Abstract
The conditionally-lethal pso4-1 mutant allele of the spliceosomal-associated PRP19 gene allowed us to study this gene’s influence on pre-mRNA processing, DNA repair and sporulation. Phenotypes related to intron-containing genes were correlated to temperature. Splicing reporter systems and RT–PCR showed splicing efficiency in pso4-1 to be inversely correlated to growth temperature. A single amino acid substitution, replacing leucine with serine, was identified within the N-terminal region of the pso4-1 allele and was shown to affect the interacting properties of Pso4-1p. Amongst 24 interacting clones isolated in a two-hybrid screening, seven could be identified as parts of the RAD2, RLF2 and DBR1 genes. RAD2 encodes an endonuclease indispensable for nucleotide excision repair (NER), RLF2 encodes the major subunit of the chromatin assembly factor I, whose deletion results in sensitivity to UVC radiation, while DBR1 encodes the lariat RNA splicing debranching enzyme, which degrades intron lariat structures during splicing. Characterization of mutagen-sensitive phenotypes of rad2Δ, rlf2Δ and pso4-1 single and double mutant strains showed enhanced sensitivity for the rad2Δ pso4-1 and rlf2Δ pso4-1 double mutants, suggesting a functional interference of these proteins in DNA repair processes in Saccharomyces cerevisiae.
INTRODUCTION
The radiation-sensitive pso4-1 mutant of Saccharomyces cerevisiae, originally isolated as xs9 (1), shows a pleiotropic phenotype, including sensitivity to DNA cross-linking agents, nearly blocked sporulation, reduced mutability, and is a thermoconditional mutant with no viability at 36°C (2,3). Thus, pso4-1 is phenotypically similar to the recA mutant of Escherichia coli, as it combines mutagen and radiation sensitivity with a block in recombination and in induced mutagenesis (2,3). Heterologous expression with a multi-copy vector containing the E.coli recA gene restored the induced mutagenesis in the pso4-1 genetic background (4). The combination of the deficiency phenotypes in recombination and error-prone repair made PSO4 a very interesting candidate for further understanding of the interconnection between these DNA repair processes.
Cloning of the PSO4 gene by complementation of the pso4-1 mutant’s lack of UV-induced mutability and inability to sporulate revealed its allelism to the known yeast gene PRP19 (5). PRP19 encodes an essential splicing factor associated with a protein complex that is required for the first cleavage–ligation step in the splicing reaction (6–9). Up to now, excluding Prp19p, seven components of the Prp19p-associated complex [Ppr19-AC] are known: Cef1p (also known as Ntc85p), Ntc90p (also known as Syf1p) and Ntc77p (also known as Clf1p or Syf3p) are all encoded by essential genes and were identified by direct sequencing of purified components of the complex (10,11). The other four components, Snt309p (also named Ntc25p), Ntc20p, Ntc30p (also named Isy1p) and Ntc31p are all encoded by non-essential genes without any functional motifs in their protein sequences (10–14). Together, they form the Prp19-AC that associates with the spliceosome as a functional integral complex (11). Not bound tightly to small nuclear RNAs, Prp19p is, by definition, not an intrinsic component of the spliceosome, suggesting that its transient association (with the spliceosome) possibly mediates conformational rearrangements or stabilizes the structure of the spliceosome during U4-snRNA dissociation (11,14).
The recent characterization of the yeast PSO4/PRP19 human ortholog hNMP200 gene revealed a high evolutionary conservation of this new protein among metazoans, plants and parasites (15). Its extensive homology (up to 80%) and its abundance as a component of the nuclear matrix suggest that hNMP200 may have two functions: one as a structural protein of the nuclear scaffold and a second as a support for spliceosome binding and activity (15). However, databases lack metazoan or plant cell counterparts of other known members of the Prp19-AC and thus the existence of a similar complex in higher eukaryotes is not proven (11). The conditionally-lethal pso4-1 mutant allele of PRP19 (5), the presence of the human ortholog hNMP200 (15) and the recent progress made in characterization of the Prp19-AC (9–14) allow us to study this gene’s pleiotropic influence on metabolic processes in yeast (as diverse as pre-mRNA processing, DNA repair and sporulation) and hence, to better define the cellular functions of the PSO4/PRP19-encoded protein.
