Abstract
We have studied hybridisation affinities and fluorescence behaviour of intercalator-modified oligonucleotides. The phosphoramidite of (S)-1-O-(4, 4′-dimethoxytriphenylmethyl)-3-O-(1-pyrenylmethyl)glycerol, an intercalating pseudo-nucleotide (IPN), was synthesised and by standard methods inserted into 7mer and 13mer oligodeoxyribonucleotides (ODNs) to generate intercalating nucleic acids (INAs). INAs showed greatly increased affinity for complementary single-stranded DNA (ssDNA), as determined by a thermal stabilisation of the formed DNA/INA duplex of up to 10.9°C per modification when the IPN was added as a dangling end and up to 6.7°C per modification when the IPN was inserted as a bulge. There was a positive stabilisation effect of the formed DNA/INA duplex on introducing a second IPN in the INA strand, when the two IPNs were separated by at least 1 bp. The effect is more pronounced the larger the separation of the two IPNs. Contrary to the enhanced affinity for ssDNA, the IPNs lower the affinity for complementary single-stranded RNA (ssRNA), giving rise to a difference in melting temperature of up to 25.8°C for two IPN insertions in an RNA/INA duplex when compared with the corresponding DNA/INA duplex. In this way INA is able to discriminate ssDNA over ssRNA with identical sequences. Fluorescence measurements show a stronger interaction of the pyrene moiety with DNA than with RNA, indicating intercalation as the stabilising factor in DNA/INA duplexes.
INTRODUCTION
Although it has been known for some time that there are relatively large differences in the three-dimensional structure of DNA/DNA duplexes (1) and DNA/RNA duplexes (2), and that some enzymes like RNase H are able to recognise one from the other (3), there have been only a few reported modified oligonucleotides able to differentiate between ssRNA and ssDNA. Some modifications in the sugar ring, like d-hexitol nucleic acid (HNA) and locked nucleic acid (LNA), have led to oligonucleotide analogues with increased affinity for ssDNA and ssRNA in general. These modifications preferentially stabilise hybridisation to ssRNA [melting temperature (ΔTm) +3 to +5°C and +4 to +8°C for HNA and LNA, respectively] over ssDNA (ΔTm +1 to +3°C and +3 to +5°C for HNA and LNA, respectively) (4–6). Some modifications are reported to be totally RNA selective, meaning that these oligonucleotide analogues will hybridise only with RNA and not with DNA. Unfortunately, most of the modifications that introduce selectivity for RNA do that by lowering the affinity for both RNA and DNA, the latter being most pronounced (7–10). On the other hand, there are few data showing that modified oligonucleotides can be DNA selective. However, these data are embedded in publications where discrimination between ssDNA and ssRNA is of little interest (11,12). In only one paper has the focus been on DNA/RNA discrimination using cholic acid and its deoxy derivatives linked to the 5′-termini of short oligonucleotides (13).
Nucleoside analogues with fluorescent labels have attracted interest for the last couple of decades for the development of new methods of distinguishing and detecting specific nucleic acid sequences (14–18). Among many different fluorescent probes pyrene is commonly used (11,19–24) and among these there are several acyclic pseudo-nucleotide analogues (19,22,23).
Here, we describe the insertion into a DNA strand of a new intercalating pseudo-nucleotide (IPN), namely the phosphoramidite of (S)-1-O-(4,4′-dimethoxytriphenylmethyl)-3-O-(1-pyrenylmethyl)glycerol (5). Insertion of IPNs into oligodeoxynucleotides produces oligodeoxyribonucleotide (ODN) analogues which we call intercalating nucleic acids (INAs). In the above-mentioned references there are examples of ODNs that can be considered as INAs. The INAs presented here have a higher affinity for complementary ssDNA and are able to differentiate between ssDNA and ssRNA, preferring to hybridise with ssDNA. How the affinity for and discriminating efficiency between DNA and RNA of INA depend on the number of insertions of IPNs and their positions in the INA have also been investigated.
MATERIALS AND METHODS
ODN and INA synthesis, purification and measurement of melting temperatures
The ODN and INA synthesis was carried out on a Pharmacia LKB Gene Assembler Special using Gene Assembler Special software v.1.53. The pyrene amidite 5 was dissolved in a 1:1 mixture of dry acetonitrile and dry dichloromethane, as a 0.1 M solution, and inserted into the growing oligonucleotides chain using the same conditions as for normal nucleotide coupling (2 min coupling). The coupling efficiency of the modified nucleotides was >99%. The ODNs were synthesised with DMT on and purified on a Waters Delta Prep 3000 HPLC with a Waters 600E controller and a Waters 484 detector on a Hamilton PRP-1 column. Buffer A, 950 ml of 0.1 M NH4HCO3 and 50 ml of MeCN, pH 9.0; buffer B, 250 ml of 0.1 M NH4HCO3 and 750 ml MeCN, pH 9.0. Gradients, 5 min 100% A, linear gradient to 100% B in 40 min (product peak at ∼37 min). The ODNs were DMT deprotected in 925 µl of H2O and 75 µl of CH3COOH and purified by HPLC, again using the same column, buffer system and gradients (product peak at ∼26 min). To get rid of the salts, the ODNs were redissolved in 1 ml of water and concentrated in vacuo three times. The yields after purification for the synthesis of INAs were for the self-complementary sequences from 2.6 to 6.6 OD, for the mono inserted IPN in the 13mer INA 17.0 OD and for the double IPN inserted INAs from 7.4 to 15.2 OD.
