Abstract
Guanine (G)-rich oligodeoxyribonucleotides (ODNs) can form undesired complexes by self association through non-Watson–Crick interactions. These aggregates can compromise performance of DNA probes and make genetic analysis unpredictable. We found that the 8-aza-7-deazaguanine (PPG), a pyrazolo[3,4-d]pyrimidine analog, reduces guanine self association of G-rich ODNs. In the PPG heterocycle, the N-7 and C-8 atoms of G are interposed. This leaves the ring system with an electron density similar to G, but prevents Hoogsteen-bonding associated with N-7. ODNs containing multiple PPG bases were easily prepared using a dimethylformamidine-protected phosphoramidite reagent. Substitution of PPG for G in ODNs allowed formation of more stable DNA duplexes. When one or more PPGs were substituted for G in ODNs containing four or more consecutive Gs, G aggregation was eliminated. Substitution of PPG for G also improved discrimination of G/A, G/G and G/T mismatches in Watson–Crick hybrids. Use of PPG in fluorogenic minor groove binder probes was also explored. PPG prevented aggregation in MGB probes (MGBTM is a trademark of Epoch Biosciences) and allowed use of G-rich sequences. An increased signal was observed in 5′-PPG probes due to reduced quenching of fluorescein by PPG. In summary, substitution of PPG for G enhances affinity, specificity, sensitivity and predictability of G-rich DNA probes.
INTRODUCTION
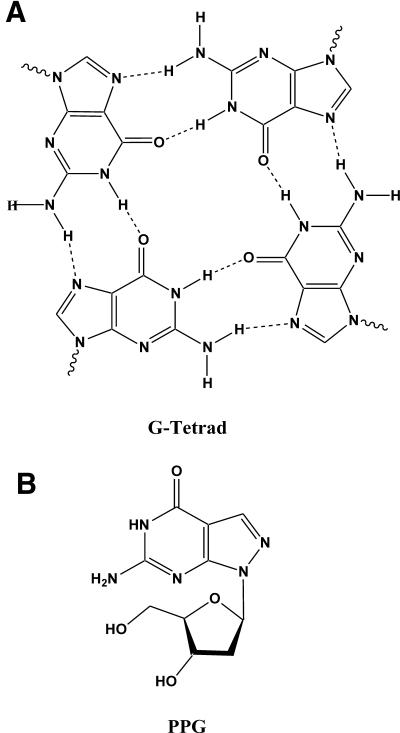
Predictable and reliable sequence-specific detection of nucleic acid sequences is central to genetic analysis. However, many sequences are not accessible to DNA probe interrogation due to intra- or inter-strand complex formation of either target or probe (1). Guanine (G)-rich oligodeoxyribonucleotides (ODNs) are especially problematic since ODNs containing one or more appropriately spaced runs of Gs can associate to form intra- or intermolecular complexes composed of G–G base pairs (2). Self-association of G-rich ODNs usually involves Hoogsteen or reverse Hoogsteen H-bonding via the N-7 of G (3,4). For example, ODNs containing three or more contiguous Gs can form tetrastranded parallel complexes in the presence of alkali cations (5) (Fig. 1A). Other abnormal structures can be formed by self-association through non-Watson–Crick interactions. G:G base-paired structures in ODNs have been reported to occur in telomeric ODNs (6–8) and in long terminal track ODNs (9). The importance of N-7 in complex formation has also been demonstrated by its inaccessibility to dimethyl sulfate methylation when involved in stable H-bonding in multi-stranded, G-rich DNA structures (10). Hoogsteen type base pairing with N-7 can also stabilize mismatches and the thermodynamic stabilities of mismatches involving G have been investigated (11).
Figure 1.
PPG nucleoside prevents aggregation in G-rich ODNs. (A) Structure of a G-tetrad showing Hoogsteen H-bonds to N-7 of G. (B) Structure of the PPG nucleoside [6-amino-1-(4-hydroxy-5-hydroxy methyl-tetrahydro-furan-2-yl)-1,5-dihydro-pyrazolo[3,4-d]pyrimidin-4-one, a substitute for G].
The modified base 7-deazaguanine has been used with some success as a substitute in G-rich ODNs. When used in DNA sequencing reactions, 7-deazaguanine reduces G self-association due to the absence of the N-7 (12). Unfortunately, 7-deazaguanine lowers DNA affinity in comparison with natural G (13). PPG has been explored previously as a substitute for G in ODNs but no major advantages were described (14,15). In the PPG heterocycle, the N-7 and C-8 atoms of G are interposed (Fig. 1B), thus retaining the electron density of the purine ring system. We had reported previously that PPG-rich triplex forming ODNs (TFOs) and alkylating TFOs had faster binding kinetics and increased gene-specific alkylation (16). We hypothesized that this improved performance was due to elimination of aggregates known to form in the G/A motif probes (17).
