Abstract
Nitrosation of guanine in DNA by nitrogen oxides such as nitric oxide (NO) and nitrous acid leads to formation of xanthine (Xan) and oxanine (Oxa), potentially cytotoxic and mutagenic lesions. In the present study, we have examined the repair capacity of DNA N-glycosylases from Escherichia coli for Xan and Oxa. The nicking assay with the defined substrates containing Xan and Oxa revealed that AlkA [in combination with endonuclease (Endo) IV] and Endo VIII recognized Xan in the tested enzymes. The activity (Vmax/Km) of AlkA for Xan was 5-fold lower than that for 7-methylguanine, and that of Endo VIII was 50-fold lower than that for thymine glycol. The activity of AlkA and Endo VIII for Xan was further substantiated by the release of [3H]Xan from the substrate. The treatment of E.coli with N-methyl-N′-nitro-N-nitrosoguanidine increased the Xan-excising activity in the cell extract from alkA+ but not alkA– strains. The alkA and nei (the Endo VIII gene) double mutant, but not the single mutants, exhibited increased sensitivity to nitrous acid relative to the wild type strain. AlkA and Endo VIII also exhibited excision activity for Oxa, but the activity was much lower than that for Xan.
INTRODUCTION
DNA that stores genetic information of cells shows certain structural instability. The DNA bases bearing an exocyclic amino group [adenine (A), guanine (G), cytosine (C)] undergo deamination. The rate of hydrolytic deamination is the highest for C under physiological conditions (1), and it is estimated that approximately 100 C residues are deaminated per human cell per day (2). If unrepaired the resulting uracil (U) induces CG→TA transitions (3). Deamination of A gives rise to hypoxanthine (Hx), which pairs with C and induces AT→GC transitions (4). Since U and Hx are mutagenic, organisms are equipped with repair mechanisms for these lesions. Uracil in DNA is removed by uracil-DNA glycosylase, which is highly conserved in prokaryotic and eukaryotic organisms (5). Hypoxanthine is generally excised from DNA by a family of methyl purine DNA glycosylases (6) and Escherichia coli has an extra enzyme for Hx, namely endonuclease (Endo) V, which incises the second phosphodiester bond on the 3′ side of Hx (7).
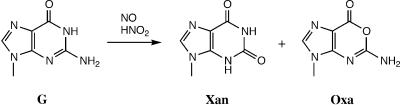
Guanine undergoes spontaneous hydrolytic deamination to yield xanthine (Xan), but the spontaneous deamination rate of G is lower than that of C and A (8). Therefore, so far as spontaneous deamination is concerned, deamination of G is biologically less important than that of C or A. However, nitrogen oxides such as nitric oxide (NO) and nitrous acid (HNO2) induce deamination of DNA bases with significant rates. In contrast to spontaneous hydrolytic deamination, G is the most sensitive of the three bases to nitric oxide and nitrous acid. NO has been characterized primarily as a second messenger exerting various physiological activities (9). In humans, 1 mmol of NO is constitutively generated per body per day and the amount increases by 10–100 times upon bacterial infection or inflammation. NO overproduced by activated macrophages in chronically inflamed tissues has been implicated as carcinogenic by virtue of its ability to cause DNA damage (10,11). Nitrous anhydride (N2O3), formed by autoxidation of NO, is a powerful nitrosating agent, and induces deamination of C, A and G in nucleosides and DNA, generating U, Hx and Xan, respectively (12). Nitrous acid that is formed by protonation of sodium nitrite (NaNO2), present in food intake, also induces base deamination via N2O3. Although nitrosation of C and A leads exclusively to U and Hx, respectively, that of G gives rise to not only Xan but also oxanine (Oxa) with a molar ratio of 3 (Xan):1 (Oxa) (Fig. 1) (13). Recently, it has been shown that Xan and Oxa are formed by nitroso group transfer to G from N-nitrosoindoles (14). Xan and Oxa are produced via the common precursor (a diazoate derivative of the 2-amino group of G), but formation of Oxa involves deamination of the 2-amino group followed by rearrangement of the ring atoms (15). The biochemical and genotoxic effects of Xan and Oxa have been assessed previously by several studies. Substitution of Xan or Oxa for G in duplex DNA results in large decreases in the helix stability (16). Xan in template DNA directs incorporation of T as well as C during DNA replication (17,18). Similarly, 2′-deoxyribonucleoside 5′-triphosphates of Xan (dXTP) and Oxa (dOTP) are incorporated opposite template T as well as C by DNA polymerase, though their incorporation efficiencies are much lower than dGTP (19). In addition, Xan but not Oxa can be spontaneously converted to a cytotoxic and mutagenic AP (apurinic/apyrimidinic) site due to its labile N-glycosidic bond (16). Thus, Xan may show additional biological effects after conversion to an AP site. Recently, Oxa has been shown to form a covalently bound adduct with glycine, suggesting a novel cytotoxic/genotoxic mechanism for this lesion (20). These data combined together indicate the necessity of repair capacities for Xan and Oxa in cells. However, studies on the enzymatic repair of these lesions are extremely limited (21).
Figure 1.
Products formed by the reaction of guanine (G) with nitrogen oxides (NO and HNO2).
In the present work, we have examined the repair capacity of E.coli base excision repair enzymes for Xan and Oxa using defined oligonucleotide substrates containing these lesions. We report here that, among the enzymes tested, AlkA and Endo VIII exhibit repair activities for Xan and Oxa in a paired base-dependent manner and the repair efficiency for Oxa is much lower than for Xan. Induction of the alkA gene encoding AlkA protein in E.coli cells results in an increased releasing activity of Xan. Furthermore, E.coli deficient in both AlkA and Endo VIII, but not either of them alone, exhibits increased sensitivity to nitrous acid.
MATERIALS AND METHODS
Strains, enzymes and chemicals
Escherichia coli MV1161 (thr1, ara14, leuB6, Δ(gpt-proA)62, lacY1, tsx33, supE44, galK2, hisG4, rfbD1, mgl51, rpsL31, kdgK51, xyl5, mtl1, argE3, thi1, rfa550) and MV1571 [alkA51::Mud1 (AmpRlac) in MV1161] were laboratory stocks (22). To construct Endo VIII mutants, MV1161 and MV1571 were infected with P1 phage carrying the Δnei::KmR allele derived from NKJ1003 (23) and were selected for the kanamycin (Km)-resistant phenotype. The resultant nei and nei alkA phenotypes were confirmed by measuring Endo VIII and Endo VIII/AlkA activities and the strains were designated KY100 (MV1161 + nei) and KY101 (MV1571 + nei), respectively. Endo III, Endo VIII, Endo IV, formamidopyrimidine DNA glycosylase (Fpg/MutM) and AlkA were purified from E.coli strains that overproduced these enzymes (24,25). Escherichia coli uracil DNA glycosylase (Ung) and exonuclease (Exo) III were purchased from New England Biolabs. Escherichia coli DNA polymerase I Klenow fragment (Pol I Kf) and T4 polynucleotide kinase were obtained from Life Technologies. 2′-Deoxycytidine 5′-triphosphate (dCTP), thymidine 5′-triphosphate (dTTP) and [γ-32P]adeno sine 5′-triphosphate (ATP) (110 TBq/mmol) were from Amersham Biosciences. 2′-[8-3H]Deoxyguanosine 5′-triphosphate (dGTP) (37 MBq/244 pmol) was purchased from NEN Life Science Products. Guanine and Xan were purchased from Sigma.