In this study, we addressed two questions: first, can the pleiotropic phenotype of the pso4-1 mutant be explained solely by impairment of the PRP19-AC? And second, does Prp19p/Pso4p, as suggested by the role of its human ortholog, interact with proteins other than the spliceosomal factors? The influence of the Prp19p/Pso4p on splicing could be measured by exploiting the temperature influence in the conditionally-lethal pso4-1 mutant. Possible binding partners different from the spliceosomal factors were collected via a two-hybrid screening (16) using the full-length LexA-PSO4/PRP19 DNA-binding domain fusion gene as bait and a genomic S.cerevisiae library. Amongst the potential interactors, we isolated a collection of different fusion proteins that could be grouped into four functional classes: cell cycle, chromatin structure and chromosome dynamics, DNA repair, and mRNA splicing.
MATERIALS AND METHODS
Yeast strains
The yeast strains used in this study are listed in Table 1. The pso4-1 strain MB1620-5A was mated with the strain L5 (cup1Δ::ura3) (17), Y00787 (rad14::kanMX4) (EURO SCARF, Frankfurt), Y05437 (rlf2::kanMX4) (EUROSCARF, Frankfurt) and SX46ARAD2° (rad2::TRP1) (courtesy of E. C. Friedberg, University of Texas, Dallas). Phenotypes of the haploid ascospores were determined by establishing the respective deletion markers: geneticin resistance (rad14:: kanMX4; rlf2::kanMX4), sensitivity to temperature (37°C-pso4-1), tryptophan prototrophy (rad2::TRP1) and sensitivity to copper (cup1Δ::ura3) as described previously (18).
Table 1. List of S.cerevisiae strains used in this study.
| Strain | Relevant genotype | Source |
|---|---|---|
| MB1620 5A | Matα ura3-52 ade2 adeX LYS2 his3 trp1 leu2 pso4-1 | This work |
| MB1620 5D | Matα ura3-52 ADE lys2 HIS3 TRP1 LEU2 PSO4 | This work |
| W303 | Mata/Matα ura3-1/ura3-1 ade2-1/ade2-1 trp1-1/trp1-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 can1-100/can1-100 PSO4/PSO4 | (41) |
| MG5128 | Mata/Matα ura3-52/ura3 ade2/ADE can1/CAN pso4-1/pso4-1 | (5) |
| MG5101 | Mata/Matα ura3-1/ura3-1 ade2-1/ADE trp1-1/TRP leu2-3,112/LEU his3-11,15/HIS can1-100/CAN pso4::HIS3/pso4-1 | (5) |
| LF1 1A | ade2 adeX ura3-52 leu2 trp1 his3 lys2 cup1::ura3 pso4-1 | This work |
| LF1 2A | ade2 adeX ura3-52 leu2 trp1 his3 lys2 cup1::ura3 PSO4 | This work |
| LF2 9A | Mata ade2 adeX ura3 leu2 his3 met15Δ0 TRP1 rad14::kanMX4 PSO4 | This work |
| LF2 9B | Mata ade2 adeX ura3 leu2 his3 met15Δ0 trp1 rad14::kanMX4 pso4-1 | This work |
| LF2 9C | Matα ADE2 ura3 leu2 his3 MET15 trp1 RAD14 pso4-1 | This work |
| LF2 9D | Matα ADE2 ura3 leu2 his3 MET15 TRP1 RAD14 PSO4 | This work |
| LF3 1A | Mata ADE2 ura3 leu2 his3 MET15 trp1 rlf2::kanMX4 PSO4 | This work |
| LF3 1B | Matα ADE2 ura3 leu2 his3 met15Δ0 TRP1 RLF2 pso4-1 | This work |
| LF3 1C | Mata ade2 adeX ura3 leu2 his3 MET15 TRP1 RLF2 PSO4 | This work |
| LF3 1D | Matα ade2 adeX ura3 leu2 his3 met15Δ0 trp1 rlf2::kanMX4 pso4-1 | This work |
| LF4 1A | Mata ADE2 ura3-52 his3 leu2 rad2::TRP1 pso4-1 | This work |
| LF4 1B | Mata ade2 adeX ura3-52 his3 leu2 trp1 RAD2 pso4-1 | This work |
| LF4 1C | Matα ADE2 ura 3–52 his3 LEU rad2::TRP1 PSO4 | This work |
| LF4 1D | Matα ade2 adeX ura3-52 his3 LEU trp1RAD2 PSO4 | This work |
| Y00787 | Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad14::kanMX4 | EUROSCARF |
| Y05437 | Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rlf2::kanMX4 | EUROSCARF |
| SX46A RAD2° | Mata ade2 his3-532 trp1-289 ura3-52 rad2::TRP1 | E. C. Friedberga |
| L5 | Mata cup1::ura3 leu2 trp1 ura3-52 lys2 his3 ade | (20) |
| EGY48 | Mata ura3 his3 trp1 LexAOp(6)::LEU2 | R. Brentb |
aLaboratory of Molecular Pathology, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, Texas, USA.