In this manuscript we report improved performance of PPG-containing ODNs in DNA hybridization assays. PPG, when substituted for G, reduced or eliminated unwanted aggregates in ODNs containing multiple contiguous Gs. PPG-containing ODNs were easily prepared using standard phosphoramidite coupling chemistry. Thermal denaturation studies of a series of 13–17mers showed sequence-specific increases in duplex melting temperature (Tm). Substitution of PPG for G in short, self-complementary ODNs improved mismatch discrimination. We also explored the use of PPG in fluorogenic MGB™ probes (18) and found similar improvements in hybridization performance. Use of PPG in MGB probes eliminated aggregate formation and allowed use of G-rich probes in 5′-nuclease (TaqMan®) real-time PCR assays. We showed further that 5′-fluorescein-PPG probes have improved quantum yield in fluorogenic assays.
MATERIALS AND METHODS
Synthesis of ODN conjugates
6-Carboxyfluorescein phosphoramidite (6-FAM) was obtained from Glen Research (Sterling, VA). ODN synthesis was performed on an ABI 394 synthesizer according to the protocol supplied by the manufacturer, except that 0.015 M (instead of the standard 0.2 M) iodine solution was utilized in the oxidation step to avoid iodination of the minor groove binder (MGB) moiety. The 3′-MGB-Q-ODN conjugates were synthesized from a novel controlled pore glass support containing a DPI3 (MGB) group and non-fluorescent quencher chromophore (E.A.Lukhtanov, I.V.Kutyavin, S.Lokhov, T.Afonina, V.V.Gorn and M.W.Reed, manuscript in preparation). PPG phosphoramidite (Super G™ is a trademark of Epoch Biosciences) is available from Glen Research. The synthesis of PPG phosphoramidite has been described previously (14). Fluorogenic MGB™ TaqMan® probes with this 3′-Q-MGB structure are available from Applied Biosystems (Foster City, CA). After ammonia deprotection, all ODNs were purified by reverse-phase HPLC and isolated as the sodium salt by butanol concentration/sodium perchlorate precipitation (19). In the case of probes containing more than three Gs in a row, multiple peaks were observed for the conjugate, due to self-association (similar to that observed for probe GGGGTTGGGG in Fig. 2). In these cases, the first peaks, where pure compound usually elutes, were collected and processed as described above. The pellets were dissolved in water and concentrations were determined spectrophotometrically. A nearest-neighbor model (20) was applied to calculate extinction coefficients (ε260) of ODNs. A260 measurements were made in PBS pH 7.2 at ambient temperature and assumed a random coil DNA structure in solution. For each PPG base substitution, a ε260 correction of +1200 M–1cm–1 was used. MGB-ODN concentrations were determined from the unique A340 absorbance of the Q-MGB chromophore (ε340 = 75 000 M–1cm–1). All ODNs for UV-melting studies were purified by denaturing PAGE.
Figure 2.
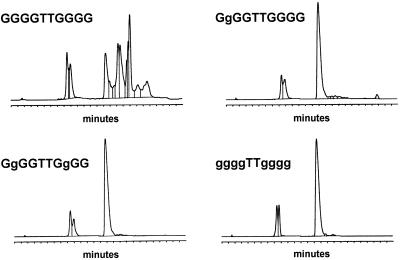
Reverse phase HPLC purification of trityl-on ODNs containing both G and PPG. g represents PPG in the indicated sequences. Chromatography was performed on a 7 × 300 mm Hamilton PRP-1 column in 0.1 M triethylammonium acetate with a gradient of 0–60% acetonitrile over 20 min, flow rate = 2 ml/min.
Tm determination
Thermodynamic parameters of duplex formation were derived by the van’t Hoff analysis methods. The shape of each melting curve was fitted to the two-state model with linear base lines (21) using a non-linear least-square program (22). Unless otherwise stated, Tm of DNA duplexes were measured in 1× PCR buffer at a concentration of 5 × 10–7 M as described earlier (18).
Synthesis and properties of fluorescein-nucleotide monomers
The fluorescein-nucleotide monomers were prepared on an ABI394 DNA synthesizer. Each of the nucleotide monomers (dA, dC, dG, dT and PPG) was added first to 1 µmol of hexanol-CPG (23) using the appropriate phosphoramidite reagent. 6-FAM (Glen Research) was added at the final coupling step, and each FAM-nucleotide was isolated by reverse phase HPLC. The homogeneous products were collected, dried in vacuo and redissolved in water. Concentration of each FAM-nucleotide monomer was determined in 1× PBS (9.2 mM disodium phosphate, 0.8 mM monosodium phosphate, 0.131 M sodium chloride, pH 7.2) using an extinction coefficient for fluorescein of 73 000 M–1cm–1 at 495 nm. Fluorescence of 0.02 µM solutions of the monomers were measured in 1× PBS using a Perkin Elmer LS-50 luminescence spectrometer equipped with a red-sensitive photomultiplier tube. Excitation was at the absorbance max. (∼494 nm) and slit width was 2.5 nm. Emission max. (∼518 nm) was measured using a slit width of 5.5 nm.