All modified ODNs were confirmed by MALDI-TOF analysis on a Voyager Elite Biospectrometry Research Station from PerSeptive Biosystems (Table 1). The transition state analyses were carried out on a Perkin Elmer UV/VIS spectrometer Lambda 2 with a PTP-6 temperature programmer using PETEMP rev. 5.1 software and PECSS software package v.4.3. Melting temperature measurements of the self-complementary sequences were made in 1 M NaCl, 10 mM sodium phosphate, pH 7.0, 1.5 µM each DNA strand. All other ODNs were measured in 150 mM NaCl, 10 mM sodium phosphate, 1 mM EDTA, pH 7.0, 1.5 µM each strand. All melting temperatures are with an uncertainty ±1.0°C as determined by repetitive experiments.
Table 1. Calculated and found masses of modified ODNs.
| Sequences | Calculated m/z | Found m/z |
|---|---|---|
| 5′-XCG CGC G | 2160 | 2161 |
| 5′-XTC GCG CGA | 2777 | 2778 |
| 5′-CTC AAG XCA ACC T | 3981 | 3979 |
| 5′-CTC AAG XXC AAC CT | 4348 | 4347 |
| 5′-CTC AAX GXC AAC CT | 4348 | 4348 |
| 5′-CTC AXA GXC AAC CT | 4348 | 4353 |
| 5′-CTC AXA GCA XAC CT | 4348 | 4349 |
| 5′-CTC AAG YCA ACC T | 3736 | 3742 |
| 5′-CTC AAG ZCA ACC T | 3750 | 3751 |
X represents the inserted compound 5, Y represents the flexible, abasic ethylene glycol backbone linker and Z the 1,3-propane-diol backbone linker.
Molecular calculations
The molecular calculations used the Amber force field (25) done in MacroModel 6.0 and 7.0 with water as solvent and minimisation was by the conjugant gradient method. Lam and Au-Yeung solved the structure of the self-complementary sequence by NMR and their data were used in this work (26). Their structure was extended with the pyrene nucleoside at the 5′ end of each strand and was used for the structural calculations. The 13mer sequence is a highly conserved HIV-1 long terminal repeat region (27). Using the coordinates from the corresponding duplex from the Brookhaven Protein Databank, G7 was replaced by the pyrene nucleotide and calculations for the duplex were made with and without an opposite C nucleotide. The pyrene was placed in the interior of the duplex from the beginning. All bonds were free to move and to rotate.
Fluorescence measurements
Fluorescence measurements were carried out in a Perkin-Elmer MPF-3 fluorescence instrument in 2 ml plastic cuvettes with excitation at 340 nm and detection at 360–600 nm. All measurements were conducted in 150 mM NaCl, 10 mM sodium phosphate, 1 mM EDTA, pH 7.0, with a concentration of 1.5 µM each strand. The plastic cuvettes did not interfere with the measurements.
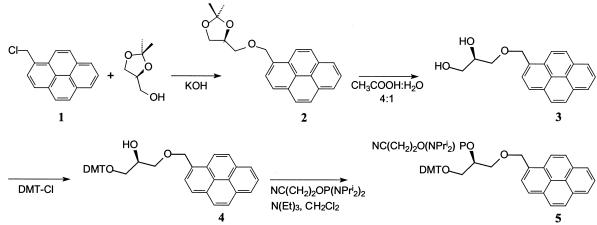
(S)-2,2-Dimethyl-4-(1-pyrenylmethoxymethyl)-1,3-dioxalane (2)
Pulverised KOH (25 g) and 1-(chloromethyl)pyrene (1, 6.0 g, 23.9 mmol) were added to a solution of (S)-(+)-2,2-dimethyl-1,3-dioxalane-4-methanol (2.6 g, 19.7 mmol) in dry toluene (250 ml). The mixture was refluxed under Dean–Stark conditions for 16 h, cooled to room temperature and H2O added (150 ml). The organic phase was washed with H2O (3 × 100 ml), dried (MgSO4 and Na2SO4) and concentrated under reduced pressure to give a viscous oil. The residue was purified by silica gel chromatography with CH2Cl2 as eluent, which afforded the pure compound 2 (6.1 g, 90%) as an oil. 1H NMR (CDCl3): δ 1.39 (s, 3H, CH3), 1.46 (s, 3H, CH3), 3.57–3.75 (m, 2H, H-5′), 3.88–4.06 (m, 2H, CHCH2OCH2), 4.32 (m, 1H, H-4′), 5.27 (m, 2H, OCH2pyrenyl), 7.99–8.39 (m, 9H, Harom). 13C NMR (CDCl3): δ 25.25 (CH3), 26.68 (CH3), 66.82 (C-1′), 70.96 (C-2′), 72.06 (OCH2pyrenyl), 74.70 (C-3′), 109.44 [C(CH3)2], 123.44, 124.46 (pyrene CH), 124.73, 124.97 (pyrene Cquat), 125.29 (2 × pyrene CH), 125.99, 127.14, 127.42, 127.52, 127.78 (pyrene CH), 129.46, 130.85, 131.02, 131.27, 131.43 (pyrene Cquat).
