Abstract
The RNA interference (RNAi) phenomenon is a recently observed process in which the introduction of a double-stranded RNA (dsRNA) into a cell causes the specific degradation of a mRNA containing the same sequence. The 21–23 nt guide RNAs, generated by RNase III cleavage from longer dsRNAs, are associated with sequence-specific mRNA degradation. Here, we show that dsRNA specifically suppresses the expression of HIV-1 genes. To study dsRNA-mediated gene interference in HIV-1-infected cells, we have designed six long dsRNAs containing the HIV-1 gag and env genes. HIV-1 replication was totally suppressed in a sequence-specific manner by the dsRNAs in HIV-1-infected cells. Especially, E2 dsRNA containing the major CD4-binding domain sequence of gp120, as the target of the HIV-1 env gene, dramatically inhibited the expression of the HIV-1 p24 antigen in PBMCs for a relatively long time. The dsRNA interference method seems to be a promising new strategy for anti-HIV-1 gene therapeutics.
INTRODUCTION
RNA interference (RNAi) occurs in a variety of organisms, including Caenorhabditis elegans, Trypanosoma brucei, plants, Drosophila, planaria, zebrafish and mouse embryos. In most of these organisms, the injection of a double- stranded RNA (dsRNA) longer than 500 bp specifically suppresses the expression of the gene with the corresponding DNA sequence, but has no effect on genes with unrelated sequences (1–10).
RNAi is initiated by the RNase III-like nuclease Dicer, which promotes processive cleavage of long dsRNAs into 21–23-nt short interfering RNAs (siRNAs) with 2-nt 3′ overhangs (11–15). Subsequently, the siRNAs are incorporated into an RNA-induced silencing complex (RISC), identified in Drosophila, and the protein–RNA effector nuclease complex recognizes and destroys the target mRNAs (16–18).
The use of this RNAi in mammalian cells appeared to be of limited utility, because dsRNAs longer than 30 bp trigger interferon responses through the activation of dsRNA- dependent protein kinase (PKR) and 2′,5′-oligoadenylate (2–5A) synthetase (19–21). To overcome this non-specific effect in mammalian cells, it has recently been found that 21–23-nt siRNAs with 2-nt 3′ overhangs exhibit an RNAi effect (22). Transfection of synthetic siRNAs into mammalian cells effectively inhibits the endogenous genes in a sequence-specific manner (22–24). Lee et al. have reported the inhibition of HIV-1 replication by the expression of small interfering RNAs targeted against HIV-1 rev in human cells (25).
Several RNA-based antiviral therapies are being studied for HIV treatments, using ribozyme and antisense approaches (26–28). However, dsRNA provides a more reliable method of viral and reporter gene inactivation than antisense RNA (29). Furthermore, the longer dsRNA-mediated inhibition of gene expression is still being studied in mammalian systems. Microinjection of dsRNA into mouse oocytes or early embryos results in the specific inhibition of the activities of both maternally and zygotically expressed proteins (9,10). Recently, Billy et al. reported that the RNAi response could be induced effectively by long dsRNA in non-differentiated mouse cells grown in culture (30). In addition, Billy et al. demonstrated that extracts prepared from mouse embryonal carcinoma (EC) cells catalyzed the processing of dsRNA into ∼23-nt RNAs and that the processing enzyme, Dicer, is localized in the cytoplasm of EC and HeLa cells.
In this study, we used a long dsRNA, in place of antisense DNA (31,32), to interfere with the expression of HIV-1 genes. We found that the greatest inhibitory effects were detected with 531 bp E2-dsRNA containing the major CD4-binding domain sequence of gp120, as the target of the HIV-1 env gene. We also found that the long dsRNA caused the sequence-specific inhibition of HIV-1 replication. Especially, the E2-dsRNA dramatically inhibited the expression of the HIV-1 p24 antigen in PBMCs for a relatively long time.