Oligonucleotides
Oligonucleotides without modified bases were purchased from Espec Oligo Service and purified by reversed phase HPLC. Preparation of 25MG and 19TG which contain a single 7-methylguanine (7mG) and thymine glycol (Tg), respectively, were reported previously (26,27). The sequences of the oligonucleotides used in this study are shown in Table 1.
Table 1. List of oligonucleotides used in this study.
| Abbreviation | Sequencea |
|---|---|
| 19T | 5′-ACAGACGCCATCAACCAGG |
| 19TG | 5′-ACAGACGCCATgCAACCAGG |
| COM19A | 3′-TGTCTGCGGTAGTTGGTCC |
| PRIM15 | 5′-CATCGATAGCATCCT |
| 25G | 5′-CATCGATAGCATCCTGCCTTCTCTC |
| 25MG | 5′-CATCGATAGCATCCTMCCTTCTCTC |
| 25XAN | 5′-CATCGATAGCATCCTXCCTTCTCTC |
| 25OXA | 5′-CATCGATAGCATCCTOCCTTCTCTC |
| COM25A | 3′-GTAGCTATCGTAGGAAGGAAGAGAG |
| COM25T | 3′-GTAGCTATCGTAGGATGGAAGAGAG |
| COM25G | 3′-GTAGCTATCGTAGGAGGGAAGAGAG |
| COM25C | 3′-GTAGCTATCGTAGGACGGAAGAGAG |
| COM30C | 3′-AAGTCGTAGCTATCGTAGGACGGAAGAGAG |
aTg, thymine glycol; M, 7-methylguanine; X, xanthine; O, oxanine. Base lesions and paired bases are underlined.
Preparation of dXTP and dOTP and their incorporation into oligonucleotides
2′-Deoxyxanthosine 5′-triphosphate (dXTP) and 2′-deoxyoxanosine 5′-triphosphate (dOTP) were synthesized from dGTP. dGTP (1 mM) was incubated with 100 mM NaNO2 at 37°C in 3 M acetate buffer (pH 3.7) for 2 h, and the resulting dXTP and dOTP were purified by reversed phase HPLC. The detailed procedures and characterization of dXTP and dOTP were reported previously (19). [3H]dXTP was prepared from [3H]dGTP in an essentially similar manner. [3H]dGTP (37 MBq/244 nmol, diluted with cold dGTP) was incubated with 100 mM NaNO2 in 1 M acetate buffer (pH 3.7, 100 µl) at 37°C for 2 h. [3H]dXTP was purified by reversed phase HPLC. Oligonucleotide substrates containing Xan and Oxa were prepared by DNA polymerase reactions. PRIM15 was 5′-end labeled using [γ-32P]ATP and T4 polynucleotide kinase and purified by a Sep-pak cartridge (Waters) (28). PRIM15 (50 pmol) and COM25C (100 pmol, 2-fold molar excess) in 10 mM Tris–HCl (pH 7.5) and 25 mM NaCl were heated at 90°C for 10 min and then annealed at room temperature. PRIM15/COM25C (25 pmol) in the polymerase buffer (200 µl) were incubated with dXTP or dOTP (both 20 nmol) and Pol I Kf (25 U) at 25°C for 5 min to introduce a single residue of Xan or Oxa opposite C (16th position from the 3′ terminus in COM25C). The polymerase buffer consisted of 66 mM Tris–HCl (pH 7.0), 1.5 mM 2-mercaptoethanol and 6.6 mM MgCl2. Then dCTP and dTTP (10 nmol each) were added to the reaction mixture to extend the primer fully. The oligonucleotides containing Xan and Oxa were designated 25XAN and 25OXA, respectively. The duplexes (25XAN/COM25C and 25OXA/COM25C) were purified by phenol extraction, ethanol precipitation and gel filtration on a Sephadex G-25 column (1 ml). The site-specific introduction of Xan and Oxa into 25XAN and 25OXA, respectively, was confirmed by the heat/acid and ammonia/piperidine treatments (see Results). For preparation of oligonucleotide substrates containing Xan:N and Oxa:N pairs (N = A, G, C, T), the DNA polymerase reaction was performed in a similar manner except that COM30C was used as a template in place of COM25C. This facilitated the subsequent electrophoretic separation of 25XAN and 25OXA from the template (COM30C). After the reaction, 25XAN and 25OXA were purified by 16% denaturing polyacrylamide gel electrophoresis (PAGE), extracted from the gel and desalted by a Sep-pak cartridge. Purified 25XAN and 25OXA were annealed with COM25N (N = A, G, C, T). 25XAN containing [3H]Xan was prepared as described above using PRIM15/COM25C, [3H]dXTP and Pol I Kf for the initial polymerization reaction. 25G containing [3H]G in place of [3H]Xan was also prepared using [3H]dGTP.
Chemical cleavage reactions of oligonucleotides
25XAN, 25OXA and 25G (all 32P-labeled at the 5′ end, 100 fmol) were heated at 90°C for 30 min under acidic conditions (pH 3.5, adjusted by acetic acid) and the solution was evaporated to dryness. Alternatively, the oligonucleotides were treated first by 30% ammonia at 65°C for 4 h. After removing ammonia by evaporation, the samples were treated by 10% piperidine at 90°C for 30 min and evaporated to dryness. The products from both treatments were mixed with gel loading buffer [95% (v/v) formamide, 20 mM EDTA, 0.1% (w/v) xylene cyanol and 0.1% (w/v) bromophenol blue] and analyzed by 16% denaturing PAGE.
Nicking assays for the activity to Xan and Oxa
In the reaction with Fpg, Endo III and Endo VIII (all N-glycosylase/AP lyase), 25G/COM25C, 25XAN/COM25C and 25OXA/COM25C (100 fmol) were incubated with the enzyme (600 fmol) in buffer A (10 µl) at 37°C for 30 min. Buffer A comprised 10 mM Tris–HCl (pH 7.5), 1 mM EDTA and 100 mM NaCl. The reactions with Endo IV (120 fmol) and Exo III (0.1 U) (both AP endonucleases) were similarly carried out in buffer B (10 µl) and C (10 µl), respectively. The composition of buffer B was 10 mM Tris–HCl (pH 7.5), 1 mM EDTA and 50 mM NaCl and that of buffer C was 10 mM Tris–HCl (pH 7.5), 2 mM CaCl2 and 1 mM EDTA. In the reaction with simple DNA glycosylases, the substrates (100 fmol) were incubated with AlkA (600 fmol) and Ung (170 µU) in buffer D (10 µl) and buffer E (10 µl), respectively, at 37°C for 30 min. The composition of buffer D was 50 mM HEPES-KOH (pH 7.5), 1 mM EDTA and 5 mM 2-mercaptoethanol and that of buffer E was 20 mM Tris–HCl (pH 8.0), 1 mM EDTA and 1 mM dithiothreitol. After incubation, the substrates were purified by phenol extraction and ethanol precipitation. The purified substrates were incubated further with Endo IV (120 fmol) at 37°C for 30 min to incise abasic sites. The sample was mixed with gel loading buffer and separated by 16% denaturing PAGE. The radioactivity of products was analyzed on a BAS2000 phosphorimaging analyzer (Fuji Film). Paired base effects on the activity of AlkA and Endo VIII for Xan and Oxa were measured using 25XAN/COM25N and 25OXA/COM25N (N = A, G, C or T) as substrates. The reactions were performed in a manner essentially similar to those described above. The amounts of the substrates (25XAN and 25OXA) and the enzymes (AlkA and Endo VIII) were 100 fmol and 300 fmol–3 pmol, respectively.