bMassachusetts General Hospital, Boston, MA, USA.
Genetic and molecular biological methods
Techniques in yeast genetics were according to Rose et al. (19) and standard molecular techniques were performed according to Sambrook et al. (20) and Ausubel et al. (21).
Escherichia coli strains and plasmids
Escherichia coli strains XL1-blue (Stratagene), KC8 (22) and TOP10 (Invitrogen) were used as recipients for cloning procedures. The following vectors were used: pLexAPRP19 (contains the PSO4/PRP19-coding ORF fused to the LexA DNA-binding domain of pEG202); ActSNT309 (contains the SNT309-coding ORF fused to the GAL4-activation domain of pACT2) and ActPRP19 (contains the PSO4/PRP19-coding ORF fused to the GAL4-activation domain of pACT2) were a gift from Soo-Chen Cheng (Institute of Molecular Biology, Academia Sinica, Nankang, Taiwan). pSB18 (TRP1, CEN/ARS, AmpR), a single copy plasmid containing the ACT1- lacZ splicing reporter construct, was kindly provided by C. Columbano (Instituto de Química, Universidade de São Paulo, SP, Brasil). pJU83 (LEU2, 2µ, AmpR), a multicopy plasmid containing the ACT1-CUP1 splicing reporter construct, was kindly provided by C. Guthrie (Department of Biochemistry and Biophysics, University of California, San Francisco). Single copy plasmid pMG470 (5), containing a functional PRP19 gene, was employed to complement pso4-1 in mutability studies.
Media and growth conditions
Yeast strains were grown in YEPD medium (1% yeast extract, 2% peptone, 2% glucose) at 23, 25, 28, 30 or 33°C, according to the needs of the experiments. For selective growth, either YEPD plus geneticin (G418, Calbiochem, 0.2 mg/ml) or SynCo (SC) (0.67% yeast nitrogen base from DIFCO/USB, 2% glucose, 1% ammonium sulfate) supplemented with the appropriate essential nutrients (40 µg/ml) was used. For detection of canavanine-resistant mutants, canavanine sulfate (Sigma) was added at 40 µg/ml to appropriately supplemented SynCo media. The sporulation medium (KAC) contained 1% potassium acetate, 0.1% yeast extract and 0.05% glucose. Diploids were sporulated on KAC agar for 3–5 days. Sporulation efficiency was calculated by determining asci in a counting chamber. The medium for plates was solidified with 2% agar.
Treatments with NaCl, calcofluor white, benomyl and caffeine
Stationary cultures were serially diluted in 1:10 steps and 5–10 µl aliquots were then spotted onto YEPD medium containing NaCl (900 mM), benomyl (40 µg/ml), caffeine (0.5 µg/ml) and calcofluor white (30 µg/ml), prepared as described previously (23–25).
Mutagen treatments and forward mutation analysis
UVC. Different concentrations of cells in the exponential phase of growth (∼107 cells/ml) were spread immediately in triplicate on YEPD plates and, after drying, irradiated with UVC (Stratalinker, Stratagene) with doses ranging from 0 to 60 J/m2. For drop tests, stationary cultures were serially diluted in 1:10 steps and 5–10 µl aliquots were then spotted onto YEPD medium. After drying, the plates were UVC-irradiated as described above.
8-MOP + UVA. Suspensions of 5 × 106 cells/ml in exponential phase were treated with photoactivated 8-methoxypsoralen (Sigma), 8-MOP + UVA according to Henriques and Moustacchi (26). After all treatments, the plates were incubated for 3–5 days at the appropriate temperature in the dark. Survival data represent the average of at least three experiments.