MATERIALS AND METHODS
Plasmid and PCR template used for RNA synthesis
Plasmid pNL4-3 was used as the PCR template for the synthesis of dsRNAs corresponding to the gag and env genes. PCR templates for dsRNA synthesis targeting the firefly luciferase (luc) reporter gene (Photinus pyralis, Luc, 1032–1562, 531 bp) were amplified from pEGFPLuc (Clontech). The transcription templates were linear double-stranded DNA (dsDNA) with blunt or 5′-protruding ends. The templates used for dsRNA synthesis were the products of PCRs with primers designed to amplify limited regions of the cDNA sequence for each gene. Each primer contained an SP6 promoter sequence (CATACGATTTAGGTGACACTATAG) and a T7 promoter sequence (TAATACGACTCACTATAG) on its 5′ end. Sense and antisense RNAs were synthesized in vitro with the SP6 and T7 polymerases, using the AmpliScribe Transcription Kit (Epicentre Technologies). DNA templates were then removed by DNase I treatment. The RNA products were extracted with phenol/chloroform and then precipitated in ethanol. The in vitro transcription products were resolved by electrophoresis in a 1% agarose gel prepared with denaturing buffer. For annealing, equimolar quantities of sense and antisense RNAs were mixed in annealing buffer (10 mM Tris pH 7.4, 0.1 mM EDTA), heated to 90°C for 1 min and incubated at 37°C for 12–16 h. The formation of dsRNA was confirmed by its migration on a 1% agarose gel in TBE.
Cell culture and RNA interference assays
COS, HeLa-CD4+ and ACH2 cells were grown in RPMI 1640 medium (Sigma) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM l-glutamine at 37°C under a 5% CO2 atmosphere. Human peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation, grown in RPMI 1640 medium supplemented with 10% FBS, and activated with 1 µg/ml phytohemaggulutinin (PHA) (Seikagaku Corp.) for 3 days in the presence of IL-2 (100 U/ml; Shionogi) at 37°C. For the p24 accumulation assays, PHA-stimulated PBMCs were infected with the HIV-1NL4-3 virus at a multiplicity of infection (MOI) of 0.001 in the presence of various concentrations of the dsRNAs. At 4, 7, 10 or 14 days after infection, culture supernatants containing the viruses were removed and the amount of p24 was measured by an HIV-1 p24 CLEIA assay (Lumipulse; Fujirebio Inc.) (33). Fresh medium was then added with one-half volume of culture medium and the cells were transfected with the indicated amounts of dsRNA by using the TransMessenger Transfection Reagent (Qiagen), as described by the manufacturer. The plasmid pNL4-3 is an HIV-based infectious vector, which was purified with the plasmid Maxi-Prep Kit (Sigma), followed by phenol extraction and ethanol precipitation. Twenty-four hours before transfection, adherent cells were seeded in 6-well multiwell plates at a density of 2 × 105 cells/well. COS cells were transfected with 2 µg dsRNA and 2 µg pNL4-3, using 2 µl of Lipofectin (Gibco BRL) and 3 µl of FuGENE 6 (Roche), according to the manufacturers’ optimized protocols. After a 72 h incubation, the virus replication was monitored in the culture supernatants with the HIV-1 p24 CLEIA assay. For the infection of HeLa-CD4+ cells, 2 × 105 cells were seeded into 6-well multiwell plates 24 h before infection and then were infected with the HIV-1NL4-3 virus at a MOI of 0.5. After a 16 h infection, the cells were washed and treated with dsRNA at a 2 µg concentration in the culture medium. HeLa-CD4+ cells were transfected at ∼24 h intervals and were asssayed 7 days after infection.
RNA isolation and RT–PCR assay
The reverse transcription and the PCR analysis were performed with total RNA from COS cells (prepared with the GenElute Mammalian Total RNA kit; Sigma) as the template (1 µg/reaction). These templates were amplified for one cycle of 60°C for 30 min and 94°C for 2 min, 40 cycles of 94°C for 1 min and 50°C for 1.5 min and one cycle of 50°C for 1 min with env gene-specific primers. The primers were 5′-ACAGCTGAACACATCTGTAGAAATTAATTG-3′ and 5′-GTTGTTATTACCACCATCTCTTGTTAATAG-3′. The ampli fied DNA was subjected to electrophoresis in a 1% agarose gel and was visualized by ethidium bromide staining.