For analysis of the kinetic parameters of AlkA, 25XAN/COM25C and 25MG/COM25C (50–1000 fmol) were incubated with 300 fmol of AlkA at 37°C for 10 min in buffer D (10 µl), and then the products were treated with Endo IV as described above. For Endo VIII, 25XAN/COM25C and 19TG/COM19A (50–1000 fmol) were incubated with Endo VIII (300 fmol) at 37°C for 15 min (25XAN/COM25C) and 2 min (19TG) in buffer A (10 µl). Reaction products were separated and quantified as described above. The parameters Vmax and Km were evaluated from Michaelis–Menten plots using a hyperbolic curve fitting program.
Release assays of Xan with AlkA and Endo VIII
To measure the N-glycosylase activity of AlkA, 25XAN/COM25C and 25G/COM25C (both 2.25 pmol) containing [3H]Xan and [3H]G, respectively, were incubated with AlkA (3 pmol) at 37°C for 30 min in buffer D (20 µl). The reaction with Endo VIII (6 pmol) was performed in a similar manner using buffer A. After the reaction, the sample was loaded onto a Sephadex G-25 fine gel filtration column (φ 3 × 300 mm) to separate the released product and the oligonucleotides. The column was eluted by water and the eluent was collected every 100 µl. Aliquots of the collected fractions were subjected to liquid scintillation counting. The oligonucleotides and the released product were eluted in fractions 12–15 and 20–25, respectively, under these conditions. The pooled fractions containing the released product were evaporated to dryness and resuspended in a small volume of water. The sample was analyzed by reversed phase HPLC. The HPLC system consisted of Hitachi L-6200 pumps equipped with a reversed phase WS-DNA column (φ 4.6 × 150 mm, Wako) and a Hitachi L-4200H UV-VIS detector. Elution was carried out with a linear gradient of acetonitrile (0–20%, v/v) in 20 mM sodium phosphate buffer (pH 5.0) at a flow rate 0.8 ml/min. The column eluent was collected every 20 s and was subjected to liquid scintillation counting.
Release assays of Xan with crude cell extracts
Escherichia coli MV1161 (wild type) and MV1571 (alkA) were cultivated in LB media (250 ml) in the absence (MV1161) and presence (MV1571) of 50 µg/ml ampicillin at 37°C for 3 h under aeration. Depending on the experiments, N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (final concentration 20 µM) was added into the culture media and incubation was continued for further 1.5 h. After recovering cells by centrifugation, the cell pellet was suspended in 10 ml of 300 mM Tris–HCl (pH 8.0), 5 mM EDTA and 1 mg/ml lysozyme, and was lyzed by repeated freeze/thaw cycles. The lyzed cells were centrifuged at 43 000 g for 1 h at 4°C and the supernatant was recovered. The proteins in the supernatant were collected by ammonium sulfate precipitation (60% saturation) and were resuspended in 10 mM Tris–HCl (pH 7.5) and 1 mM EDTA (TE buffer). After dialysis against TE buffer at 4°C, the cell extract was used for the release assay of [3H]Xan. The protein concentration was determined by a BCA protein assay reagent (Pierce) using BSA as a standard. The release assay was performed as described for AlkA using 5 µg of protein.
Nitrous acid sensitivity of alkA and nei E.coli strains
A modified procedure by Miller (29) was used. Escherichia coli strains, MV1161 (wild type), MV1571 (alkA), KY100 (nei) and KY101 (alkA nei) were cultured overnight in 1 ml of LB media with 50 µg/ml of appropriate antibiotics (ampicillin for MV1571, kanamycin for KY100, and ampicillin and kanamycin for KY101) at 37°C. These cultures were diluted into 100 ml of LB media with or without antibiotics and cells were grown to a logarithmic phase (OD600 = 0.6) at 37°C. After washing in acetate buffer (0.1 M, pH 4.6), cells were incubated in NaNO2 (0–30 mM) in 5 ml of acetate buffer (0.1 M, pH 4.6) at 37°C for 10 min. To stop incubation, an equal volume of minimal A medium [60.3 mM K2HPO4, 33.1 mM KH2PO4, 7.6 mM (NH4)2SO4 and 1.7 mM trisodium citrate] was added. After washing in minimal A medium twice, the cell suspension was appropriately diluted and incubated overnight on LB plates at 37°C. The colonies were scored for surviving fractions. It is noted that the viability of all the test strains decreased to ∼10% after incubation with acetate buffer (0.1 M, pH 4.6) alone. Thus, variations in viability depending on the strain were virtually negligible in the incubation with acetate buffer alone.
RESULTS
Site-specific introduction of Xan and Oxa into oligonucleotides
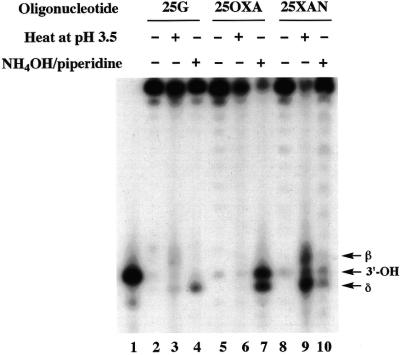
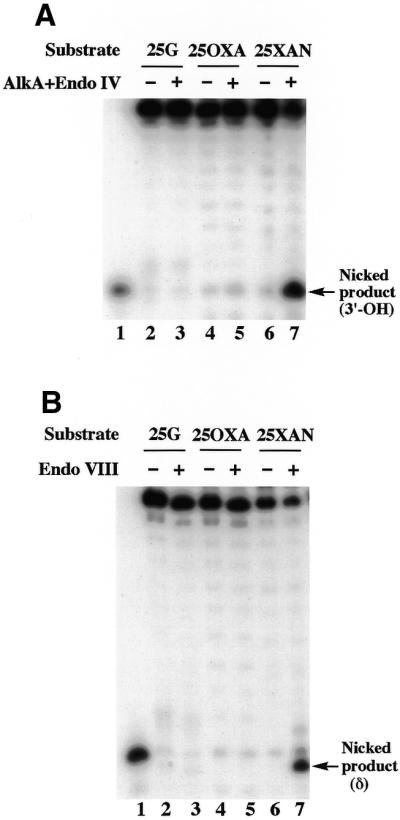
Xan and Oxa were site-specifically incorporated into the oligonucleotide substrates (25XAN and 25OXA, Table 1) by the DNA polymerase reaction using dXTP and dOTP as substrates. The N-glycosidic bond of Xan is fairly labile so that heat/acid treatment causes depurination, generating an AP site. On the other hand, the N-glycosidic bond of Oxa is as stable as that of G to the heat/acid treatment. However, Oxa contains an O-acylisourea structure in the six-membered ring (1,3-oxazine ring) that undergoes a ring-opening reaction at alkaline pH (pKa = 9.4) (16). An extensive treatment of the ring-opened structure of Oxa induces decomposition of the Oxa ring, though the final decomposition product has not been identified yet. Knowing the chemistry of Xan and Oxa, 25XAN and 25OXA prepared by the DNA polymerase reaction were treated by heat/acid or ammonia/piperidine as described in Materials and Methods. PAGE analysis of the treated oligonucleotides showed that 25XAN was specifically cleaved by the heat/acid treatment (Fig. 2, lane 9) but not the ammonia/piperidine treatment (Fig. 2, lane 10). According to the gel mobility of the products, the cleaved products were 15mers with different 3′ terminal modifications resulting from breakdown of an AP site (a mixture of β-elimination, δ-elimination and 3′-OH products). Conversely, 25OXA was stable in the heat/acid treatment (Fig. 2, lane 6) but was specifically cleaved by the ammonia/piperidine treatment (Fig. 2, lane 7). The products were 15mers bearing 3′-phosphate (δ-elimination product) and 3′-OH groups. 25G was virtually intact in both treatments (Fig. 2, lanes 3 and 4). These results indicate that Xan and Oxa were site-specifically introduced into 25XAN and 25OXA, respectively, by the DNA polymerase reaction.