(R)-1-O-(1-Pyrenylmethyl)glycerol (3)
(S)-2,2-Dimethyl-4-(1-pyrenemethoxymethyl)-1,3-dioxalane (2, 6.1 g, 17.6 mmol) was added to a mixture of CH3COOH and H2O (100 ml, 4/1 v/v) and stirred at room temperature for 19 h. The mixture was concentrated under reduced pressure, giving an oil in quantitative yield. 1H NMR (CDCl3): δ 3.56–3.62 (m, 4H, H-1′, H-3′), 3.88 (m, 1H, H-2′), 5.14 (s, 2H, CH2), 7.89–8.26 (m, 9H, Harom). 13CNMR: δ (CDCl3) 63.79 (C-3′), 70.05 (C-2′), 71.45 (CH2), 72.0 (C-1′), 123.08, 124.44 (pyrene CH), 124.65, 124.89 (pyrene Cquat), 125.33 (2 × pyrene CH), 126.00, 127.12, 127.33, 127.56, 127.92 (pyrene CH), 129.36, 130.73, 131.20, 131.47 (pyrene Cquat). Maldi HRMS, found m/z 329.1137; calculated for C20H18O3 (M + Na+), 329.1148. Found, C 76.64%, H 5.84%; calculated for C20H18O3·0.35H2O, C 76.61%, H 6.05%.
(S)-1-O-(4,4′-Dimethoxytriphenylmethyl)-3-O-(1-pyrenylmethyl)glycerol (4)
(R)-1-O-(1-Pyrenylmethyl)glycerol (760 mg, 2.48 mmol) was dissolved in dry pyridine (20 ml) and dimethoxytrityl chloride (DMT-Cl) added (920 mg, 2.72 mmol). The reaction mixture was stirred for 24 h and concentrated under reduced pressure. The residue was purified by silica gel chromatography with EtOAc/cyclohexane/triethylamine (49/49/2 v/v/v) as eluent, affording compound 4 (1.20 g, 80%) as a white foam. 1H NMR(CDCl3): δ 2.8 (s, 1H, OH), 3.20 (m, 2H, H-3′), 3.68–3.72 (m, 6H, OCH3. H-1′, H-2′), 3.80 (s, 3H, OCH3), 5.26 (s, 2H, OCH2pyrenyl), 6.72–8.33 (m, 22H, Harom). 13C NMR (CDCl3): δ 55.06 (2 × CH3), 64.50 (C-1′), 70.00 (C-2′), 71.89 (CH2), 72.00 (C-3′), 86.08 (OCPh3 ), 113.00 (4 × trityl CH), 113.00 (pyrene CH), 123.35, 124.40 (pyrene CH), 124.67, 124.88 (pyrene Cquat), 125.21, 125.26, 125.94, 126.70, 127.06, 127.38 (pyrene CH), 127.42 (trityl CH), 127.75 (2 × trityl CH), 128.06 (2 × trityl CH), 129.31 (pyrene Cquat), 129.98 (4 × trityl CH), 130.75, 131.07, 131.19, 131.32 (pyrene Cquat), 135.90 (2 × trityl Cquat), 144.81 (trityl Cquat), 158.37 (2 × trityl COme). Maldi HRMS, found m/z 631.2448; calculated for C41H36O5 (M + Na+) 631.2455. Found, C 79.36%, H 6.07%; calculated for C41H36O5·0.7H2O, C 79.26%, H 6.06%.
Phosphoramidite of (S)-1-O-(4,4′-dimethoxytriphenyl methyl)-3-O-(1-pyrenylmethyl)glycerol (5)
(S)-1-O-(4,4′-Dimethoxytriphenylmethyl)-3-O-(1-pyrenyl methyl)glycerol (4, 458 mg, 753 µmol), 2-cyanoethyl N,N,N′,N′-tetraisopropylphosphorodiamidite (453 mg, 429 µl, 1.51 mmol) and diisopropylammonium tetrazolide (193 mg, 1.13 mmol) were mixed in dry CH2Cl2 (7 ml) and stirred under a nitrogen atmosphere for 6 days. The product was purified by silica gel chromatography with EtOAc/cyclohexane/triethylamine (49/49/2 v/v/v) and dried under reduced pressure. Yield 438 mg (72%) as a white foam. 31P NMR (CDCl3): δ 150.17, 150.21. Maldi HRMS, found m/z 815.3810; calculated for C41H36O5 (M + Li+) 815.3801.