RESULTS AND DISCUSSION
The efficient cellular uptake of the dsRNAs is a critical step to enable targeting to the mRNA. Using COS cells, we examined the cellular uptake of fluorescently labeled E2-dsRNA with several kinds of transfection reagents (FuGENE 6, Lipofectin and Gene PORTER 2) at 37°C for 24 and 72 h. The cells were washed twice with PBS, resuspended in PBS containing 1% paraformaldehyde and analyzed by the FACS Calibur and CellQuest software. When we used Lipofectin as the transfection reagent, after 24 h, 94% of the COS cell-associated fluorescence signal was found, while after 72 h it had only decreased to 84%, which might explain the enhanced RNAi effect (data not shown). To test for silencing of the HIV-1 gene, we chose Lipofectin as the transfection reagent.
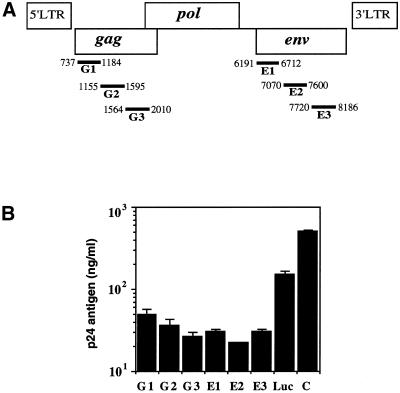
To test for silencing, we designed six longer dsRNAs containing the HIV-1 gag (G1–G3) and env genes (E1–E3) (Fig. 1A). We also designed a control long dsRNA, which included the firefly luciferase gene (Photinus pyralis, Luc dsRNA, 1032–1562, 531 bp). Sense (ss) and antisense (as) RNAs were transcribed in vitro from PCR products and annealed to each other to produce the dsRNA. The ssRNAs, asRNAs and dsRNAs were each tested for their ability to suppress HIV-1 expression specifically. Using Lipofectin (Gibco), dsRNA duplexes were co-transfected into COS cells with the HIV-1 expression vector pNL4-3. The virus production in the culture supernatant was assessed by the HIV-1 p24 antigen assay at 3 days, and was related to the amount produced in the absence of dsRNA. The six longer dsRNAs containing the HIV-1 gag (G1–G3) and env sequences (E1–E3) specifically reduced the p24 expression by ∼10- to 23-fold as compared to the untreated control cells. Notably, the three dsRNAs targeted to the HIV-1 env gene (E1–E3) inhibited p24 expression more effectively than those targeted to the HIV-1 gag gene (G1–G3) (Fig. 1B). We also investigated the sequence-specific inhibition of HIV-1 gene expression, using the Luc-dsRNA as the control dsRNA. The Luc-dsRNA showed higher expression of the p24 antigen than the six dsRNAs targeted to the HIV-1 gag (G1–G3) and env genes (E1–E3) (Fig. 1B). However, the Luc-dsRNA exhibited very low inhibition of HIV-1 replication. Some inhibition by the control Luc-dsRNA might have occurred if it caused the activation of a dsRNA-dependent enzyme, such as PKR or 2–5A synthetase. The greatest inhibitory effects were detected with the 531 bp E2-dsRNA containing the major CD4-binding domain sequence of gp120, as the target of the HIV-1 env gene.
Figure 1.
(A) The target sites of the dsRNAs used in this study and described in the text. The target regions for the dsRNAs are indicated as black bars below the coding region of the HIV-1 genes. Double-stranded RNAs were generated corresponding to the HIV-1 gag (G1, 737∼1184, 448 bp; G2, 1155∼1595, 441 bp; G3, 1564∼2010, 447 bp) and env (E1, 6191∼6712, 522 bp; E2, 7070∼7600, 531 bp; E3, 7720∼8186, 467 bp) genes. The Luc-dsRNA, as a non-specific control, corresponds to the following positions in the luciferase gene from the firefly Photinus pyralis (Luc, 1032∼1562, 531 bp). (B) Specific RNAi by longer dsRNAs. The dsRNAs (2 µg) encapsulated with Lipofectin (2 µl) were transfected into COS cells first, followed by encapsulated pNL4-3 proviral DNA (2 µg) after 5 h. At 3 days post-transfection, p24 antigen production was detected by the HIV-1 p24 CLEIA assay (C, positive control cells transfected with pNL4-3 proviral DNA). The plotted data were averaged from duplicate experiments and the bars represent ± SD.