Figure 2.
Chemical cleavage of oligonucleotides containing G (25G), Xan (25XAN) and Oxa (25OXA). 5′-32P-labeled 25G, 25XAN and 25OXA were heated at 90°C for 30 min in an acidic aqueous solution (pH 3.5, adjusted by acetic acid) (lanes 3, 6 and 9). Alternatively they were incubated first in 30% (v/v) ammonia at 65°C for 4 h, and then in 10% (v/v) piperidine at 90°C for 30 min as described in Materials and Methods (lanes 4, 7 and 10). The samples were separated by 16% denaturing PAGE. Lane 1 shows a 5′-32P-labeled marker (PRIM15). The bands of β-elimination, δ-elimination and 3′-OH products are indicated by arrows.
Repair activities of AlkA and Endo VIII for Xan and Oxa
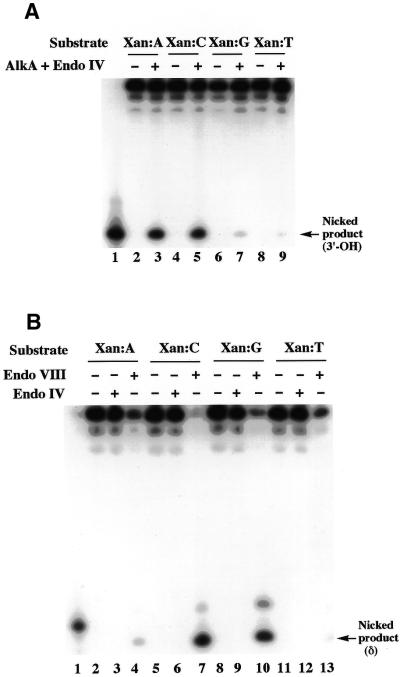
The repair activities of various E.coli DNA repair enzymes for Xan and Oxa were examined using the nicking assay with 25XAN/COM25C and 25OXA/COM25C as substrates. The enzymes tested included three bifunctional glycosylases containing N-glycosylase/AP lyase activities (Fpg, Endo III and Endo VIII), two monofunctional N-glycosylases (AlkA and Ung) and two AP endonucleases (Endo IV and Exo III). Among the enzymes tested, AlkA and Endo VIII recognized Xan. The results of PAGE analysis of the reaction products of AlkA and Endo VIII are shown in Figure 3. The treatment of 25XAN by AlkA (followed by Endo IV) resulted in a clear band of a 3′-OH product arising from cleavage at the introduced Xan site (Fig. 3A, lane 7). Similarly, the treatment with Endo VIII resulted in a δ-elimination product (Fig. 3B, lane 7). The observed incision products were consistent with the known catalytic modes of AlkA (+ Endo IV) and Endo VIII. Although incision products of 25OXA by AlkA (+ Endo IV) and Endo VIII were barely detected with the same amount (600 fmol) of the enzymes as used for 25XAN (Fig. 3A and B, lane 5), treatment with an excessive amount of AlkA and Endo VIII (both 3 pmol) revealed their activity for Oxa (see Paired base effects on the repair activity for Xan and Oxa).
Figure 3.
Reaction products formed in the treatment of substrates containing Xan (25XAN) and Oxa (25OXA) by AlkA and Endo VIII. (A) AlkA reactions. 100 fmol of 25G/COM25C, 25XAN/COM25C and 25OXA/COM25C (25G, 25XAN and 25OXA were 5′-32P labeled) were incubated with 600 fmol of AlkA at 37°C for 30 min. After incubation, the sample was extracted with phenol and DNA was recovered by ethanol precipitation. The sample was treated further with 120 fmol of Endo IV at 37°C for 30 min. (B) Endo VIII reactions. 100 fmol of 25G/COM25C, 25XAN/COM25C and 25OXA/COM25C were incubated with 600 fmol of Endo VIII at 37°C for 30 min. In panels (A) and (B), the sample was separated by 16% denaturing PAGE. The substrates and enzymes used are indicated on the top of the gels. Lane 1 shows a 5′-32P-labeled marker (PRIM15).
Since Xan appeared to be a fair substrate of AlkA and Endo VIII, their kinetic parameters for Xan were determined from Michaelis–Menten plots and the results were compared with those for 7mG (AlkA) and Tg (Endo VIII). The Km values of AlkA for 7mG and Xan were 31 and 53 nM, respectively (Table 2). The Vmax values were 2.1 and 0.72 nM/min for 7mG and Xan, respectively. These results indicated that AlkA had almost comparable affinities (Km) for 7mG and Xan, but the catalytic rate for Xan was somewhat lower than for 7mG. The difference in the reaction efficiencies (Vmax/Km) between 7mG and Xan was only 5-fold. Considering that 7mG is a physiological substrate of AlkA (30) and the relatively small difference in the reaction efficiencies, Xan is a fairly good substrate for AlkA. In contrast, Endo VIII showed a significantly lower reaction efficiency for Xan than for Tg, a physiological substrate of Endo VIII (31–33). The difference in the Vmax/Km values was ∼50-fold. In terms of affinity (Km), Xan and Tg differed by 2.6-fold (124 versus 47 nM) and of catalytic rate (Vmax) by 19-fold (0.94 versus 18 nM/min). Thus, Xan is a relatively poor substrate among those recognized by Endo VIII.
Table 2. Kinetic parameters of AlkA and Endo VIII for Xana.
| Enzyme | Substrate | Km(nM) | Vmax(nM/min) | Vmax/Kmb (min–1) |
|---|---|---|---|---|
| AlkA | Xanthine | 53 ± 24 | 0.72 ± 0.04 | 1.4 × 10–2 (1) |
| 7-Methylguanine | 31 ± 2 | 2.1 ± 0.4 | 6.8 × 10–2 (4.9) | |
| Endo VIII | Xanthine | 124 ± 33 | 0.96 ± 0.26 | 7.6 × 10–3 (1) |
| Thymine glycol | 47 ± 10 | 18 ± 5 | 3.8 × 10–1 (50) |
aThe values are derived from three independent experiments and standard deviations are also indicated.
bThe values in parentheses indicate relative values of Vmax/Km.
N-glycosylase activity of AlkA and Endo VIII for Xan
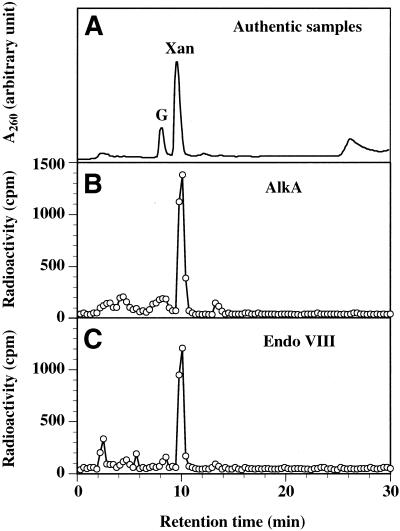
To confirm the activity of AlkA and Endo VIII for Xan further, the N-glycosylase activity of both enzymes was examined by the release assay using 25XAN/COM25C containing [3H]Xan. After enzymatic treatment, the released product was separated from the substrate by gel filtration. Analysis of the radioactivity of the fraction from the gel filtration column showed that there was significant release of the tritium count when 25XAN/COM25C was incubated with AlkA and Endo VIII. However, the released tritium count was negligible when control 25G/COM25C containing [3H]G instead of [3H]Xan was incubated with AlkA and Endo VIII. This was also the case when 25XAN/COM25C was incubated with the reaction buffer alone. The pooled gel filtration fractions containing the released product were further analyzed by reversed phase HPLC. In the HPLC analysis of the product released by AlkA and Endo VIII, the major peak of the tritium count was eluted at 10.4 min (Fig. 4B and C) where authentic Xan (unlabeled) was eluted (Fig. 4A). Authentic G was eluted at 8.7 min under these conditions. Accordingly, AlkA and Endo VIII acted as N-glycosylases for 25XAN/COM25C, releasing a free base of Xan from the substrate.
