to two rounds of selection using IVC as described (9) using HhaI restriction endonuclease to digest unmethylated DNA (Fig. 1). After each round of selection the DNA was amplified with primers which nested inside the previous pair. In each case the biotinylated primer appended four HhaI R/M sites. Enrichment for genes encoding active methyltransferases was monitored by in vitro translating unselected DNA and DNA amplified after the first and second rounds of selection and then incubating the translated protein with a substrate DNA containing one HhaI R/M site and four HaeIII R/M sites (Fig. 4). When the substrate DNA was subsequently digested with HaeIII restriction endonuclease (R.HaeIII) the DNA was cut to completion, regardless of whether the DNA was selected or not, indicating that there was no methylation of HaeIII R/M sites (5′-GGCC-3′) before or after selection. In contrast, when the unselected library DNA was incubated with HhaI restriction endonuclease (R.HhaI), 5% of the DNA remained uncut after digestion, suggesting the presence of a low level of methylase activity even before selection. After one round of selection 87% of the substrate DNA was not digested with HhaI restriction endonuclease (R.HhaI) and after two rounds of selection 96% remained undigested, indicating an enrichment of genes encoding enzymes which methylate HhaI R/M sites (5′-GCGC-3′). However, as these assays were intended simply to detect enrichment for methyltransferase activity and were not carried out under initial velocity conditions, the level of activity in the unselected library is greatly overestimated. This is reflected in the percentage of active clones in the unselected versus the selected libraries (see below).
Figure 4.
Enrichment for genes encoding active methyltransferases by selection of the loop II library. Protein translated in vitro from unselected DNA or DNA amplified after round 1 and round 2 selection was incubated with a substrate DNA containing a single HhaI R/M site (5′-GCGC-3′) and the level of methylation of the substrate DNA assayed by measuring the percentage of DNA remaining undigested after incubating with HhaI restriction endonuclease (R.HhaI). As a control the substrate DNA was also digested with HaeIII restriction endonuclease (R.HaeIII), for which there are four R/M sites (5′-GGCC-3′).
DNA from the unselected target recognition loop II library and DNA amplified after round 1 and round 2 of selection was subcloned and individual clones with inserts of the expected size were in vitro translated and assayed for the ability to methylate a substrate containing the HhaI R/M site as described above. The number of clones showing detectable methyltransferase activity increased from 0% (0 of 32 clones tested) in the unselected library to 10% (3 of 31 clones tested) after one round of selection, and to 23% (8 of 35 clones tested) after the second round of selection, indicating an enrichment in each round of selection. None of the clones tested showed any detectable ability to methylate HaeIII R/M sites. The modest further enrichment in the second round mainly results from the high proportion of active clones after the first round of selection; assuming an average droplet size of 2.6 µm diameter (9), the 0.5 nM DNA concentration used in the selection corresponds to an average of ∼2.5 genes per compartment and methylation of co-compartmentalised genes would mean that the frequency of active clones would not exceed 40%.
Characterisation of active mutants selected from the library
Eleven selected clones showing methyltransferase activity towards the HhaI R/M site (5′-GCGC-3′) (three after one round of selection and eight after two rounds) were analysed. None of the mutants showed any detectable ability to methylate R/M target sites of related sequences, for example, HaeIII sites (5′-GGCC-3′), HpaII sites (5′-CCGG-3′) or AciI sites (5′-CCGC-3′) (HpaII and AciI sites would also be blocked by CpG methylation) when assayed by digestion with the cognate restriction endonuclease. Moreover, bisulphite modification and sequencing of methylated DNA (22) only revealed methylation at HhaI R/M sites. However, as relatively low methylation levels would not be detected by bisulphite sequencing, it is still possible that some mutations could confer a broader (relaxed) specificity, but difficulties expressing and purifying the mutants prevented further characterisation of specificity.