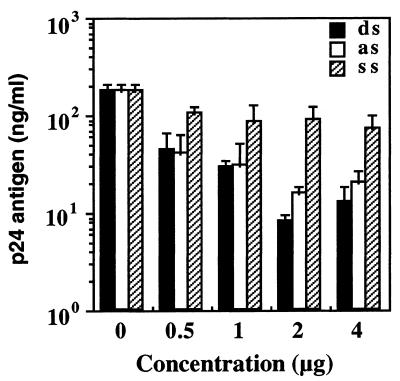
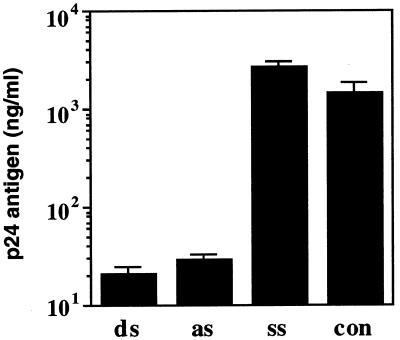
We also assessed whether the RNAi was dose dependent. COS cells were transduced with 0.5–4 µg of the E2-ssRNA, asRNA or dsRNA and the HIV-1 expression vector pNL4-3, using Lipofectin. As expected, the E2-dsRNA exhibited the HIV-1-dependent dsRNA-mediated inhibition in a dose-dependent manner (Fig. 2). With this result, we confirmed the effectiveness of the dsRNAs by their dose dependence, with the best response at 2 µg. Interestingly, the E2-asRNA also reduced the p24 expression, with a similar dose- dependent efficiency. The E2-ssRNA was incapable of inhibiting HIV-1 gene expression, even when transfected with an excess of the minimum saturating dsRNA concentration.
Figure 2.
Dose dependence of E2-dsRNA in COS cells. ssRNA (ss), asRNA (as) or dsRNA (ds) (0.5–4 µg), encapsulated with Lipofectin, was transfected into COS cells first, followed by encapsulated pNL4-3 (2 µg) after 5 h. At 3 days post-transfection, p24 antigen production was detected by the HIV-1 p24 CLEIA assay. The plotted data were averaged from three independent experiments and the bars represent ± SD.
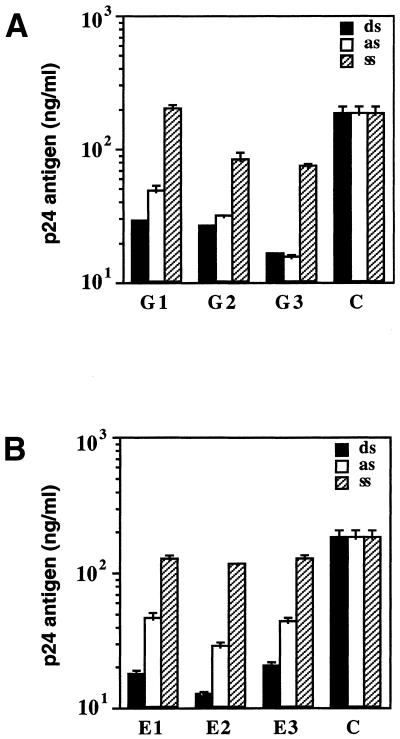
To characterize the relationship between RNAi and asRNAs further, we compared the inhibitory effects of six longer dsRNAs targeted to the HIV-1 gag and env genes with those of the corresponding ssRNAs and asRNAs. A comparison of the effects of the ss-, as- and dsRNAs containing the HIV-1 gag sequences (G1–G3) is shown in Figure 3A. In comparison, there was no dramatic difference in the p24 antigen suppression levels between the dsRNAs and asRNAs (G1–G3), as shown in Figure 3A. The G1-ssRNA was ineffective in reducing the amount of p24 antigen. In addition, the G2- and G3-ssRNAs reduced the amount of p24 antigen by only 2- to 3-fold. Figure 3B shows a comparison of the effects of ss-, as- and dsRNAs containing the HIV-1 env sequences (E1–E3). The three dsRNAs targeted to the HIV-1 env genes (E1–E3) effectively inhibited HIV-1 gene expression more than 2- to 3-fold relative to the asRNAs. In contrast, the three control ssRNAs suppressed the p24 expression by only ∼2-fold. Although the asRNA was able to inhibit HIV-1 gene expression, the results presented in Figure 3 indicate that the dsRNA is a more potent inhibitor than the asRNA. These results strengthen the conclusion that the dsRNA is more effective in targeting HIV-1 genes than the asRNA. The dsRNA acted in a sequence-specific manner and did not affect the expression from the non-complementary control templates.