Figure 4.
N-glycosylase activity assays of AlkA and Endo VIII for Xan. (A) HPLC separation of authentic guanine (G) and Xan. Analysis was performed as described in Materials and Methods. (B) HPLC analysis of [3H]Xan released by AlkA. 2.25 pmol of 25XAN/COM25C containing [3H]Xan was incubated with 3 pmol of AlkA at 37°C for 30 min. The released 3H-labeled material was separated from DNA by a Sephadex G-25 column. The column fractions containing the released 3H-labeled material were pooled and evaporated. The sample was resuspended in a small volume of water and was subjected to HPLC analysis. HPLC analysis was performed as described in panel (A). (C) HPLC analysis of [3H]Xan released by Endo VIII. The experiment was performed in a similar manner using 6 pmol of Endo VIII.
Repair capacity of E.coli cell extracts for Xan
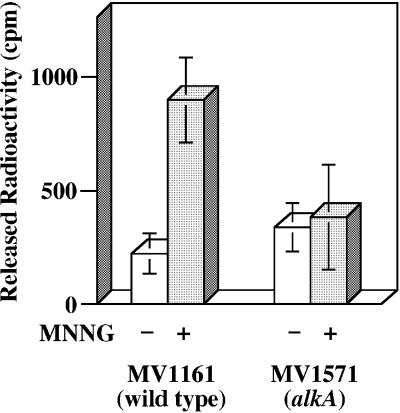
The repair capacity of the E.coli cell extracts for Xan was measured by the release assay. 25XAN/COM25C containing [3H]Xan was incubated with the cell extracts from E.coli MV1161 (wild type) and MV1571 (alkA), and the released tritium count was measured after separation on a gel filtration column. However, no significant difference in released radioactivity was observed between wild type and alkA mutant cells (Fig. 5). It is possible that the basal level of AlkA protein in the wild type cell was so low that the activity difference between the wild type and alkA mutant cells could be within an experimental fluctuation. The alkA gene is inducible by alkylating agents (34). Therefore, MV1161 and MV1571 cells were treated with MNNG, and the cell extracts from the two strains were subjected to the release assay. After MNNG treatment, the wild type cell extract showed a 4-fold increase in Xan-releasing activity in comparison with the cell extract without the MNNG treatment (Fig. 5). In contrast, such induction was not observed with the alkA mutant treated with MNNG (Fig. 5). The results obtained with MNNG-treated cells were consistent with the data obtained with purified AlkA.
Figure 5.
Xan-releasing activity of the extracts from wild type and alkA mutant E.coli cells. The cell extracts were prepared from E.coli MV1161 (wild type) and MV1571 (alkA) treated without and with MNNG, respectively, as described in Materials and Methods. 25XAN/COM25C containing [3H]Xan (2.25 pmol) was incubated with 5 µg of the cell extracts at 37°C for 30 min. After incubation, the sample was loaded on to a Sephadex G-25 column and eluted with water. The amount of [3H]Xan released was counted on a liquid scintillation counter. Open columns, cells without MNNG treatment; closed columns, cells with MNNG treatment. The data are averages of three independent experiments. Standard deviations are indicated with error bars.
Paired base effects on the repair activity for Xan and Oxa
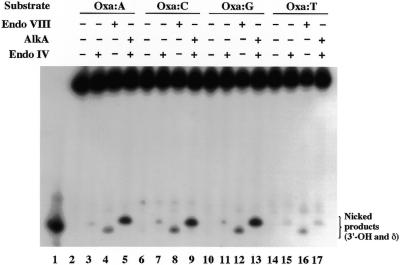
Xan and Oxa are potentially mutagenic lesions and pair with C and T during DNA replication (16–18). Thus, the ability of AlkA and Endo VIII to recognize Xan and Oxa paired with different bases was examined. The activity was measured by the nicking assay using 25XAN/COM25N and 25OXA/COM25N (N = A, G, C, T) as substrates. AlkA recognized not only a Xan:C pair (Fig. 6A, lane 5) but also a Xan:A pair (Fig. 6A, lane 3). The activities for both pairs were comparable. The Xan:G and Xan:T pairs were very poor substrates of AlkA (Fig. 6A, lanes 7 and 9). The order of activity for Xan was C = A >> G > T with respect to the paired base. Unlike AlkA, Endo VIII recognized Xan:C and Xan:G pairs with comparable efficiency (Fig. 6B, lanes 7 and 10). The Xan:A and Xan:T pairs were very poor substrates. The order of activity was C = G >> A > T with respect to the paired base. Compared to the reaction with Xan, a large excess of the enzymes (5- or 10-fold) and a long incubation time (1 h) were required to detect the activity for Oxa. For Oxa, AlkA and Endo VIII exhibited a similar preference of the paired bases. The pairs of Oxa:A (Fig. 7, lanes 4 and 5), Oxa:C (Fig. 7, lanes 8 and 9) and Oxa:G (lanes 12 and 13) were comparable substrates for AlkA and Endo VIII, whereas an Oxa:T pair was recognized poorly by the enzymes (Fig. 7, lanes 16 and 17). Therefore, the order of the activity of both enzymes was C = G = A > T with respect to the paired base.
Figure 6.
Activities of AlkA and Endo VIII for Xan paired with different bases. (A) AlkA reactions. 25XAN/COM25N (N = A, G, C, T) (100 fmol) was incubated with 300 fmol of AlkA at 37°C for 20 min followed by 120 fmol of Endo IV at 37°C for 30 min. (B) Endo VIII reactions. 25XAN/COM25N (N = A, G, C, T) (100 fmol) was incubated with 600 fmol of Endo VIII at 37°C for 1 h or 120 fmol of Endo IV at 37°C for 30 min. In (A) and (B), products were analyzed by 16% denaturing PAGE. The substrates (base pairs) and enzymes used are indicated on the top of the gels. Lane 1 shows a 5′-32P-labeled marker (PRIM15). The 3′-OH and δ-elimination products are indicated by arrows.
Figure 7.
Activities of AlkA and Endo VIII for Oxa paired with different bases. 25OXA/COM25N (N = A, G, C, T) (100 fmol) was incubated with 3 pmol of AlkA or Endo VIII for 1 h and products were analyzed as described in Figure 6. The substrates (base pairs) and enzymes used were indicated on the top of the gels. Lane 1 shows a 5′-32P-labeled marker (PRIM15).