RESULTS AND DISCUSSION
Synthesis of the building block
1-Pyrenemethanol is commercially available, but it is also easily prepared from pyrene by Vilsmeier–Haack formylation (28) followed by reduction with sodium borohydride. Subsequent conversion of the alcohol with thionyl chloride affords 1-(chloromethyl)pyrene (29).
The acyclic amidite 5 was prepared from (S)-(+)-2,2-dimethyl-1,3-dioxalane-4-methanol and 1-(chloromethyl)pyrene in 52% overall yield (Scheme 1). The synthesis of 5 was accomplished using KOH in the first step for the alkylation reaction (30) and then using 80% aqueous acetic acid (31) to give the diol 3. Protection by DMT-Cl (32) and finally reaction with 2-cyanoethyl N,N,N′,N′-tetraisopropylphosphorodiamidite affords the target compound 5 where the yield in the latter step was 72% (21). This yield decreased from 72 to 14% if 2-cyanoethyl N,N-diisopropylchlorophosphoramidite was used as the phosphitylating reagent.
Scheme 1. Synthesis of the phosphoramidite 5.
To increase the coupling yield in oligonucleotide synthesis, 5 was dissolved in a 1:1 mixture of acetonitrile and dichloromethane instead of in pure acetonitrile, in which 5 is poorly soluble. The phosphoramidite 5 was inserted into the growing chain of an ODN using standard nucleotide coupling conditions (2 min couplings). The coupling efficiency was over 98% as determined by detritylation measurement.
Self-complementary INAs with an intercalating pseudo-nucleotide as a dangling end
To investigate the stacking ability of 5, it was added to the 5′ end of two different self-complementary strands (5′-XCGCGCG and 5′-XTCGCGCGA, X = 5). The melting temperatures of modified and unmodified self-complementary oligonucleotides are shown in Table 2. Incorporation of the pyrene moiety at the 5′ end to give a dangling end stabilises the DNA duplex by 17.2 or 21.8°C (8.6 or 10.9°C per modification) depending on the underlying base pair. The stabilisations of the duplexes due to incorporation of 5 at the 5′-termini of the nucleic acid strands are similar to those found by Guckian et al. (33), who added a 2-deoxyribose pyrene nucleotide at the 5′ terminus of one of the very same self-complementary ODNs.
Table 2. Melting temperatures of self-complementary INAs with the 5′ modification.
| Duplex | Tm (°C) | ΔTm (°C) |
|---|---|---|
| 5′-CGCGCG | ||
| 3′-GCGCGC | 41.0 | |
| 5′-TCGCGCGA | ||
| 3′-AGCGCGCT | 46.9 | |
| 5′-XCGCGCG | ||
| 3′-GCGCGCX | 62.8 | 21.8 |
| 5′-XTCGCGCGA | ||
| 3′-AGCGCGCTX | 64.1 | 17.2 |
X = 5.
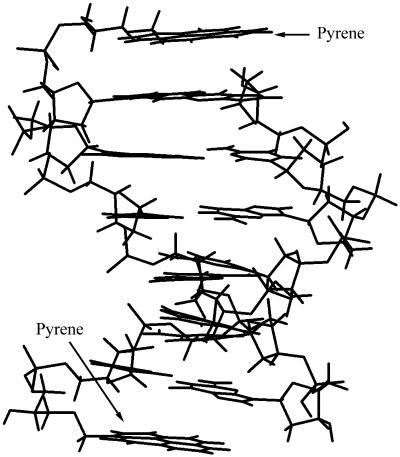
The stabilisation can be explained by MacroModel calculations, which confirm a possible structure where the pyrene moiety interacts with both nucleosides in the underlying base pair (Fig. 1).
Figure 1.
The structure of a self-complementary INA duplex with the sequence 5′-XCGCGCG-3′, X = 5, found by calculations in MacroModel. The pyrene moiety is coaxially stacked with the underlying base pair.
DNA/INA duplex where a nucleotide is replaced by an intercalating pseudo-nucleotide
UV melting temperature measurements (Table 3) where a G nucleotide is replaced by the flexible, abasic linker ethylene glycol or 1,3-propandiol show a large decrease in duplex stability compared to the unmodified, fully complementary sequence (Table 3, entry 5 versus entries 1 and 2). The required DMT-protected cyanoethyl N,N-diisopropylphosphoramidites of ethylene glycol and 1,3-propandiol were synthesised by standard methods (34). The pyrene-containing IPN prepared from 5 was inserted in the same position of the hybrid instead of the abasic diols. This increased the melting temperature by 16.4–18.0°C, indicating that the pyrene is coaxially stacked with both sides of the duplex (Table 3, entry 3 versus entries 1 and 2), as the stabilisation per modification exceeds the effect of placing the pyrene IPN at the end of a duplex (Table 2).