Figure 3.
Inhibition effects of ssRNAs (ss), asRNAs (as) or dsRNAs (ds) on HIV-1 gene expression in COS cells. COS cells were transfected with pNL4-3 (2 µg) alone or in the presence of different ss-, as- or dsRNAs (2 µg) as indicated, using Lipofectin. After a 72 h incubation, the virus replication was monitored in the culture supernatants with the HIV-1 p24 CLEIA assay (C, positive control cells transfected with pNL4-3 proviral DNA). (A) Comparison of the effect of ss-, as- and dsRNAs containing the HIV-1 gag gene (G1–G3). (B) Comparison of the effect of ss, as and dsRNAs containing the HIV-1 env gene (E1–E3). Bars represent means ± SE of triplicate experiments.
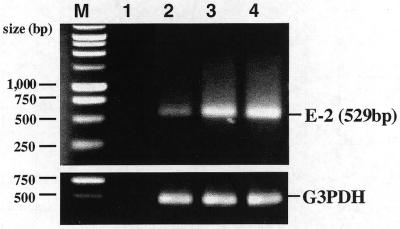
Moreover, we examined the HIV-1 mRNA levels to identify the contribution of the dsRNA-mediated specific RNAi. RT–PCRs were used to establish the level of uncleaved HIV-1 mRNA. Equal amounts of total RNA from COS cells, transfected with E2-dsRNA or ssRNA and pNL4-3 at a weight (µg) ratio of 1:1, were subjected to RT–PCR analysis. HIV-1 env-specific DNA primers (sense, 7072–7101; antisense, 7571–7600) were used to amplify a 529 bp (7072–7600) fragment in the transcripts. The COS cells transfected with the E2-dsRNA showed an 85% reduction in HIV-1 transcripts. However, the E2-ssRNA did not reduce the abundance of HIV-1 mRNA (Fig. 4). The reduction in the functional full-length HIV-1 mRNA is consistent with a dsRNA-mediated interference effect at the post-transcriptional level. The results of these experiments indicate that dsRNA causes a specific reduction in the target mRNA levels.
Figure 4.
RT–PCR analysis of HIV-1 mRNA expression. RT–PCR analysis of uncleaved HIV-1 mRNA was carried out using HIV-1 env-specific primers with concomitant amplification of G3PDH. Total RNA was extracted from COS cells transfected with E2-ss- or dsRNAs first, followed by pNL4-3. The relative amounts of HIV-1 transcripts were determined by RT–PCR, as described in Materials and Methods. PCR primers were used to amplify a 529 bp (7072–7600) fragment in HIV-1 transcripts. Lane 1, negative control (no template added); lane 2, relative amount of transcripts in cells transfected with E2-dsRNA; lane 3, relative amount of transcripts in cells transfected with E2-ssRNA; lane 4, relative amount of transcripts in positive control cells transfected with pNL4-3 proviral DNA. The G3PDH RT–PCR was run in parallel to normalize the levels of mRNA in the samples.