Electrophoretic studies
Electrophoresis of ODNs was conducted in 8% polyacrylamide gels run in 1× TBE buffer for 45 min at 40°C. Samples (70 µM) were loaded in 1× TBE buffer containing 40% glycerol. Gels were stained with Daiichi 2D Silver Stain II® and Rm values for the stained ODN bands were determined using two control ODNs as standards.
Real-time PCR
MGB-ODN conjugates with sequences shown in Table 3 were used as fluorogenic probes in the 5′-nuclease PCR assay (24). Amplification was conducted in an Idaho Technologies LC-24 LightCycler® with real-time fluorescence monitoring (25). The amplicon d(CACCTCAGCCTCCCAAGTAACTTTT AACCCCCCCCCATTTGTTGTGCTGTTTTCATACCTGTAATCCTGGCACTTT) was used as a template with underlined portions of the target sequence corresponding to the primers. The bold portion indicates complementary to probes 11–18. Amplification reactions contained 105 copies of this 76mer target, 500 nM of each primer, 50 nM fluorescent probe, 20 mM Tris–HCl pH 7, 40 mM NaCl, 5 mM MgCl2, 0.05% BSA, 125 nM each dNTP, 0.38 U/reaction JumpStart Taq polymerase (Sigma, St. Louis, MO) and 0.01 U/reaction uracil-N-glycosylase. The cycling program was one cycle of 50°C for 3 min, then 95°C for 2 min, followed by 50 cycles of 95°C for 2 s, then 60°C for 30 s. Fluorogenic curves were measured in the FAM channel and data was processed using the software provided with the instrument.
Table 3. Effect of PPG substitution on electrophoretic mobility of MGB-ODN conjugates.
| aSequence | GC content | bRm | Rm | |
|---|---|---|---|---|
| |
|
|
X = G |
X = PPG |
| A | d(ACCTGTATTCCTTGCC) | 8/16 | 1.00 | |
| B | d(XTACAXCAAATXXAA) | 6/15 | 1.00 | |
| 11 | d(CAAATXXXXXXXXX) | 10/14 | 0.58 | 0.96 |
| 12 | d(ACAAATXXXXXXXX) | 9/14 | 0.42 | 0.97 |
| 13 | d(AACAAATXXXXXXX) | 8/14 | 0.37 | 0.95 |
| 14 | d(CAACAAATXXXXXX) | 8/14 | 0.35 | 0.95 |
| 15 | d(ACAACAAATXXXXX) | 7/14 | 0.32 | 0.98 |
| 16 | d(CACAACAAATXXXX) | 7/14 | 0.29 | 1.03 |
| 17 | d(CACAACAAATXXX) | 6/13 | 0.96 | 0.98 |
| 18 | d(AGCACAACAAATXX) | 6/14 | 0.96 | 0.96 |
aMGB-ODNs contained 5′-fluorescein, 3′-Q-MGB as shown in Figure 3. In sequence B, X = PPG. Sequences 11–18 contained either G or PPG at the indicated X positions.
bMobility relative to control (Rm) of G- and PPG-containing ODNs were measured by non-denaturing PAGE. Rm values were measured separately for G- and PPG-containing ODNs with respect to control ODNs A and B, respectively.
RESULTS AND DISCUSSION
Duplex stability
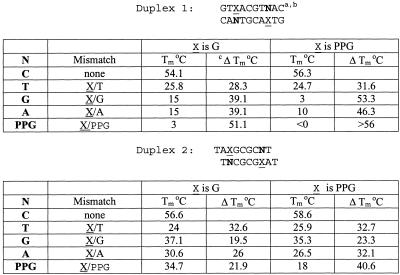
The effect of substituting G bases with PPG on DNA duplex stability is shown in Table 1. Ten different ODN sequences (13–17mers) were synthesized with either G or PPG at the indicated X positions. DNA synthesis was routine, and no problems with instability of the formamidine-protecting group were observed (14). ODNs that contained from 2 to 6 Gs were studied by forming DNA duplexes with their respective complements. Hyperchromicity at 260 nm was measured in UV-melting experiments to determine Tm. While these data do not give a comprehensive picture of duplex stability, several trends are clear. ΔTm per PPG substitution is between 0.5 and 1.5°C, and has certain sequence dependencies. The largest increases in Tm are seen when the Gs are separated, as in sequences 5 and 8, while smaller Tm increases are seen with ODNs that have clusters of two or three PPGs in a row (sequences 7 and 10). These enhancements in DNA duplex stability are significant for a modified base that bears no other exocyclic substituents and have been observed earlier in a limited sequence context (15,26).