Table 3. DNA/INA and RNA/INA duplexes where a G nucleotide is replaced with an intercalating pseudo-nucleoside 5.
MacroModel calculations showed that IPN only makes a minor distortion of the double helix when intercalated into the duplex, having interaction with nucleobases both to the 5′ side and to the 3′ side of the intercalation site (Fig. 2). The stabilisation of the duplex by coaxial stacking of the pyrene moiety is not large enough to compensate for the loss in binding affinity due to the reduced number of hydrogen bonds by substitution of G with the IPN, the duplex with the INA being less stable than the unmodified fully complementary duplex by 8.6°C (Table 3, entry 3 versus entry 5). The same trend is found for DNA/RNA duplexes, although these have lower melting temperatures in general than the corresponding DNA/DNA duplexes. The stabilisation of the pyrene moiety is only 8.2°C for the DNA/RNA duplex when compared with ethylene glycol whereas the stabilisation is 16.4°C for the DNA/DNA duplex. Thus the pyrene insertion results in an improved discrimination between ssDNA and ssRNA giving a 9.0°C difference in the melting temperatures of their corresponding duplexes (Table 3).
Figure 2.
A slice of the calculated structure of the DNA/INA duplex 5′-AGCTTGCCTTGAG-3′ + 5′-CTCAAGXCAAGCT-3′, X = 5. The pyrene is coaxially stacked with both the upper and lower neighbouring nucleobases of the opposite strand.
INAs with intercalating pseudo-nucleosides forming bulges when hybridised
Normally the introduction of a bulge into the double helix decreases the melting temperature (35). This is also observed here when inserting a flexible, abasic linker into one of the strands (Table 4, entries 3–7). If the pyrene IPN, however, is inserted as the bulge, the melting temperature of the DNA hybrid is increased by 3°C. Insertions of non-Watson–Crick binding intercalators normally tend to cause a decrease in the melting temperature, but Ossipov et al. found that introduction of a bulge in such a case could prevent a large destabilisation (12). Insertion of 5 as a bulge stabilises the duplex by 11.2°C compared to the flexible ethylene glycol linker, indicating that the pyrene moiety is intercalated into the duplex (Table 4, entry 3).
Table 4. Melting temperatures of duplexes with bulging INAs hybridised to either DNA or RNA.
The difference in melting temperature between the DNA/INA duplex and the corresponding RNA/INA duplex is 12.6°C when 5 is inserted as a bulge. This difference is 7.4°C larger than in the unmodified duplexes and much larger than the differences between the duplexes containing a natural nucleotide or a flexible ethylene glycol as a bulge (Table 4). This means that the pyrene moiety is selective and only able to stabilise DNA/INA duplexes and not RNA/INA duplexes. The latter duplex has nearly the same melting temperature regardless of whether the bulge insertion contains the pyrene moiety or not, indicating that the pyrene does not intercalate in the RNA/INA duplex.
Structural calculation (MacroModel) of the pyrene- modified DNA/INA structure (Fig. 3) shows that the pyrene moiety makes only a minor distortion in the duplex and that the linker introduces enough flexibility in the backbone to have a distance of 3.4 Å between the pyrene moiety and the nucleobases of the same strand. The nucleobases in the opposite strand have a little shorter spacing between the pyrene moiety and the nucleobases than the optimum 3.4 Å (36).
Figure 3.
A slice of the calculated structure in MacroModel of a 12mer/13mer DNA/INA duplex with the sequence 5′-AGCTTGCTTGAG-3′ + 5′-CTCAAGXCAAGCT-3′, X = 5. The pyrene moiety is able to interact with both the upper and lower neighbouring nucleobases in the opposite strand.
To investigate the discrimination and stabilisation phenomena further, some INAs with two pyrene insertions were prepared. The results (Table 4, entries 4–7) show that, depending on the distance between two insertions and their neighbouring base pairs, it is possible to stabilise the double inserted pyrene DNA/INA duplex by up to 13.4°C (6.7°C per modification) compared to the natural DNA duplex. Both the stabilisation of the DNA/INA duplex and the destabilisation of the RNA/INA duplex is somewhat additive, so that the difference in melting temperature between the DNA/INA duplex and the RNA/INA duplex is up to 25.8°C when two pyrene moieties are inserted. For both stabilisation and discrimination the best results are obtained when the insertions are separated by 4 bp (Table 4, entry 7). When two insertions of the pyrene moieties are placed next to each other in the INA there is a decrease in melting temperature of 5.2°C compared to the unmodified DNA/DNA duplex and a decrease of 8.2°C compared to the mono modified DNA/INA duplex (Table 4, entry 4 versus entry 3). It is noteworthy that only one nucleobase pair between two IPN insertions in the DNA/INA duplex is sufficient to improve stabilisation and discrimination between DNA and RNA, when compared with the duplex where the INA strand is only modified with one insertion (Table 4, entry 5 versus entry 3).