Forward mutation analysis. Drops (20 µl) of cell suspension (2 × 108/ml) were spotted on SynCo + Can media and, after drying, UVC-irradiated at doses ranging from 0 to 60 J/m2. Incubation was for 5 days at temperatures of 23, 28 and 33°C. Survival was determined by plating appropriate dilutions on SynCo media. UVC irradiation and growth procedures were as described above.
Two-hybrid analysis
Two-hybrid analysis was carried out essentially as described by Gyuris et al. (22). Yeast strain EGY48, containing the reporter plasmid pSH18-34 and the bait plasmid pLexA-PSO4, was transformed with a yeast genomic library cloned into the prey plasmid pJG4-5 (27). Plasmids were isolated from yeast that survived selection for leucine prototrophy on galactose and showed lacZ expression on X-Gal-galactose plates. Escherichia coli strain K12 KC8 pyrF::Tn5, hsdR, leuB600, trpC9830, lacD74, strA, galK, hisB436 was used for the rescue of the plasmids as described by Gyuris et al. (22). Plasmid DNA was sequenced with an Applied Biosystems (Foster City, CA) sequencer. All sequences obtained were submitted to a BLAST search at MIPS (Munich Information Center for Protein Sequences) (28).
Construction of pLexAPso4-1
PCRs of genomic DNA using specific primers [PSO4N (5′-GCCAAGAAAGCCAACTAGGG-3′) and PSO4C (5′-GAAAGTACAAACGTGTCAGCG-3′)] and a high-fidelity thermostable DNA polymerase (Pwo®, Roche Molecular Biochemicals) were used to amplify the sequence of the pso4-1 mutant allele. Standard molecular techniques were used to replace a 645 bp internal fragment (BanII to BstEII) within the PRP19 coding region of pLexAPRP19 with the fragment containing the L45S mutation, yielding pLexAPso4-1. The expression of the LexA fusion baits was analyzed by western blotting using a monoclonal antibody against LexA (Clontech Laboratories Inc.).
Sequencing of the pso4-1 mutant allele
PCRs of genomic DNA, using specific primers and a high-fidelity thermostable DNA polymerase (Pwo® from Roche Molecular Biochemicals), were performed to amplify the sequence of the mutant allele. The PCR product was cloned in the pZero-2 vector (Invitrogen) and five individual clones were completely sequenced on both strands in an automated sequencer (Abi Prism A-377).
RT–PCR analysis
Total RNA was purified from exponential phase cultures using the QIAGEN RNeasy kit. Equal amounts of total RNA (1 µg) were subjected to cDNA synthesis using the specific antisense primer for the S.cerevisiae ECM33 gene (5′-TTATCTACATATAAAATTCAGTGATGAACC-3′) and SUPERSCRIPT II reverse trancriptase from Gibco-BRL. PCR was carried out with 10% of the first strand cDNA synthesis reaction [with the sense primer (5′-ATGCAATTCAAGAACGCTTTGACTG-3′) and antisense primer (5′-TTATCTACATATAAAATTCAGTGATGAACC-3′), with the sense primer beginning at the ATG start codon] which was carried out according to the manufacturer’s protocol.
β-Galactosidase assay
Exponential phase cultures (∼2 × 107 cells/ml) were used to assay the activity of the ACT1-lacZ splicing reporter (pSB18) and from the pSH18–34 two-hybrid reporter plasmid. β-Galactosidase activity was assayed and quantified according to Ausubel et al. (21). Three to four individual transformants were assayed in liquid SC lacking the appropriate nutrients. When using the ACT1-CUP1 fusion construct, stationary cultures were serially diluted by 1:10 steps and 5–10 µl aliquots were spotted on SynCo media plates containing 10 mM CuSO4, prepared as described by Lesser and Guthrie (18). In ACT1-lacZ and ACT1-CUP1 splicing reporters the lacZ and CUP1 coding ORFs are fused in frame with the initial part of the second exon of the ACT1 coding ORF, thus allowing β-galactosidase activity or CuSO4 resistance if the pre-mRNA transcript is correctly processed.