Table 1. PPG increases substitution on Tm of ODNs.
| 5′-Probe Sequence-3′ | g content | aTm°C | Tm°C | bΔTm/base | |
|---|---|---|---|---|---|
| |
|
|
X = PPG |
X = G |
|
| 1 | CCAACACTCXTXAA | 2/14 | 54.9 | 52.9 | 1.0 |
| 2 | CCACCCXCCTCAX | 2/13 | 60.7 | 59.7 | 0.5 |
| 3 | CTXTAAXTAXATATAAC | 3/17 | 51.8 | 49.5 | 0.8 |
| 4 | XATTACCTXXATTT | 3/14 | 50.6 | 48.8 | 0.6 |
| 5 | XTAAXTAXACACAXC | 4/15 | 59.5 | 53.5 | 1.5 |
| 6 | XTAAXTAXXCATAAC | 4/15 | 55.7 | 53.2 | 0.6 |
| 7 | CXXCTACATXCTXX | 5/14 | 61.1 | 58.8 | 0.5 |
| 8 | XTAAXTAXACXAXC | 5/15 | 62.9 | 56.7 | 1.2 |
| 9 | XTAAXTAXXCXCAXC | 6/15 | 65.5 | 58.2 | 1.2 |
| 10 | CAXXXAXCTTTXXA | 6/14 | 59.9 | 56.4 | 0.6 |
aMelting temperature (Tm) of DNA duplexes formed with ODNs containing G or PPG at the X positions. The accuracy of the Tm determination is ±0.2°C according to the instrument manufacturer (Cary 400 Bio UV/Vis Spectrophotometer, Varian, Palo Alto, CA).
bΔTm/base is the average increase in Tm per base when G is substituted with PPG.
Reduced aggregation with PPG ODNs
Aggregation of G-rich ODNs can complicate DNA synthesis performance as well as hybridization performance. Predictable performance typically requires that probes be designed with G/C content in the 20–80% range, and runs of four or more Gs should especially be avoided (Primer Express® Software 2.0, Applied Biosystems, Foster City, CA). These ‘guidelines’ are helpful, but problems with aggregation of G-rich probes can be unpredictable. To determine how PPG substitution in a run of Gs affects aggregation, the ODNs shown in Figure 2 were designed. The exceptionally G-rich 10mer ODN in Figure 2 was shown previously to form large superstructures spontaneously (27). Reverse phase HPLC purification profiles of this ODN with all natural dG shows the presence of multiple complexes where a single peak for full-length ODN was expected. This behavior of the trityl-on ODN in a denaturing organic solvent was unexpected, but proved to be a sensitive assay for studying aggregation. In a kinetic study of G-tetrad formation it was shown that G-rich oligonucleotides at concentrations >1 mM occurred rapidly (28). Once formed, these higher order structures are exceedingly stable with t0.5 in the order of 60 days. Since all the oligonucleotide and oligonucleotide conjugates in this study involve a similar concentration step, which yields concentrations >1 mM, it is assumed that solutions of oligonucleotides containing more than four Gs in row are primarily associated in higher order structures. The strong self-association observed with the all dG 10mer was eliminated with substitution of one, two or all Gs with PPG, showing single HPLC peaks where full-length product was expected.
Improved mismatch discrimination with PPG ODNs
The effect of substitution of PPG for G on mismatch detection was investigated in a series of self-complementary ODNs (Table 2). The difference in stability (ΔTm) between matched mismatched duplexes of G/T, G/G, G/A and G/PPG mismatches in Duplexes 1 were all lower than ΔTm of the corresponding PPG/T, PPG/G, PPG/A and PPG/PPG mismatches. In these sequences, the mismatch was surrounded by two A/T nearest-neighbor base pairs. A similar trend was seen with the same mismatches between two A/G nearest-neighbor base pairs (Duplex 2). The G/T mismatch was discriminated equally well with G or PPG. All other mismatches were discriminated substantially better when PPG was in the mismatch position. The relatively sharp decrease in Tm, observed in a G/G mismatch when PPG was substituted, suggests that the nitrogen in the 2 position in PPG (equivalent to the purine 8-position) may not be as readily available for H-bonding as N-7 in G. This is in agreement with a previous report by Pyeret et al. (11).
Table 2. PPG substitution improves mismatch discrimination (ΔTm) in self-complementary duplexes.
aBold nucleotides (N) opposite X (G or PPG) in the self-complementary. ODNs give the indicated mismatch.
bODN concentration was 5 × 10–6 M in 1 M NaCl, 10 mM Na cacodylate, pH 7.0.
cΔTm = Tm(match) – Tm(mismatch), a measure of probe specificity.