Fluorescence of INAs
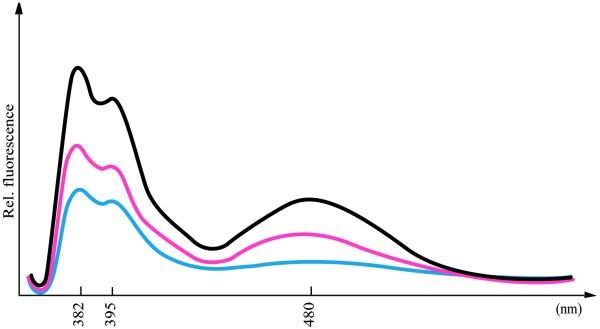
A decreased fluorescence emission upon excitation at 340 nm of the monopyrene-modified INA strands upon binding to the complementary strands indicates pyrene intercalation into the double helix (Fig. 4) (37,38). Double pyrene insertions in the oligonucleotides give the same results for all of the different INAs investigated (data not shown). The quenching effect upon hybridisation is more pronounced when the INA is hybridised with complementary ssDNA than when hybridised with complementary ssRNA (Figs 4 and 5), indicating less intercalation of pyrene into the RNA/INA duplexes. This supports the conclusion from the thermal melting experiments about the failure of pyrene intercalation into the bulged RNA/INA duplexes. The latter was deduced from the nearly identical melting temperatures with the abasic flexible ethylene glycol linker and pyrene-containing bulges when hybridised to ssRNA (Table 4).
Figure 4.
Fluorescence measurements of a 13mer, mono pyrene IPN-modified ssINA (black), its duplex with complementary 12mer RNA (red) and its duplex with complementary 12mer DNA (blue). The sequences are as shown in Table 4, entry 3.
Figure 5.
Fluorescence measurements of a 14mer ssINA with two pyrene IPN modifications separated by 1 nt (black), its duplex with complementary 12mer RNA (red) and its duplex with complementary 12mer DNA (blue). The sequences are as shown in Table 4, entry 5.
Two pyrene moieties separated by only one nucleotide generates a third peak at 480 nm, due to excimer formation of the pyrene residues (39). However, this band is almost extinguished when this type of INA hybridises to a complementary DNA strand (Fig. 5). This indicates intercalation around an intact base pair preventing the two pyrene moieties being placed in the physical distance of ∼3.4 nm needed for excimer formation. When a double pyrene IPN-modified INA hybridises to a complementary RNA, the two pyrene moieties are still able to interact since a substantial excimer band is found.
CONCLUSION
Here we have described the easy synthesis of an acyclic pyrene phosphoramidite 5, which can be inserted as an IPN into oligonucleotides giving INAs in good yields using normal oligonucleotide synthesis procedures. INAs have a higher affinity for complementary ssDNA than unmodified DNA, and the duplex formed has a melting temperature that is up to 6.7°C higher per modification when the IPN is inserted internally in the formed duplex and up to 10.9°C when added as a dangling end. Furthermore, the pyrene-containing IPN introduces selectivity to INAs for complementary ssDNA over complementary ssRNA. Two appropriately placed pyrene IPNs increase the differences in melting temperature between the two types of duplexes in an additive manner, giving a difference in melting temperature for the DNA/INA duplex that is up to 25.8°C higher than for the corresponding RNA/INA duplex. Quenching of pyrene fluorescence is observed upon hybridisation as a consequence of pyrene intercalation. This phenomenon is most pronounced for the hybridisation to DNA when compared to the same sequence of RNA. The thermal melting data for duplexes support the idea of pyrene intercalation in the DNA/INA duplex and not in the RNA/INA duplex. It is believed that INAs can become an important tool for discrimination between DNA and RNA with identical sequences and that the INA technology can be used as a versatile tool in oligonucleotide applications when enhanced affinities towards DNA are required.
Acknowledgments
ACKNOWLEDGEMENT
The Nucleic Acid Center is funded by The Danish National Research Foundation for studies on nucleic acid chemical biology.