All 11 clones had Gly at position 257, as in wild-type M.HhaI (Table 1). At position 254 (Tyr in wild-type M.HhaI) there appeared to be little or no bias towards any particular amino acid (Table 1). At position 252, which is Ser in wild-type M.HhaI, there seems to be some bias towards certain amino acids, with Thr found five times and Val and Cys twice each (Table 1).
Table 1. Sequences and activities of active methyltransferases selected from the target recognition loop II library.
| Methyltransferase | Amino acid residue | Relative rate | ||
|---|---|---|---|---|
| 252 | 254 | 257 | ||
| Wild-type | S | Y | G | 1.00 |
| R2N20 | C | A | G | 0.49 |
| R1N4 | C | A | G | 0.29 |
| R2N34 | L | R | G | 0.26 |
| R1N19 | T | G | G | 0.20 |
| R2N21 | T | K | G | 0.13 |
| R2N32 | T | Q | G | 0.10 |
| R2N8 | T | A | G | 0.06 |
| R2N3 | T | S | G | 0.06 |
| R2N31 | V | S | G | 0.08 |
| R1N15 | V | F | G | 0.06 |
| R2N29 | A | T | G | 0.08 |
The amino acid residues found at positions 252, 254 and 257 are shown for the wild-type HhaI methyltransferase (M.HhaI) and for active mutants isolated from the target recognition domain loop II library after one (prefix R1) or two rounds (prefix R2) of selection. Difficulties in expressing and purifying the mutant methyltransferases prevented a full kinetic characterisation. However, after in vitro translation, the initial rates of the selected methyltransferases were compared to the wild-type under conditions (10 nM R/M sites) under which at least the wild-type M.HhaI [kcat = 1.3 per min and Km = 2.3 nM (31)] should be working close to Vmax. The rates of DNA methylation are shown relative to the wild-type.
The crystal structure indicates that Gly257 makes an interaction with G-4′ of the DNA through a main chain atom (N) (16) (Fig. 2). However, the complete conservation of Gly at position 257 in the active mutants suggests that it is crucial for methyltransferase activity (Table 1). The presence of a side chain may directly interfere with the interaction with DNA or indirectly disrupt the interaction with the DNA by changing the conformation of loop II. The latter seems likely as examination of the crystal structure revealed that Gly257 has φ–ψ angles incompatible with the presence of a side chain, i.e. it lies in a disallowed region for non-glycine residues on a Ramachandran plot (23).
At position 254, the sequences seemed to show quite a mix of different amino acids and did not reveal any obvious bias (Table 1). None, however, contained Tyr (the wild-type amino acid) at this position. As it is a main chain atom (O) of Tyr254 that is thought to be involved in the base-specific interaction with C-4 of the DNA, it is understandable that a wide range of amino acid side chains might be compatible with methylase activity.
The side chain hydroxyl of Ser252 (Oγ) is thought to form a hydrogen bond with N7 of G-3 of the DNA through a water molecule (16). Perhaps surprisingly, none of the sequenced, active clones had Ser at this position (Table 1). Indeed, five different amino acids were found at this position in the active mutants. However, no residues with charged or bulky side chains were observed, suggesting there may be some side chain preference. The relative activities of the mutants seems to correlate with the amino acid at position 252. The most active (29–49% of the activity of the wild-type enzyme) were the two clones with Cys252. Cys differs from Ser only in the replacement of a hydroxyl group (-OH) with a thiol (-SH) and since thiol groups are common hydrogen bond donors in biological systems (24) it is perhaps not surprising that Cys can substitute quite effectively for Ser. Likewise, five of the 11 active mutants sequenced had Thr252, the hydroxyl group of which could potentially also make a hydrogen bond with N7 of G-3 of the DNA through a water molecule. The activity of all the mutants with Thr252 was, however, lower than the mutants with Cys252. Indeed, one mutant (R2N8) had an otherwise identical sequence to the two clones with Cys252 yet ∼5-fold lower activity, suggesting that Cys is a better substituent at position 252. Surprisingly, the remaining active mutants had Leu, Val or Ala at position 252. These aliphatic side chains could certainly not be involved in any hydrogen bonding interaction. Indeed, one of the mutants (R2N34), with Leu252, was 26% as active as the wild-type methylase and more active than all the Thr252 mutants, throwing into some doubt the functional significance of the proposed water-mediated hydrogen bonding interaction between Ser252 and G-3 (16).