Synthesis and properties of minor groove binder—PPG probes
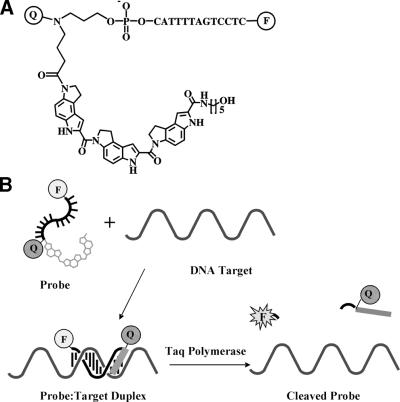
We have described previously the synthesis and improved performance of 3′-fluorogenic MGB quencher oligonucleotide conjugates synthesized from the DPI3 class of MGBs (18). These MGB-probes bind tightly with complementary DNA strands to give B-form duplexes (Fig. 3) (29). Their short length, high Tm and predictable performance makes MGB-probes especially useful for high temperature applications such as in PCR probes. The MGB probes were prepared with 5′-reporter dye, Eclipse™ internal quencher (Epoch Biosciences, Bothell, WA) and a 3′-DPI3 group.
Figure 3.
(A) The structure of a fluorogenic MGB-probe conjugate. Q is a diazine analog, F is fluorophore. (B) Illustration of the hybridization of the MGB-Q-ODN-F conjugate to its target. The MGB binds in the minor groove to stabilize the duplex. Taq polymerase cleaves the 3′-MGB-Q-ODN-F conjugate during PCR to generate a fluorescent signal.
The non-fluorescent quencher chromophore was optimized for FRET-based PCR assays with fluorescein (E.A.Lukhtanov, R.O.Dempcy, A.Gall, I.Afonina, V.V.Gorn and M.W.Reed, manuscript in preparation). We have demonstrated that 3′-MGB probes with a 5′-fluor and 3′-quencher have excellent performance as fluorogenic probes in the 5′-nuclease assay (18). G-rich MGB-ODNs can suffer from the same aggregation problems as other G-rich probes. We therefore studied the effect of PPG in fluorogenic MGB probes in the hope of further improving their hybridization performance and predictability. The results were analogous to findings for the non-MGB probes described above.
Reduced aggregation of MGB-PPG probes
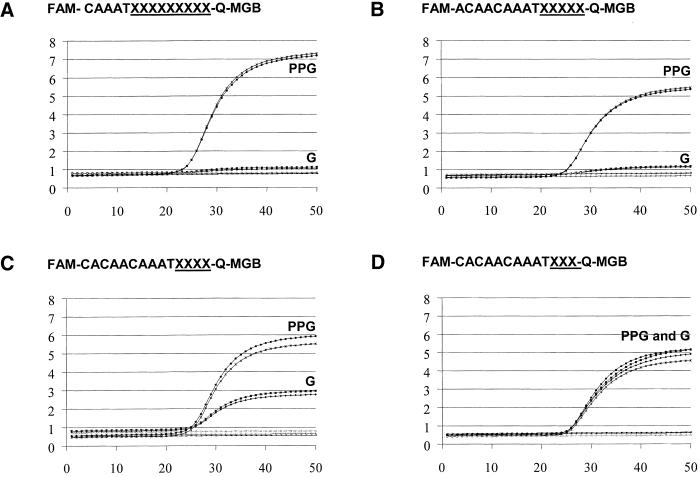
The MGB probes in Table 3 were designed for a model 5′-nuclease PCR assay. The ability of PPG to eliminate mobility shifts, reflective of aggregation, caused by runs of contiguous Gs in an ODN is shown in Table 3. This series of fluorogenic probes was designed to hybridize near the same site on the amplicon. Two sets of probes, one with natural G nucleotides and the other in which the Gs were replaced by PPG, were synthesized. Probe sequences 11–18 contain two to nine consecutive Gs at the 3′-terminus. The mobility values (Rm) were measured separately for the G- and PPG-containing ODNs relative to control ODNs A and B, respectively.
Self association of the G-rich probes was observed during non-denaturing PAGE. The reference ODNs A contained two isolated G residues and B contained four isolated PPG residues, and they migrated an essentially identical distance under the PAGE conditions. G-containing ODNs exhibited diffuse bands upon electrophoresis (the Rm values for these ODNs were determined by measuring from the center of the band). ODNs containing four or more G residues (11–16, Table 3) show significant reduction in Rm when compared with same length ODNs containing two G residues or fewer (e.g. 17, 18 and A), indicating aggregation of G-rich oligonucleotides. In contrast, ODNs containing two to nine PPG residues have similar Rms to one another and to the controls. This PAGE assay shows that little or no aggregation is seen in MGB-ODNs containing up to nine consecutive PPG residues.