Nitrous acid sensitivities of E.coli strains deficient in AlkA and Endo VIII
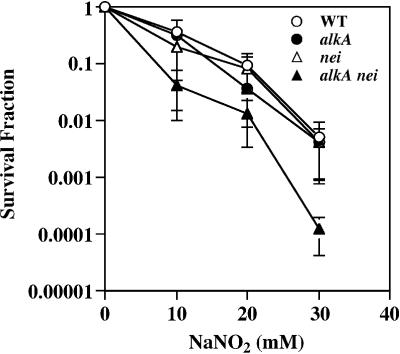
To assess the role of AlkA and Endo VIII in the repair of Xan and Oxa in E.coli cells, the sensitivity of E.coli strains MV1161 (wild type), MV1571 (alkA), KY100 (nei) and KY101 (alkA nei) to nitrous acid was determined by treating them with sodium nitrite (NaNO2) pH 4.6 (29) under selective conditions (i.e. in the presence of appropriate antibiotics). The single mutants defective in either the alkA or nei gene showed nitrous acid sensitivities similar to those of the wild type cell. On the other hand, the alkA nei double mutant exhibited increased sensitivity (Fig. 8), implying a degenerative role of AlkA and Endo VIII in counteracting the cytotoxic effect of nitrous acid. In the above NaNO2 treatment, E.coli strains MV1161, MV1571 and KY100 were grown in the absence or presence of one antibiotic (ampicillin or kanamycin), while strain KY101 was grown in the presence of two antibiotics (ampicillin and kanamycin). The differences in growth conditions (i.e. selective pressure) might have created an advantage for the strains MV1161, MV1571 and KY100 over the strain KY101 with respect to survival. Thus, NaNO2 treatment was also performed in the absence of antibiotics by taking advantage of the fact that the chromosomal markers for antibiotic resistance (ampicillin and kanamycin in this study) are rarely lost even in the absence of selection pressure. However, the results obtained without antibiotics were essentially similar to those obtained with antibiotics, hence ruling out the effects of selection pressure on survival measurements.
Figure 8.
Sensitivity of E.coli strains to nitrous acid. Escherichia coli cells proficient and deficient in AlkA (encoded by the alkA gene) or Endo VIII (encoded by the nei gene) were treated with the indicated concentrations of NaNO2 in acetate buffer (pH 4.6) and the survival fractions were determined as described in Materials and Methods. Open circles, MV1161 (wild type); closed circles, MV1571 (alkA); open triangles, KY100 (nei); closed triangles, KY101 (alkA nei). The data are averages of three independent experiments. Standard deviations are indicated with error bars.
DISCUSSION
In the present study, we have screened the repair activity of E.coli enzymes for Xan and Oxa formed from G by nitrogen oxides such as NO and nitrous acid. For this purpose, we prepared defined oligonucleotide substrates containing Xan and Oxa by DNA polymerase reactions with dXTP and dOTP. It has been reported that dXTP is a poor substrate of DNA polymerase I (35). Our previous study has also shown that dOTP as well as dXTP are poorly utilized by DNA polymerase I relative to dGTP (19). However, the low but significant usability of dXTP and dOTP by DNA polymerase I was good enough for targeted incorporation of the nucleotides under appropriate DNA polymerase reaction conditions (Fig. 2).
Among the seven enzymes tested, AlkA and Endo VIII recognized Xan and Oxa, though their activity for Oxa was lower than for Xan. Analysis of enzymatic parameters revealed that the activity (Vmax/Km) of AlkA for Xan is one-fifth of that for 7mG, a physiological substrate of AlkA (Table 2). AlkA recognizes a variety of alkylated purine and pyrimidine lesions (34). It has been shown that 5-formyluracil and fragmented thymine residues are also substrates of AlkA (24,36–38). Thus, AlkA accepts structurally diverse base lesions as substrates. Common features of the substrates recognized by AlkA are weak N-glycosidic bonds and explicit or implicit positive charges induced in the base (24). A plausible catalytic mechanism of AlkA has been proposed based on these features and the three-dimensional structure of AlkA (39–41). In view of the weak N-glycosidic bond of Xan and the capacity of the active site of AlkA, it seems reasonable that AlkA recognizes Xan as a substrate. Oxa was also a substrate of AlkA but recognized poorly. The relative hydrolysis rate of the N-glycosidic bond of Oxa is 44-fold lower than that of Xan (16). Therefore, it is very likely that such a difference affected the hydrolysis of the N-glycosidic bond by AlkA and slowed down the reaction of Oxa relative to Xan.
The cell extracts from the E.coli strains proficient and deficient in the alkA gene showed no significant difference in Xan-releasing activity. Possibly, the basal level of AlkA protein present in wild type cells was too low to exhibit detectable activity over the experimental fluctuation in the present assay. Although the attempt to show the difference in Xan-releasing activity between wild type and alkA mutant cells was unsuccessful, the repair activity for Xan in the wild type cell was clearly increased when the cell was treated with MNNG (Fig. 5). Such an increase was not observed with the alkA mutant when it was treated in the same manner, indicating that the induction of the repair activity for Xan in cells was dependent on the alkA gene. These results suggest that AlkA can at least participate in the repair of Xan when it is present at the induced level. The alkA gene is under the control of the ada gene and inducible by the alkylating agent such as MNNG (34). However, no information is currently available on whether NO or nitrous acid induces the alkA gene through this pathway.
In this study, it has also been shown that Endo VIII has repair activity for Xan. The activity of Endo VIII for Xan measured by enzymatic parameters was low relative to that for its physiological substrate (Tg) (Table 2). However, the low but significant activity may be crucial for repair of Xan when the level of AlkA is not sufficiently high, e.g. in uninduced cells. Endo VIII was originally found as a repair enzyme for Tg and urea residues (31), and later it was shown to recognize other oxidative pyrimidine damage such as 5-hydroxycytosine, 5-hydroxyuracil and uracil glycol (33,42). Thus, substrate specificity of Endo VIII is similar to that of Endo III. However, the amino acid sequence of Endo VIII shows no homology to that of Endo III. Rather, the N-terminal and C-terminal regions of Endo VIII are similar to those of Fpg involved in repair of oxidative purine lesions in E.coli (32). Furthermore, Endo VIII has been shown recently to excise oxidative purine lesions including 7,8-dihydro-8-oxoguanine and formamidopyrimidine (25,43). Thus, Endo VIII has a potential capacity to accept purine lesions as well as pyrimidine lesions, though the activities may vary. In this context, it may not be surprising that Endo VIII recognized Xan and Oxa. However, questions remain as to why Xan was a better substrate than Oxa for Endo VIII and why Xan and Oxa were substrates for Endo VIII but not for Fpg. Concerning the latter, the three-dimensional structures of Endo VIII and Fpg complexed covalently to DNA have recently been solved (44,45). Although the overall structures of Endo VIII and Fpg resemble each other, there are three dissimilar patches where amino acids are not conserved between the two enzymes. One of them coincides with putative binding pockets for the everted base. Thus, interactions in the dissimilar binding pocket are likely to account for the distinct damage specificity of Endo VIII and Fpg.