In addition, to clarify the ability of dsRNA to inhibit viral replication in HIV-1-infected cells, we used the HeLa-CD4+ cell line and the E2-dsRNA. HeLa-CD4+ cells were infected with the HIV-1NL4-3 virus at a MOI of 0.5. After a 16 h infection, the cells were washed and co-transfected with E2-ss-, as- or dsRNA at a 2 µg concentration, using Lipofectin. In the cells treated with the E2-dsRNA, the virus replication was inhibited by ∼70-fold, as compared to the untreated control cells (Fig. 5). The long single-stranded asRNA also had highly inhibitory effects, but the ssRNA had virtually no effect. In this infection experiment, we also observed sequence-specific, dsRNA-mediated silencing.
Figure 5.
Effects of ssRNA (ss), asRNA (as) or dsRNA (ds) in HIV-1 infected HeLa-CD4+ cells. Two micrograms of liposomally encapsulated E2-ss-, as- or dsRNAs were transfected in HIV-1NL4-3-infected HeLa-CD4+ cells (MOI 0.5). At 7 days post-infection, p24 antigen production was detected by the HIV-1 p24 CLEIA assay (con, positive control cells infected with HIV-1NL4-3). The plotted data were averaged from three independent experiments and the bars represent ± SD.
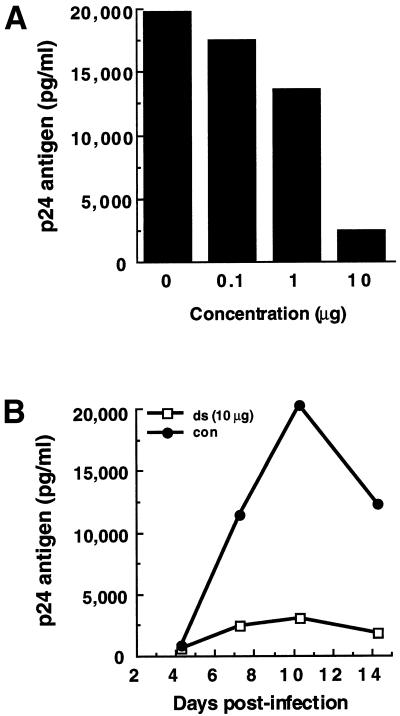
Next, the inhibitory activity of the E2-dsRNA against HIV-1NL4-3 replication in PBMCs was also demonstrated by a p24 accumulation assay of culture supernatants from virus-infected cells. The dose dependence of the dsRNA effect was investigated in PBMCs, using E2-dsRNA. PHA-stimulated PBMCs were infected with the HIV-1NL4-3 virus at a MOI of 0.001, in the presence of various concentrations of the dsRNAs. At 10 days post-infection, the amount of p24 antigen was measured by an HIV-1 p24 CLEIA assay. As expected, the E2-dsRNA (0.1–10 µg) reduced the p24 antigen levels in a dose-dependent manner (Fig. 6A). We next measured the persistence of the dsRNA silencing effect in PBMCs over a 14 day period. PHA-stimulated PBMCs were infected with the HIV-1NL4-3 virus at a MOI of 0.001 in the presence of the E2-dsRNA (10 µg). At 4, 7, 10 and 14 days after infection, culture supernatants containing the viruses were removed and the amount of p24 was measured by an HIV-1 p24 CLEIA assay. As shown in Figure 6B, the E2-dsRNA (10 µg) was also effective against HIV-1NL4-3 replication for a relatively long time (14 days). These findings highlight the general utility of RNAi technology in suppressing HIV-1 expression.
Figure 6.