5′-Nuclease real-time PCR assays
The fluorogenic probes shown in Table 3 were compared for their ability to detect amplicon in a 5′-nuclease assay (Fig. 4). For each sequence, real-time PCR performance curves were compared between normal Gs and the corresponding PPG substituted probes. Similar fluorescent signal between G and PPG was observed only with probes containing three Gs (Fig. 4D) or fewer in a row. The probe with four consecutive Gs detected amplified target ∼50% as effectively as that of the corresponding PPG-containing probe. No detection of amplicon was observed with any probes containing more than four Gs in a row. Figure 4A and B show only the detection of amplicon with nine and five consecutive PPGs, respectively. This is analogous to the four-G performance cut-off results shown in the PAGE assay and indicates that aggregation of G-rich probes can limit performance. In contrast, successful amplification was observed with all PPG probes containing four to nine uninterrupted PPGs (probes 11–16). The real-time PCR detection data for probes with six to eight successive Gs are not shown.
Figure 4.
Real-time PCR curves showing the effect of PPG substitution on the fluorescent signal. Fluorogenic MGB-probes were prepared with either G or PPG at the underlined sequence and evaluated in the 5′-nuclease PCR assay. (The probes in A, B, C and D contain nine, five, four and three Gs or PPG, respectively.) Assays were performed in duplicate. The y-axis shows fluorescence units and the x-axis shows the number of PCR cycles. FAM is a fluorescein reporter group. Q is a non-fluorescent diazine analog. The Q-MGB linker structure is shown in Figure 3.
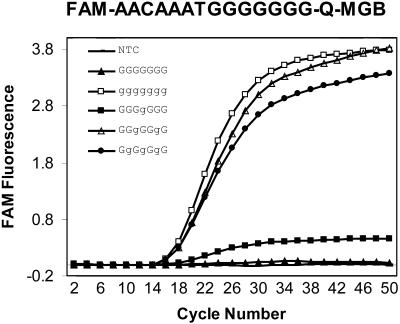
As shown in Figure 2, substitution of a single PPG in a run of Gs was sufficient to eliminate aggregation in normal ODNs. We therefore investigated the minimum number of PPGs required in a MGB-probe with seven consecutive Gs to achieve satisfactory detection of amplified target in a 5′-nuclease amplification assay (Fig. 5). The probe with seven Gs did not detect amplification. Substitution of one G with a PPG (g) in the middle of the row of seven Gs (GGGgGGG) barely detects amplification above background. Replacement of two Gs (GGgGgGG), three Gs (GgGgGgG) or all Gs with PPG in these probes, however, allowed robust detection of amplification. In general, self-association in probes with long runs of G can usually be prevented by PPG substitution at every third G.
Figure 5.
Relationship between MGB-probe performance and number of PPGs. G-rich MGB probes were prepared with either G or PPG as indicated and evaluated in the 5′-nuclease PCR assay. g represents PPG in the sequences.
Reduction of guanine-induced fluorescence quenching in probes for the 5′-exonuclease assay
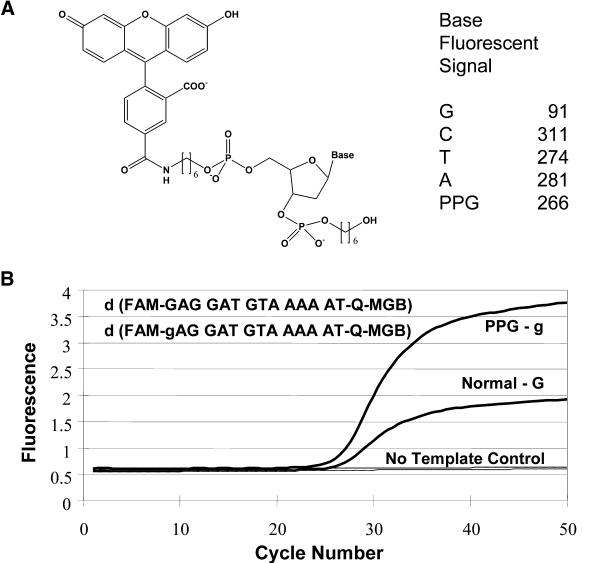
In the 5′-nuclease (TaqMan) embodiment of the real-time PCR assay, the Taq polymerase digests the fluorogenic probe primarily at the first internuceotide linkage, releasing the fluorophore attached to a 5′-terminal fragment. It has been observed that G adjacent to 5′-fluorescein in these probes quenches fluorescence. Sequence-specific fluorescence phenomena are well known and can plague quantitative PCR assays. The 5′-fluorescein in a probe for the 5′-exonuclease assay is quenched by a 5′-G because the G remains attached to the fluoroscein after exonucleolytic cleavage. In this study, we compared the fluorescence of fluorescein coupled to each of G, C, T, A or PPG monomers via their 5′-phosphates (Fig. 6A) and the fluorescence of 20 nM solutions of these compounds was determined. Excitation was at 494 nm and fluorescence emission was measured at 522 nm. Fluorescence emission of the FAM-G conjugate was 91 U, while the fluorescence emission of the FAM-PPG conjugate was 266 U. The other FAM-nucleotide monomers did not significantly influence the FAM emission signal. Thus, quenching of the fluorophore by G was substantially relieved when PPG was substituted for G, leading to an increase in fluorescence yield of the FAM-PPG of >2-fold, compared with FAM-G conjugate. This increase in quantum yield of FAM-PPG was also observed as increased signal of the fluorogenic MGB-probe as shown in the 5′-nuclease PCR assay (Fig. 6B). Probes containing either a G or a PPG adjacent to the 5′-fluorescein in a 5′-nuclease assay were compared. The PPG-containing probe shows approximately double the fluorescence of the G-containing probe after 5′-nuclease cleavage, similar to that seen in the mononucleotide model study.