REFERENCES
- 1.Watson J.D. and Crick,F.H.C. (1953) Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature, 171, 737–738. [DOI] [PubMed] [Google Scholar]
- 2.Milman G., Langridge,R. and Chamberlin,M.J. (1967) Structure of a DNA-RNA hybrid. Proc. Natl Acad. Sci. USA, 57, 1804–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhlmann E. and Peyman,A. (1990) Structure of a DNA-RNA hybrid. Chem. Rev., 90, 543–584. [Google Scholar]
- 4.Van Aerschot A., Verheggen,I., Hendric,C. and Herdewijn,P. (1995) 1,5-Anhydrohexitol nucleic acids, a new promising antisense construct. Angew. Chem. Int. Ed. Engl., 34, 1338–1339. [Google Scholar]
- 5.Hendrix C., Rosemeyer,H., Verheggen,I., Seela,F., Van Aerschot,A. and Herdewijn,P. (1997) 1′,5′-Anhydrohexitol oligonucleotides: synthesis, base pairing and recognition by regular oligodeoxyribonucleotides and oligoribonucleotides. Eur. J. Org. Chem., 3, 110–120. [Google Scholar]
- 6.Koshkin A.A., Singh,S.K., Nielsen,P., Rajwanshi,V.K., Kumar,R., Meldgaard,M., Olsen,C.E. and Wengel,J. (1998) LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerization and unprecedented nucleic acid recognition. Tetrahedron, 54, 3607–3630. [Google Scholar]
- 7.Fujimori S., Shudo,K. and Hashimoto,Y. (1990) Enantio-DNA recognizes complementary RNA but not complementary DNA. J. Am. Chem. Soc., 112, 7436–7438. [Google Scholar]
- 8.Adams A.D., Petrie,C.R. and Meyer,R.B.,Jr (1991) Preparation and hybridization properties of oligonucleotides containing 1-α-d-arabinofuranosylthymine. Nucleic Acids Res., 19, 3647–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhan P., Bhan,A., Hong,M., Hartwell,J.G., Saunders,J.M. and Hoke,G.D. (1997) 2′,5′-Linked oligo-3′-deoxyribonucleoside phosphorothioate chimeras: thermal stability and antisense inhibition of gene expression. Nucleic Acids Res., 25, 3310–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keinicke L., Sorensen,M.D. and Wengel,J. (2002) α-l-RNA (α-l-ribo configured RNA): synthesis and RNA-selective hybridization of α-l-RNA/α-l-LNA chimera. Bioorg. Med. Chem. Lett., 12, 593–596. [DOI] [PubMed] [Google Scholar]
- 11.Yamana K., Iwase,R., Furutani,S., Tsuchida,H., Zako,H., Yamaoka,T. and Murakami,A. (1999) 2′-Pyrene modified oligonucleotide provides a highly sensitive fluorescent probe of RNA. Nucleic Acids Res., 27, 2387–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ossipov D., Zamaratski,E. and Chattopadhyaya,J. (1999) Dipyrido[3,2-a:2′,3′-c]phenazine-tethered oligo-DNA: synthesis and thermal stability of their DNA·DNA and DNA·RNA duplexes and DNA·DNA·DNA triplexes. Helv. Chim. Acta, 82, 2186–2200. [Google Scholar]
- 13.Bleczinski C.F. and Richert,C. (1999) Steroid-DNA interactions increasing stability, sequence-selectivity, DNA/RNA discrimination, and hypochromicity of oligonucleotide duplexes. J. Am. Chem. Soc., 121, 10889–10894. [Google Scholar]
- 14.Letsinger R.L. and Schott,M.E. (1981) Selectivity in binding a phenanthridinium-dinucleotide derivative to homopolynucleotides. J. Am. Chem. Soc., 103, 7394–7396. [Google Scholar]
- 15.Telser J., Cruickshank,K.A., Morrison,L.E., Netzel,T.L. and Chan,C.K. (1989) DNA duplexes covalently labeled at two sites: synthesis and characterization by steady-state and time-resolved optical spectroscopies. J. Am. Chem. Soc., 111, 7226–7232. [Google Scholar]
- 16.Oser A. and Valet,G. (1990) Nonradioactive assay of DNA hybridization by DNA-template-mediated formation of a ternary Tb(III) complex in pure liquid-phase. Angew. Chem. Int. Ed. Engl., 29, 1167–1169. [Google Scholar]
- 17.Murakami A., Nakaura,M., Nakatsuji,Y., Nagahara,S., Tran-Cong,Q. and Makino,K. (1991) Fluorescent-labeled oligonucleotide probes: detection of hybrid formation in solution by fluorescence polarization spectroscopy. Nucleic Acids Res., 19, 4097–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins Y. and Barton,J.K. (1992) A sequence-specific molecular light switch: tethering of an oligonucleotide to a dipyridophenazine complex of ruthenium(II). J. Am. Chem. Soc., 114, 8736–8738. [Google Scholar]
- 19.Yamana K., Nunota,K., Nakano,H. and Sangen,O. (1994) A new method for introduction of a pyrene group into a terminal position of an oligonucleotide. Tetrahedron Lett., 35, 2555–2558. [Google Scholar]
- 20.Korshun V.A., Stetsenko,D.A. and Gait,M.J. (2002) Novel uridin-2′-yl carbamates: synthesis, incorporation into oligodeoxyribonucleotides and remarkable fluorescence properties of 2′-pyren-1-ylmethylcarbamate. J. Chem. Soc. Perkin Trans. 1, 1092–1104. [Google Scholar]