The activities of AlkA and Endo VIII for Xan and Oxa showed dependence on the base opposite the lesions, though the activity for Oxa was consistently much lower than for Xan. For Xan, the activity decreased in the following order of the paired base: C = A >> G > T for AlkA and C = G >> A > T for Endo VIII (Fig. 6). For Oxa, the order was C = G = A > T for both AlkA and Endo VIII (Fig. 7). Considering the base pairing capacity of Xan and Oxa, C and T are the most biologically relevant paired bases. The Xan:C and Oxa:C pairs can be formed by the reaction of a G:C pair with NO and nitrous acid, and Xan:T and Oxa:T pairs by misincorporation of T opposite these lesions during DNA replication (16–18). Excision of Xan and Oxa from Xan:C and Oxa:C pairs, respectively, by AlkA and Endo VIII and subsequent repair synthesis leads to restoration of genetic information, thus error free. Conversely, excision of Xan and Oxa from Xan:T and Oxa:T pairs, respectively, results in mutation fixation and induces G:C→A:T transitions if it occurs in cells. According to the present results (Figs 6 and 7), AlkA and Endo VIII recognized Xan:C and Oxa:C pairs relatively well but Xan:T and Oxa:T pairs were poor substrates for the enzymes. Thus, the poor activity of AlkA and Endo VIII for Xan:T and Oxa:T pairs seems to be consistent with avoidance of mutation fixation in cells. This is in contrast to the preference of E.coli Endo V, which recognizes all four Xan:N pairs (N = A, G, C, T) with a preference for the Xan:T pair (21). Endo V was originally found as a deoxyinosine endonuclease that incises the second phosphodiester bond on the 3′ side of deoxyinosine (a deoxyribonucleoside form of Hx) (7). More recently it has been shown to have a broad substrate specificity including Hx, U, Xan, AP site and mismatched bases (7,21,46,47).
In the present work, it has been shown that the alkA nei double mutant, but not the alkA and nei single mutants, was sensitive to nitrous acid (Fig. 8). These results suggest a degenerative role of AlkA and Endo VIII in the repair of a certain class of nitrous acid-induced DNA lesions, for example Xan and Oxa, though involvement of other E.coli base excision repair enzymes such as MutY, Mug and TagI has not been eliminated in this study. The actual mechanisms operating in E.coli cells to counteract the lethal and mutagenic effects of nitrous acid and NO appear to be rather complicated. Several studies show cell killing and mutagenicity by nitrous acid or NO increase in the cells deficient in nucleotide excision repair (48–50) or recombination repair (51). In contrast, studies on nfi (the Endo V gene) mutants by Weiss’s group indicate the involvement of Endo V in avoidance of nitrous acid-induced mutations (52–54). Thus, at least four repair pathways including base excision repair indicated by this study, nucleotide excision repair, recombination repair, and repair initiated by Endo V have been implicated in restoration of DNA lesions formed by nitrous acid and NO. It is not clear whether such differences originate from dosage or types of nitrogen oxide species, cell conditions or accessed phenotypes (lethal or mutation effect). Thus, further studies are necessary to clarify the whole picture of the cellular mechanisms to counteract mutagenic and lethal effects associated with nitrogen oxide species.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to H.I. and K.M.).
REFERENCES
- 1.Frederico L.A., Kunkel,T.A. and Shaw,B.R. (1990) A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry, 29, 2532–2537. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 3.Duncan B.K. and Weiss,B. (1982) Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J. Bacteriol., 151, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill-Perkins M., Jones,M.D. and Karran,P. (1986) Site-specific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat. Res., 162, 153–163. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 6.Saparbaev M. and Laval,J. (1994) Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat and human alkylpurine DNA glycosylases. Proc. Natl Acad. Sci. USA, 91, 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao M., Hatahet,Z., Melamede,R.J. and Kow,Y.W. (1994) Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem., 269, 16260–16268. [PubMed] [Google Scholar]
- 8.Lindahl T. (1979) DNA glycosylases, endonuclease for apurinic/apyrimidinic sites and base excision repair. Prog. Nucleic Acid Res. Mol. Biol., 22, 135–192. [DOI] [PubMed] [Google Scholar]
- 9.Marletta M.A. (1989) Nitric oxide: biosynthesis and biological significance. Trends Biochem. Sci., 14, 488–492. [DOI] [PubMed] [Google Scholar]
- 10.Ohshima H. and Bartsch,H. (1994) Chronic infections and inflammatory processes as cancer risk factors: possible role of nitric oxide in carcinogenesis. Mutat. Res., 305, 253–264. [DOI] [PubMed] [Google Scholar]
- 11.Wink D.A., Vodovots,Y., Laval,J., Laval,F., Dewhirst,M.W. and Mitchell,J.B. (1998) The multifaceted roles of nitric oxide in cancer. Carcinogenesis, 19, 711–721. [DOI] [PubMed] [Google Scholar]
- 12.Wink D.A., Kasprzak,K.S., Maragos,C.M., Elespuru,R.K., Misra,M., Dunams,T.M., Cebula,T.A., Koch,W.H., Andrews,A.W., Allen,J.S. and Keefer,L.K. (1991) DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science, 254, 1001–1003. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T., Yamaoka,R., Nishi,M., Ide,H. and Makino,K. (1996) Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxynucleotide, and calf thymus DNA treated by nitrous acid and nitric oxide. J. Am. Chem. Soc., 118, 2515–2516. [Google Scholar]
- 14.Lucas L.T., Gatehouse,D. and Shuker,D.E. (1999) Efficient nitroso group transfer from N-nitrosoindoles to nucleotides and 2′-deoxyguanosine at physiological pH. J. Biol. Chem., 274, 18319–18326. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T., Ide,H., Yamada,M., Endo,N., Kanaori,K., Tajima,K., Morii,T. and Makino,K. (2000) Formation of 2′-deoxyoxanosine from 2′-deoxyguanosine and nitrous acid: mechanism and intermediates. Nucleic Acids Res., 28, 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki T., Matsumura,Y., Ide,H., Kanaori,K. Tajima,K. and Makino,K. (1997) Deglycosylation susceptibility and base-pairing stability of 2′-deoxyoxanosine in oligonucleotide. Biochemistry, 36, 8013–8019. [DOI] [PubMed] [Google Scholar]
- 17.Eritja R. Horowitz,D.M., Walker,P.A., Ziehler-Martin,J.P., Boosalis,M.S., Goodman,M.F., Itakura,K. and Kaplan,B.E. (1986) Synthesis and properties of oligonucleotides containing 2′-deoxynebularine and 2′-deoxyxanthosine. Nucleic Acids Res., 24, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamiya H., Shimizu,M. Inoue,H. and Ohtsuka,E. (1992) Mutation induced by deoxyxanthosine in codon 12 of a synthetic c-Ha-ras gene. Nucl. Nucl., 11, 247–260. [PubMed] [Google Scholar]
- 19.Suzuki T., Yoshida,M., Yamada,M., Ide,H., Kobayashi,M., Kanaori,K., Tajima,K. and Makino,K. (1998) Misincorporation of 2′-deoxyoxanosine 5′-triphosphate by DNA polymerases and its implication for mutagenesis. Biochemistry, 37, 11592–11598. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T., Yamada,M., Ide,H., Kanaori,K., Tajima,K., Morii,T. and Makino,K. (2000) Identification and characterization of a reaction product of 2′-deoxyoxanosine with glycine. Chem. Res. Toxicol., 13, 227–230. [DOI] [PubMed] [Google Scholar]
- 21.He B., Qing,H. and Kow,Y.W. (2000) Deoxyxanthosine in DNA is repaired by Escherichia coli endonuclease V. Mutat. Res., 459, 109–144. [DOI] [PubMed] [Google Scholar]