Figure 6.
Effect of PPG on G quenching of fluorescein. (A) Structure and relative fluorescence of FAM-nucleotide monomers. The indicated compounds (0.02 µM in 1× PBS, pH 7.2) were analyzed (excitation = 494 nm, emission = 522 nm. (B) Effect of PPG substitution in the 5′-nuclease assay on fluorescent signal. g = PPG.
CONCLUSIONS
G-rich ODNs can cause problems in DNA synthesis and performance of hybridization assays. Unwanted aggregation can plague studies that assume ODNs behave as single strands with predictable hybridization performance. DNA probe design software can alert users to potential problem sequences, but certain genetic targets (for example SNPs) can require G-rich probes.
The results show that PPG, when substituted in G-rich probes, can significantly reduce self association. When PPG was substituted for G in MGB probes containing four or more consecutive Gs, G self-association was disrupted and 5′-nuclease assay performance was enhanced. In probes containing repetitive Gs, substitution of at least one PPG for G in every three consecutive Gs eliminates G self-association. This property of PPG makes it attractive for use in detection probes especially in light of its other advantages in assay performance.
In contrast to 7-deazaguanine, which destabilizes duplexes when substituted for G (13), PPG-containing ODNs are found to have ∼0.5–1.5°C/base higher Tms than their G-containing counterparts. This allows the design of probes with similar hybridization properties to G-containing probes. Data presented here also demonstrate improved mismatch discrimination properties of PPG-containing ODNs. The substitution of G with PPG could be beneficial in the case of relatively stable mismatches where G is involved (11).
PPG eliminated G self-association when substituted for G in fluorogenic MGB probes containing consecutive Gs. This property, in addition to its ability to reduce fluorescein quenching and improve mismatch discrimination, makes PPG-containing MGB probes especially attractive in SNP detection and allelic discrimination. We are currently expanding the use of PPG containing probes in other assays for genetic analysis.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Mohammad Ahmadian for critical review of the manuscript. The careful preparation of the figures by Dave Walburger is acknowledged.
REFERENCES
- 1.Southern E.M., Case-Green,S.C., Elder,J.K., Johnson,M., Mir,K.U., Wang,L. and Williams,J.C. (1994) Arrays of complementary oligonucleotides for analyzing the hybridization behaviour of nucleic acids. Nucleic Acids Res., 22, 1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- 3.Mohanty D. and Bansal,M. (1994) Conformational polymorphism in telomeric structures: loop orientation and interloop pairing in d(G4TnG4). Biopolymers., 34, 1187–1211. [DOI] [PubMed] [Google Scholar]
- 4.Murchie A.I.H. and Lilly,D.M.J. (2000) Tetraplex folding of telomeric sequences and the inclusion of adenine bases. EMBO J., 13, 993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen D. and Gilbert,W. (1992) Novel DNA superstructures formed by telomere-like oligomers. Biochemistry, 31, 65–70. [DOI] [PubMed] [Google Scholar]
- 6.Henderson E., Hardin,C.C., Walk,S.K., Tinoco,I.,Jr and Blackburn,E.H. (1987) Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell, 51, 899–908. [DOI] [PubMed] [Google Scholar]
- 7.Marsh T.C., Vesenka,J. and Henderson,E. (1995) A new DNA nanostructure, the G-wire, imaged by scanning probe microscopy. Nucleic Acids Res., 23, 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu M., Guo,Q. and Kallenbach,N.R. (1993) Thermodynamics of G-tetraplex formation by telomeric DNAs. Biochemistry, 32, 598–601. [DOI] [PubMed] [Google Scholar]
- 9.Protozanova E. and Macgregor,R.B.,Jr (2000) Thermal activation of DNA frayed wire formation. Biophys. Chem., 84, 137–147. [DOI] [PubMed] [Google Scholar]
- 10.Poon K. and Macgregor,R.B.,Jr (2000) Formation and structural determinants of multi-stranded guanine-rich DNA complexes. Biophys. Chem., 84, 205–216. [DOI] [PubMed] [Google Scholar]