- 21.Burmeister J., Azzawi,A. and von Kiedrowski,G. (1995) Synthesis of novel phosphoramidite derivatives bearing pyrenyl and dansyl groups. Tetrahedron Lett., 36, 3667–3668. [Google Scholar]
- 22.Moran S., Ren,R.X.F., Sheils,C.J., Rumney,S.,IV and Kool,E.T. (1996) Non-hydrogen bonding ‘terminator’ nucleosides increase the 3′-end homogeneity of enzymic RNA and DNA synthesis. Nucleic Acids Res., 24, 2044–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamana K., Takei,M. and Nakano,H. (1997) Synthesis of oligodeoxyribonucleotide derivatives containing pyrene labeled glycerol linkers: enhanced excimer fluorescence on binding to a complementary DNA sequence. Tetrahedron Lett., 38, 6051–6054. [Google Scholar]
- 24.Puri N., Zamaratski,E., Sund,C. and Chattopadhyaya,J. (1997) Synthesis of 5′-polyarene-tethered oligo-DNAs and the thermal stability and spectroscopic properties of their duplexes and triplexes. Tetrahedron, 53, 10409–10432. [Google Scholar]
- 25.Cornell W.D., Cieplak,P., Bayly,C.I., Gould,I.R., Merz,K.M.,Jr, Ferguson,D.M., Spellmeyer,D.C., Fox,T., Caldwell,J.W. and Kollman,P.A. (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc., 117, 5179–5197. [Google Scholar]
- 26.Lam S.L. and Au-Yeung,S.C.F. (1997) Sequence-specific local structural variations in solution structures of d(CGXX′CG)2 and d(CAXX′TG)2 self-complementary deoxyribonucleic acids. J. Mol. Biol., 266, 745–760. [DOI] [PubMed] [Google Scholar]
- 27.Mujeeb A., Kerwin,S.M., Kenyon,G.L. and James,T.L. (1993) Solution structure of a conserved DNA sequence from the HIV-1 genome: restrained molecular dynamics simulation with distance and torsion angle restraints derived from two-dimensional NMR spectra. Biochemistry, 32, 13419–13431. [DOI] [PubMed] [Google Scholar]
- 28.Bayer O. (1954). Herstellung von Aldehyden. In Müller,E. (ed.), Methoden der Organischen Chemie—Vierte Auflage. Georg Thieme Verlag, Stuttgart, Germany, Vol. 7/1, p. 34.
- 29.Bair K.W., Tuttle,R.L., Knick,V.C., Cory,M. and McKee,D.D. (1990) [(1-Pyrenylmethyl)amino] alcohols, a new class of antitumor DNA intercalators. Discovery and initial amine side chain structure-activity studies. J. Med. Chem., 33, 2385–2393. [DOI] [PubMed] [Google Scholar]
- 30.Tirosh O., Kohen,R., Katzhendler,J., Gorodetsky,R. and Barenholz,Y. (1997) Novel synthetic phospholipid protects lipid bilayers against oxidation damage: role of hydration layer and bound water. J. Chem. Soc. Perkin Trans. 2, 383–389. [Google Scholar]
- 31.Iguchi K., Kitade,M., Kashiwagi,T. and Yamada,Y. (1993) Structure and synthesis of petrosynes, new acetylenic enol ether glycerides from the Okinawan marine sponge of the genus Petrosia. J. Org. Chem., 58, 5690–5698. [Google Scholar]
- 32.Chattopadhyaya J.B. and Reese,C.B. (1978) The 9-phenylxanthen-9-yl protecting group. J. Chem. Soc. Chem. Commun., 15, 639–640. [Google Scholar]
- 33.Guckian K.M., Schweitzer,B.A., Ren,R.X.F., Sheils,C.J., Paris,P.L. and Kool,E.T. (1996) Experimental measurement of aromatic stacking affinities in the context of duplex DNA. J. Am. Chem. Soc., 118, 8182–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seela F. and Kaiser,K. (1987) Oligodeoxyribonucleotides containing 1,3-propanediol as nucleoside substitute. Nucleic Acids Res., 15, 3113–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeBlanc D.A. and Morden,K.M. (1991) Thermodynamic characterization of deoxyribooligonucleotide duplexes containing bulges. Biochemistry, 30, 4042–4047. [DOI] [PubMed] [Google Scholar]
- 36.Lerman L.S. (1961) Structural considerations in the interaction of deoxyribonucleic acid and acridines. J. Mol. Biol., 3, 18–30. [DOI] [PubMed] [Google Scholar]
- 37.Telser J., Cruickshank,K.A., Morrison,L.E. and Netzel,T.L. (1989) Synthesis and characterization of DNA oligomers and duplexes containing covalently attached molecular labels: comparison of biotin, fluorescein, and pyrene labels by thermodynamic and optical spectroscopic measurements. J. Am. Chem. Soc., 111, 6966–6976. [Google Scholar]
- 38.Haralambidis J., Angus,K., Pownall,S., Duncan,L., Chai,M. and Tregear,G.W. (1990) The preparation of polyamide-oligonucleotide probes containing multiple non-radioactive labels. Nucleic Acids Res., 18, 501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birks J.B. (1968) Pyrene excimer. Acta Phys. Pol., 34, 603–617. [Google Scholar]