- 22.Wang G., Palejwala,V.A., Dunman,P.M., Aviv,D.H., Murphy,H.S., Rahman,M.S. and Humayun,M.Z. (1995) Alkylating agents induce UVM, a recA-independent inducible mutagenic phenomenon in Escherichia coli. Genetics, 141, 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito Y., Uraki,F., Nakajima,S., Asaeda,A., Ono,K., Kubo,K. and Yamamoto,K. (1997) Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J. Bacteriol., 179, 3783–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masaoka A., Terato,H., Kobayashi,M., Honsho,A., Ohyama,Y. and Ide,H. (1999) Enzymatic repair of 5-formyluracil. I. Excision of 5-formyluracil site-specifically incorporated into oligonucleotide substrates by AlkA protein (Escherichia coli 3-methyladenine DNA glycosylase II). J. Biol. Chem., 274, 25136–25143. [DOI] [PubMed] [Google Scholar]
- 25.Asagoshi K., Yamada,T., Okada,Y., Terato,H., Ohyama,Y., Seki,S. and Ide,H. (2000) Recognition of formamidopyrimidine by Escherichia coli and mammalian thymine glycol glycosylases. J. Biol. Chem., 275, 24781–24786. [DOI] [PubMed] [Google Scholar]
- 26.Sarker A.H., Ikeda,S., Nakano,H., Terato,H., Ide,H., Imai,K., Akiyama,K., Tsutsui,K., Bo,Z., Kubo,K., Yamamoto,K., Yasui,A., Yoshida,M.C. and Seki,S. (1998) Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J. Mol. Biol., 282, 761–774. [DOI] [PubMed] [Google Scholar]
- 27.Asagoshi K., Odawara,H., Nakano,H., Miyano,T., Terato,H., Ohyama,Y., Seki,S. and Ide,H. (2000) Comparison of substrate specificities of Escherichia coli endonuclease III and its mouse homologue (mNTH1) using defined oligonucleotide substrates. Biochemistry, 39, 11389–11398. [DOI] [PubMed] [Google Scholar]
- 28.Ide H., Okagami,M., Murayama,H., Kimura,Y. and Makino,K. (1993) Synthesis and characterization of oligonucleotides containing the alpha-anomer of deoxyadenosine to study its influence on DNA replication. Biochem. Mol. Biol. Int., 31, 485–491. [PubMed] [Google Scholar]
- 29.Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Thomas L., Yang,C.-H. and Goldthwait,D.A. (1982) Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry, 21, 1162–1169. [DOI] [PubMed] [Google Scholar]
- 31.Melamede R.J., Hatahet,Z., Kow,Y.W., Ide,H. and Wallace,S.S. (1994) Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry, 33, 1255–1264. [DOI] [PubMed] [Google Scholar]
- 32.Jiang D., Hatahet,Z., Blaisdell,J.O., Melamede,R.J. and Wallace,S.S. (1997) Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J. Bacteriol., 179, 3773–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang D., Hatahet,Z., Melamede,R.J., Kow,Y.W. and Wallace S.S. (1997) Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem., 272, 32230–32239. [DOI] [PubMed] [Google Scholar]
- 34.Lindahl T., Sedgwick,B., Sekiguchi,M. and Nakabeppu,Y. (1988) Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem., 57, 133–157. [DOI] [PubMed] [Google Scholar]
- 35.Bessman M.J., Lehman,I.R., Adler,J., Zimmerman,S.B., Simms,E.S. and Kornberg,A. (1958) Enzymatic synthesis of deoxyribonucleic acid. III. The incorporation of pyrimidine and purine analogues into deoxyribonucleic acid. Proc. Natl Acad. Sci. USA., 44, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjelland S., Birkeland,N.K., Benneche,T., Volden,G. and Seeberg,E. (1994) DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J. Biol. Chem., 269, 30489–30495. [PubMed] [Google Scholar]
- 37.Terato H., Masaoka,A., Kobayashi,M., Fukushima,S., Ohyama,Y., Yoshida,M. and Ide,H. (1999) Enzymatic repair of 5-formyluracil II. Mismatch formation between 5-formyluracil and guanine during DNA replication and its recognition by two proteins involved in base excision repair (AlkA) and mismatch repair (MutS). J. Biol. Chem., 274, 25144–25150. [DOI] [PubMed] [Google Scholar]
- 38.Privezentzev C.V., Saparbaev,M., Sambandam,A., Greenberg,M.M. and Laval,J. (2000) AlkA protein is the third Escherichia coli DNA repair protein excising a ring fragmentation product of thymine. Biochemistry, 39, 14263–14268. [DOI] [PubMed] [Google Scholar]
- 39.Yamagata Y., Kato,M., Odawara,K., Tokuno,Y., Nakashima,Y., Tatsushima,N., Yasumura,K., Tomita,K., Ihara,K., Fujii,Y., Nakabeppu,Y., Sekiguchi,M. and Fujii,S. (1996) Three dimensional structure of a DNA repair enzyme, 3-methyladenine DNA glycosylase II, from Escherichia coli. Cell, 86, 311–319. [DOI] [PubMed] [Google Scholar]
- 40.Labahn J., Scharer,O.D., Long,A., Ezaz-Nikpay,K., Verdine,G.L. and Ellenberger,T.E. (1996) Structure basis for the excision repair of alkylation-damaged DNA. Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- 41.Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) DNA bending and a flip-out mechanism for base excision by the helix–hairpin–helix DNA glycosylase, Escherichia coli AlkA. EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purmal A.A., Lampman,G.W., Bond,J.P., Hatahet,Z. and Wallace S.S. (1998) Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J. Biol. Chem., 273, 10026–10035. [DOI] [PubMed] [Google Scholar]
- 43.Blaisdell J.O., Hatahet,Z. and Wallace,S.S. (1999) A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G→T transversions. J. Bacteriol., 181, 6396–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zharkov D.O., Golan,G., Gilboa,R., Fernandes,A.S., Gerchman,S.E., Kycia,J.H., Rieger,R.A., Grollman,A.P. and Shoham,G. (2002) Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J., 21, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilboa R., Zharkov,D.O., Golan,G., Fernandes,A.S., Gerchman,S.E., Matz,E., Kycia,J.H., Grollman,A.P. and Shoham,G. (2002) Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem., 277, 19811–19816. [DOI] [PubMed] [Google Scholar]
- 46.Yao M. and Kow,Y.W. (1995) Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem., 270, 28609–28616. [DOI] [PubMed] [Google Scholar]
- 47.Yao M. and Kow,Y.W. (1997) Further characterization of Escherichia coli endonuclease V. J. Biol. Chem., 272, 30774–30779. [DOI] [PubMed] [Google Scholar]
- 48.Hartman Z., Henrickson,E.N., Hartman,P.E. and Cebula,T.A. (1994) Molecular models that may account for nitrous acid mutagenesis in organisms containing double-stranded DNA. Environ. Mol. Mutagen., 24, 168–175. [DOI] [PubMed] [Google Scholar]
- 49.Sidorkina O., Saparbaev,M. and Laval,J. (1997) Effects of nitrous acid treatment on the survival and mutagenesis of Escherichia coli cells lacking base excision repair (hypoxanthine-DNA glycosylase-ALK A protein) and/or nucleotide excision repair. Mutagenesis, 12, 23–28. [DOI] [PubMed] [Google Scholar]
- 50.Tamir S., Burney,S. and Tannenbaum,S.R. (1996) DNA damage by nitric oxide. Chem. Res. Toxicol., 9, 821–827. [DOI] [PubMed] [Google Scholar]
- 51.Spek E.J., Wright,T.L., Stitt,M.S., Taghizadeh,N.R., Tannenbaum,S.R., Marinus,M.G. and Engelward,B.P. (2001) Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. J. Bacteriol., 183, 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo G. and Weiss,B. (1998) Endonuclease V (nfi) mutant of Escherichia coli K-12. J. Bacteriol., 180, 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schouten K.A. and Weiss,B. (1999) Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res., 435, 245–254. [DOI] [PubMed] [Google Scholar]
- 54.Weiss B. (2001) Endonuclease V of Escherichia coli prevents mutations from nitrosative deamination during nitrate/nitrite respiration. Mutat. Res., 461, 301–309. [DOI] [PubMed] [Google Scholar]