- 11.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor, thermodynamics and NMR of DNA sequences with internal A·A, C·C, G·G and T·T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Rachubinski F., Eng,B., Murray,W.W., Blajchman,M.A. and Rachubinski,R.A. (1990) Incorporation of 7-deaza dGTP during the amplification step in polymerase chain reaction procedure improves subsequent DNA sequencing. DNA Seq., 1, 137–140. [DOI] [PubMed] [Google Scholar]
- 13.Seela F., Ramzaeva,N. and Becher,G. (1996) 7-Deazapurine DNA: Oligonucleotides containing 7-substituted 7-deaza-2′-deoxyguanosine and 8-aza-7-deaza-2′-deoxyguanosine. Collect Czech. Chem. Commun., 61, S258–S261. [Google Scholar]
- 14.Seela F. and Driller,H. (1988) 8-aza-7-deaza-2′-deoxyguanosine: phosphoramidite synthesis and properties of octanucleotides. Helv. Chim. Acta, 71, 1191–1198. [Google Scholar]
- 15.Seela F. and Becher,G. (1999) Oligonucleotides containing pyrazolo[3,4-d]pyrimidines: the influence of 7-substituted 8-aza-7-deaza-2′-deoxyguanosines on the duplex structure and stability. Helv. Chim. Acta, 82, 1640–1655. [Google Scholar]
- 16.Belousov E.S., Afonina,I.A., Kutyavin,I.V., Gall,A.A., Reed,M.W., Gamper,H.B., Wydro,R.M. and Meyer,R.B. (1998) Triplex targeting of a native gene in permeabilized intact cells: covalent modification of the gene for the chemokine receptor CCR5. Nucleic Acids Res., 26, 1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampe J.N., Kutyavin,I.V., Rhinehart,R., Reed,M.W., Michael,W., Meyer,R.B. and Gamper,H.B.,Jr (1997) Factors influencing the extent and selectivity of alkylation within triplexes by reactive G/A motif oligonucleotides. Nucleic Acids Res. 25, 4123–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutyavin I.V., Afonina,I.A., Mills,A., Gorn,V.V., Lukhtanov,E.A., Belousov,E.S., Singer,M.J., Walburger,D.K., Lokhov,S.G., Gall,A.A. et al. (2000) 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res., 28, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milesi D., Kutyavin,I., Luktanov,E.A., Gorn,V.V. and Reed,M.W. (1999) Synthesis of oligonucleotide conjugates in anhydrous dimethyl sulfoxide. In Phillips,M.K. (ed.), Methods In Enzymology, Vol. 313. Academic Press, Orlando, FL. pp.164–173. [DOI] [PubMed] [Google Scholar]
- 20.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide interactions. III. Circular dicroism studies of the conformation of deoxyoliognucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 21.Xia T., SantaLucia,J.,Jr, Burkard,M.E., Kierzek,R., Schroeder,S.J., Jiao,X., Cox,C. and Turner,D.H. (1998) Thermodynaic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry, 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
- 22.Lohkov S.G. and Pyshnyi,D.V. (1997) Thermodynamic and spectral properties of DNA miniduplexes with the terminal G:A mispairs and 3′ or 5′ dangling bases. FEBS Lett., 420, 134–138. [DOI] [PubMed] [Google Scholar]
- 23.Gamper H.B., Reed,M.W., Cox,M.W., Virosco,J.S., Adams,A.D., Gall,A.A., Scholler,J.K. and Meyer,R.B. (1993) Facile preparation of nuclease resistant 3′ modified oligodeoxynucleotides. Nucleic Acids Res., 21, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak K.J., Flood,S.J., Marmaro,J., Giusti,W. and Deetz,K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl., 4, 357–362. [DOI] [PubMed] [Google Scholar]
- 25.Wittwer C.T., Herrmann,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques, 22, 130–138. [DOI] [PubMed] [Google Scholar]
- 26.Seela F. and Driller,H. (1989) Alternating d(G-C)3 and d(C-G)3 hexanucleotides containing 7-deaza-2′-deoxyguanosine or 8-aza-7-deaza-2′-deoxyguanosine in place of dG. Nucleic Acids Res., 17, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh T.C. and Henderson,E. (1994) G-Wires: self-assembly of a telomeric oligonucleotide d(GGGGTTGGGG), into large superstructures. Biochemistry, 33, 10718–10724. [DOI] [PubMed] [Google Scholar]
- 28.Watt J.R. and Davis,P.W. (1996) Kinetics of G-quartet-mediated tetramer formation. Biochemistry, 35, 8002–8008. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S., Reed,M.W., Gamper,H.B.,Jr, Gorn,V.V., Lukhtanov,E.A., Foti,M., West,J., Meyer,R.B. and Schweitzer,B.I. (1998) Solution structure of a highly stable DNA duplex conjugated to a minor groove binder. Nucleic Acids Res., 26, 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]