DISCUSSION
The mechanism by which DNA cytosine-5 methyltransferases such as M.HhaI specifically recognise their DNA targets is poorly understood. Moreover, the target recognition domains of DNA methyltransferases with different sequence specificities share little sequence conservation, except for the Thr·Leu dipeptide diagnostic of most target recognition domains (25–27), making the identification of residues involved in DNA binding by sequence analysis impossible. Thus, the most important insight into DNA recognition to date has come from crystallographic studies, which indicate that in M.HhaI and another cytosine-5 methyltransferase, M.HaeIII, the small domain contains two target recognition loops (loops I and II) which contact the target DNA sequence (16) (Fig. 2). In this study we have attempted to probe the functional significance of the three residues in target recognition loop II thought, from the crystallographic study, to interact with bases in the target sequence. To this end, we created a library of M.HhaI genes with the codons for these three loop II residues randomised and used IVC to select active mutants of M.HhaI from the library. The selection was based on genes encoding active methyltransferases becoming methylated at the HhaI R/M sites carried on the same DNA molecule and rendered resistant to cleavage by HhaI restriction endonuclease (Fig. 1). A similar principle has been used to clone a large number of methyltransferases: genomic libraries were expressed in vivo, in E.coli, and plasmids encoding methyltransferases were rendered resistant to digestion with restriction endonuclease and thereby isolated (28). However, this in vivo technique has been used only rarely to select libraries of mutated methyltransferases (29,30) and never for a saturation mutagenesis study as described here. The in vitro selections described here require no cloning or transformation and thereby allow the selection of much larger libraries. They also permit much more control over selection pressure than selection in vivo, for example allowing variation in methylation time and substrate concentration.
Using IVC, active clones were successfully isolated after both the first and second rounds of selection. Ten of 11 active clones had different sequences (Table 1). It seems likely, therefore, that if more clones were analysed, more active sequences would be discovered. Many of the selected clones were only slightly less active than the wild-type enzyme and it is intriguing that, even in a region of the protein thought to be intimately involved in DNA binding, many sequences are functional. Also, none of the active clones selected had the wild-type sequence and none was as active as wild-type M.HhaI (Table 1). However, for a gene to be selected it is only necessary that each of the four HhaI R/M sites it contains be methylated. The kcat of M.HhaI is only 1.3 per min (31); however, since it has a relatively low Km of 2.3 nM (31) and the concentration of a single gene in a compartment of average size [2.6 µm diameter (9)] is 0.2 nM (i.e. 0.8 nM R/M sites), a single wild-type enzyme molecule could still methylate all four R/M sites in a matter of minutes. Hence, it is not surprising that mutants with activities up to 10–15 times less than wild-type (see Table 1) can methylate all four R/M sites in the course of a 2 h incubation, allowing the genes that encode them to be selected. In fact, the conditions within these cell-like compartments (volume, enzyme and DNA concentrations, etc.) are similar to those in a bacterial cell. It is therefore likely that these in vitro selected clones could methylate DNA in vivo and confer resistance to the cognate HhaI endonuclease.