Effects of E2-dsRNA in HIV-1-infected PBMCs. (A) E2-dsRNA dose-dependent inhibition of p24 antigen production in PBMCs infected with HIV-1NL4-3. PBMCs were freshly isolated from an individual and were stimulated with PHA. Three days later, the PHA-stimulated PBMCs were transfected with E2-dsRNA (0.1–10 µg) encapsulated with TransMessenger, and infected with HIV-1NL4-3 at a MOI of 0.001 after 4 h. At 10 days post-infection, p24 antigen production was detected by the HIV-1 p24 CLEIA assay. (B) Time course of E2-dsRNA inhibition of p24 antigen production by HIV-1NL4-3. PHA-stimulated PBMCs were infected with the HIV-1NL4-3 virus at a MOI of 0.001 in the presence of E2-dsRNA (10 µg). At 4, 7, 10 and 14 days after infection, culture supernatants containing the viruses were removed and the amount of p24 was measured by the HIV-1 p24 CLEIA assay (con, positive control cells infected with HIV-1NL4-3).
To investigate the inhibitory mechanism of dsRNA on the replication cycle of HIV-1, we examined the anti-retroviral effect of a longer E2-dsRNA on ACH2 cells. The ACH2 cell line, a model for chronic HIV-1 infection, possesses a single integrated copy of the HIV-1 strain LAI and can be induced to produce virus by a variety of stimuli (34). ACH2 cells were transfected with E2-ss-, as- or dsRNAs and were stimulated with 10 nM (final concentration in the cell culture medium) tetradecanoyl phorbol acetate (TPA). In this induction system, the E2-dsRNA was less effective in inhibiting virus replication than in the transient system described above (data not shown). These observations suggest that dsRNA may effectively prevent early steps prior to integration, rather than later post-integration steps, in the HIV-1 infection cycle. RNAi is likely to reflect natural defenses against viruses, as a self-regulating mechanism of gene expression (15,35,36). However, the mechanisms of the RNAi process in mammalian cells remain unresolved.
In this report, we have shown that dsRNA can be successfully used as a new anti-HIV-1 agent in HIV-1-infected cells. In the case of COS cells co-transfected with both the HIV-1 pNL4-3 proviral DNA and dsRNA, the dsRNA was a more potent inhibitor than the single-stranded asRNA in promoting the inhibition of HIV-1 replication. We also demonstrated that dsRNA significantly suppressed HIV-1 infection in HeLa-CD4+ cells and PBMCs for a relatively long period of time. Thus, we suggest that this longer dsRNA-mediated specific interference in HIV-1-infected cells may provide a new strategy for anti-HIV-1 gene therapy.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by a Grant-in-Aid for High Technology Research from the Ministry of Education, Science, Sports, and Culture, Japan; a Grant from the Japan Society for the Promotion of Science in the ‘Research for the Future’ program (JSPS-RFTF97L00593), and a Research Grant from the Human Science Foundation (HIV-K-1031).
REFERENCES
- 1.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z., Morris,J.C., Drew,M.E. and Englund,P.T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem., 275, 40174–40179. [DOI] [PubMed] [Google Scholar]
- 3.Ngo H., Tschudi,C., Gull,K. and Ullu,E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassenegger M. and Pelissier,T. (1998) A model for RNA-mediated gene silencing in higher plants. Plant Mol. Biol., 37, 349–362. [DOI] [PubMed] [Google Scholar]
- 5.Misquitta L. and Paterson,B.M. (1999) Targeted disruption of gene function in Drosophila by RNA interference (RNAi): a role for nautilus in embryonic somatic muscle formation. Proc. Natl Acad. Sci. USA, 96, 1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caplen N.J., Fleenor,J., Fire,A. and Morgan,R.A. (2000) dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene, 252, 95–105. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez Alvarado A. and Newmark,P.A. (1999) Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl Acad. Sci. USA, 96, 5049–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y.X., Farrell,M.J., Liu,R., Mohanty,N. and Kirby,M.L. (2000) Double-stranded RNA injection produces null phenotypes in zebrafish. Dev. Biol., 217, 394–405. [DOI] [PubMed] [Google Scholar]