From the crystal structure, it seems that two of the base-specific interactions from loop II (Tyr254 and Gly257) are main chain mediated, but the third (Ser252) involves a side chain (Fig. 2) (16). Our data indicate that most likely no side chain can be present at position 257 since Gly257 is conserved in all active mutants. As no side chain would be compatible with the φ–ψ angles observed for Gly257, this must allow loop II to take up a conformation which allows direct contacts between the main chain atoms of Tyr254 and Gly257 with C-4 and G-4′, respectively, thus providing the specificity for the last base pair (Fig. 2). Tyr254 can be substituted by a wide range of amino acid residues whilst retaining activity consistent with the side chain, which points away from the DNA, having no role in target recognition. Several residues can substitute for Ser252, two of which (Thr and Cys), like Ser252, could potentially form a hydrogen bond with G-3 through a water molecule, but three (Leu, Val and Ala) cannot. This indicates that a side chain interaction with G-3 could make some contribution, but is certainly not a requirement for DNA recognition. The specificity for position 3 could potentially be provided entirely by the interaction of the main chain oxygen of Gln237 with C-3′ from target recognition loop I (Fig. 2) (16). In another study, where Gln237 was systematically mutated to all 19 other amino acids (32), all mutants retained enzymatic activity and specificity for the 5′-GCGC-3′ target site. Hence, it is an intriguing possibility that specificity for the second half of the target site (base pairs 3 and 4) does not involve any amino acid side chains and may be mediated entirely by main chain interactions with bases.
The crystal structures only reveal one step along the reaction pathway (33). Binding of DNA by M.HhaI involves a complex dynamic equilibrium with the target cytosine in multiple states: stacked in B-DNA, flipped-out of the duplex and, finally, flipped-out and locked into the active site (34). Hence, despite the fact that the crystal structures show no major rearrangement in the target recognition domain upon binding of DNA, it is possible that the contacts in the the initial binding event that contribute to sequence specificity differ from those seen to interact with the DNA in the crystal structure of the chemically trapped reaction intermediate (16).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Helen Cuhen for bisulphite sequencing and Leo James for helping with the preparation of Figure 2. Y.-F.L. was supported by the Cambridge Commonwealth Trust (Trinity College, Cambridge) and an Overseas Research Students Award (Committee of the Vice-Chancellors and Principals of the Universities of the United Kingdom).
REFERENCES
- 1.Fersht A. (1999) Structure and Mechanism in Protein Science. W.H. Freeman and Co., New York, NY.
- 2.Cunningham B.C. and Wells,J.A. (1989) High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science, 244, 1081–1085. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez M.R., Biosca,J.A., Torres,D., Crosas,B. and Pares,X. (1999) A double residue substitution in the coenzyme-binding site accounts for the different kinetic properties between yeast and human formaldehyde dehydrogenases. J. Biol. Chem., 274, 37869–37875. [DOI] [PubMed] [Google Scholar]
- 4.Clackson T. and Wells,J.A. (1994) In vitro selection from protein and peptide libraries. Trends Biotechnol., 12, 173–184. [DOI] [PubMed] [Google Scholar]
- 5.Wahler D. and Reymond,J.L. (2001) Novel methods for biocatalyst screening. Curr. Opin. Chem. Biol., 5, 152–158. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths A.D. and Tawfik,D.S. (2000) Man-made enzymes—from design to in vitro compartmentalisation. Curr. Opin. Biotechnol., 11, 338–353. [DOI] [PubMed] [Google Scholar]
- 7.Pluckthun A., Schaffitzel,C., Hanes,J. and Jermutus,L. (2000) In vitro selection and evolution of proteins. Adv. Protein Chem., 55, 367–403. [DOI] [PubMed] [Google Scholar]
- 8.Fastrez J. (1997) In vivo versus in vitro screening or selection for catalytic activity in enzymes and abzymes. Mol. Biotechnol., 7, 37–55. [DOI] [PubMed] [Google Scholar]
- 9.Tawfik D.S. and Griffiths,A.D. (1998) Man-made cell-like compartments for molecular evolution. Nat. Biotechnol., 16, 652–656. [DOI] [PubMed] [Google Scholar]
- 10.Ghadessy F.J., Ong,J.L. and Holliger,P. (2001) Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl Acad. Sci. USA, 98, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dryden D.T.F. (1999) Bacterial DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, pp. 283–340.
- 12.Vertino P.M. (1999) Eukaryotic DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, pp. 341–372.