- 9.Svoboda P., Stein,P., Hayashi,H. and Schultz,R.M. (2000) Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development, 127, 4147–4156. [DOI] [PubMed] [Google Scholar]
- 10.Wianny F. and Zernicka-Goetz,M. (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nature Cell Biol., 2, 70–75. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- 12.Zamore P.D., Tuschi,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 13.Yang D., Lu,H. and Erickson,J.W. (2000) Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol., 10, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 15.Ketting R.F., Fischer,S.E.J., Bernstein,E., Sijen,T., Hannon,G.J. and Plasterk,R.H.A. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev., 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nykanen A., Haley,B. and Zamore,P.D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed] [Google Scholar]
- 19.Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- 20.Gil J. and Esteban,M. (2000) Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis, 5, 107–114. [DOI] [PubMed] [Google Scholar]
- 21.Player M.R. and Torrence,P.F. (1998) The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol. Ther., 78, 55–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 23.Caplen N.J., Parrish,S., Imani,F., Fire,A. and Morgan,R.A. (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA, 98, 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holen T., Amarzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee N.S., Dohjima,T., Bauer,G., Li,H., Li,M.-J., Ehsani,A., Salvaterra,P. and Rossi,J. (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol., 20, 500–505. [DOI] [PubMed] [Google Scholar]
- 26.Dorman N. and Lever,A.M. (2001) RNA-based gene therapy for HIV infection. HIV Med., 2, 114–122. [DOI] [PubMed] [Google Scholar]
- 27.Michienzi A., Conti,L., Varano,B., Prislei,S., Gessani,S. and Bozzoni,I. (1998) Inhibition of human immunodeficiency virus type 1 replication by nuclear chimeric anti-HIV ribozymes in a human T lymphoblastoid cell line. Hum. Gene Ther., 9, 621–628. [DOI] [PubMed] [Google Scholar]
- 28.Veres G., Junker,U., Baker,J., Barske,C., Kalfoglou,C., Ilves,H., Escaich,S., Kaneshima,H. and Bohnlein,E. (1998) Comparative analyses of intracellularly expressed antisense RNAs as inhibitors of human immunodeficiency virus type 1 replication. J. Virol., 72, 1894–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse P.M., Graham,M.W. and Wang,M.-B. (1998) Virus resistance and gene silencing in plants can be induced by stimultaneous expression of sense and antisense RNA. Proc. Natl Acad. Sci. USA, 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Billy E., Brondani,V., Zhang,H., Muller,U. and Filipowicz,W. (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA, 98, 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakashima H., Shoji,Y., Kim,S.G., Shimada,J., Mizushima,T., Ito,M., Yamamoto,N. and Takaku,H. (1994) Anti-human immunodeficiency virus type 1 activity of phosphorothioate analogs of oligodeoxynucleotides: penetration and localization of oligodeoxynucleotides in HIV-1-infected MOLT-4 cells. Nucleic Acids Res., 22, 5004–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuwasaki T., Hosono,K., Takai,K., Ushijima,K., Nakashima,H., Saito,T., Yamamoto,N. and Takaku,H. (1996) Hairpin antisense oligonucleotides containing 2′-methoxynucleosides with base-pairing in the stem region at the 3′-end: penetration and localization and anti-HIV activity. Biochem. Biophys. Res. Commun., 228, 623–631. [DOI] [PubMed] [Google Scholar]
- 33.Sakai A., Hirabayasi,Y., Aizawa,S., Tanaka,M., Ida,S. and Oka,S. (1999) Investigation of a new p24 antigen detection system by the chemiluminescence-enzyme-immunoassay. J. Japn. Assoc. Infect. Dis., 73, 205–212. [DOI] [PubMed] [Google Scholar]
- 34.Kim J.H., Mosca,J.D., Vahey,M.T., McLinden,R.J., Burke,D.S. and Redfield,R.R. (1993) Consequences of human immunodeficiency virus type 1 superinfection of chronically infected cells. AIDS Res. Hum. Retroviruses, 9, 875–882. [DOI] [PubMed] [Google Scholar]
- 35.Voinnet O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet., 17, 449–459. [DOI] [PubMed] [Google Scholar]
- 36.Hutvagner G., McLachlan,J., Pasquinelli,A.E., Balint,E., Tuschl,T. and Zamore,P.D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]