- 13.Laird P.W. and Jaenisch,R. (1996) The role of DNA methylation in cancer genetic and epigenetics. Annu. Rev. Genet., 30, 441–464. [DOI] [PubMed] [Google Scholar]
- 14.Schmutte C. and Jones,P.A. (1998) Involvement of DNA methylation in human carcinogenesis. Biol. Chem., 379, 377–388. [DOI] [PubMed] [Google Scholar]
- 15.Cheng X., Kumar,S., Posfai,J., Pflugrath,J.W. and Roberts,R.J. (1993) Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell, 74, 299–307. [DOI] [PubMed] [Google Scholar]
- 16.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 17.Schluckebier G., O’Gara,M., Saenger,W. and Cheng,X. (1995) Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol., 247, 16–20. [DOI] [PubMed] [Google Scholar]
- 18.Cheng X. and Blumenthal,R.M. (1999) S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific, Singapore.
- 19.Kumar S., Cheng,X., Klimasauskas,S., Mi,S., Posfai,J., Roberts,R.J. and Wilson,G.G. (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 21.Caserta M., Zacharias,W., Nwankwo,D., Wilson,G.G. and Wells,R.D. (1987) Cloning, sequencing, in vivo promoter mapping and expression in Escherichia coli of the gene for the HhaI methyltransferase. J. Biol. Chem., 262, 4770–4777. [PubMed] [Google Scholar]
- 22.Paulin R., Grigg,G.W., Davey,M.W. and Piper,A.A. (1998) Urea improves efficiency of bisulphite-mediated sequencing of 5′-methylcytosine in genomic DNA. Nucleic Acids Res., 26, 5009–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramachandran G.N. and Sasiskharan,V. (1968) Conformation of polypeptides and proteins. Adv. Protein Chem., 23, 283–437. [DOI] [PubMed] [Google Scholar]
- 24.Allen F.H., Bird,C.M., Rowland,R.S. and Raithby,P.R. (1997) Hydrogen-bond acceptor and donor properties of divalent sulfur (Y-S-Z and R-S-H). Acta Crystallogr., B53, 696–701. [Google Scholar]
- 25.Vilkaitis G., Dong,A., Weinhold,E., Cheng,X. and Klimasauskas,S. (2000) Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem., 275, 38722–38730. [DOI] [PubMed] [Google Scholar]
- 26.Lange C., Wild,C. and Trautner,T.A. (1996) Identification of a subdomain within DNA-(cytosine-C5)-methyltransferases responsible for the recognition of the 5′ part of their DNA target. EMBO J., 15, 1443–1450. [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng X. and Blumenthal,R.M. (1996) Finding a basis for flipping bases. Structure, 4, 639–645. [DOI] [PubMed] [Google Scholar]
- 28.Szomolanyi E., Kiss,A. and Venetianer,P. (1980) Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene, 10, 219–225. [DOI] [PubMed] [Google Scholar]
- 29.Kiss A., Posfai,G., Zsurka,G., Rasko,T. and Venetianer,P. (2001) Role of DNA minor groove interactions in substrate recognition by the M.SinI and M.EcoRII DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 29, 3188–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vilkaitis G., Lubys,A., Merkiene,E., Timinskas,A., Janulaitis,A. and Klimasauskas,S. (2002) Circular permutation of DNA cytosine-N4 methyltransferases: in vivo coexistence in the BcnI system and in vitro probing by hybrid formation. Nucleic Acids Res., 30, 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J.C. and Santi,D.V. (1987) Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem., 262, 4778–4786. [PubMed] [Google Scholar]
- 32.Mi S., Alonso,D. and Roberts,R.J. (1995) Functional analysis of Gln-237 mutants of HhaI methyltransferase. Nucleic Acids Res., 23, 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Gara M., Klimasauskas,S., Roberts,R.J. and Cheng,X. (1996) Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes. J. Mol. Biol., 261, 634–645. [DOI] [PubMed] [Google Scholar]
- 34.Klimasauskas S., Szyperski,T., Serva,S. and Wuthrich,K. (1998) Dynamic modes of the flipped-out cytosine during HhaI methyltransferase–DNA interactions in solution. EMBO J., 17, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]