RESULTS
Temperature-dependent splicing-related phenotypes
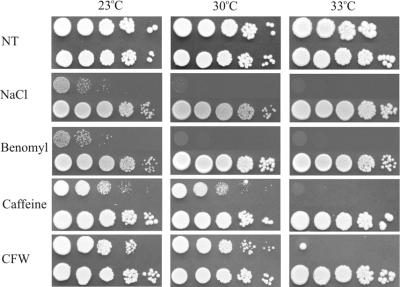
The allelism of PSO4 and PRP19 (5) suggested that an altered Pso4-1p would influence more than one cellular function, since non-effective pre-mRNA splicing could influence the expression of up to 238 intron-containing genes of S.cerevisiae (28). Therefore, we tested the temperature-dependent expression of some selected phenotypes correlated to intron-containing genes in the pso4-1 mutant. Figure 1 shows that indeed all phenotypes were influenced by changes of temperature and that there is significantly higher resistance to caffeine and to calcofluor white at the permissive temperature of 23°C. At 33°C the pso4-1 mutant is highly sensitive to all treatments.
Figure 1.
Temperature-dependent splicing-related phenotypes tested at 23, 30 and 33°C. Upper lines show pso4-1 mutant strain. Lower lines show wild-type strain. NT, no treatment; NaCl (900 mM); benomyl (40 µg × ml–1); caffeine (0.5 mg × ml–1) and CFW, calcofluor white (30 µg × ml–1).
To test if the phenotypes were most likely a consequence of impaired pre-mRNA processing, the splicing reporter system ACT1-lacZ (pSB18 plasmid) and standard RT–PCR were used to assay for splicing efficiency at the same temperatures. Figure 2A shows a good correlation between the temperature-dependent sensitivity phenotypes and ACT1-lacZ activity, where the pso4-1 mutant still has nearly 70% of pre-mRNA processing efficiency at 23°C but only ∼10% at 33°C. Additionally, another splicing reporter system, the ACT1-CUP1 fusion construct, was used to confirm splicing efficiency in the same conditions. Expressed from the multi-copy plasmid pJU83, ACT1-CUP1 allowed regular growth of the pso4-1 mutant in the presence of 10 mM CuSO4 at 23 and 30°C but not at 33°C, indicating low processing efficiency of the ACT1-CUP1 fusion reporter at the higher temperature (data not shown). Although untreated pso4-1 has a predicted splicing efficiency of only 10% at 33°C it survives reasonably well on YEPD at this temperature. Increasing the growth temperature to 34–35°C led to a lethal phenotype (data not shown).
Figure 2.
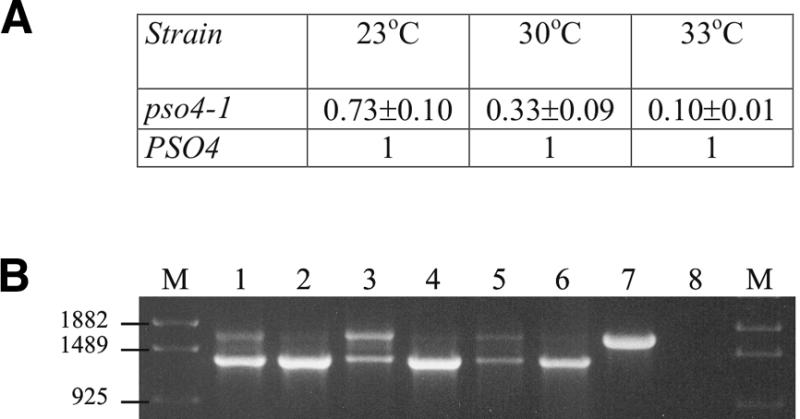
Evaluation of splicing efficiency at different temperatures. (A) Relative levels of ACT1-lacZ fusion splicing reporter system for the pso4-1 mutant strain at 23, 30 and 33°C. (B) ECM33-specific RT–PCR reactions of the pso4-1 mutant strain at 23°C (lane 1), 30°C (lane 3) and 33°C (lane 5). Lanes 2, 4 and 6 stand for the WT strain at the same temperatures, respectively. M is the molecular marker in base pairs. Lane 7 shows the positive control where total DNA was used to amplify the ECM33 gene. Lane 8 shows the control without reverse transcriptase.
The temperature-sensitive prp19 mutant accumulates unspliced precursor mRNA at non-permissive temperatures as described previously (6). We could complement our results obtained with the ACT1-lacZ splicing reporter system by determining splicing efficiency of the transcript of the intron-containing ECM33 gene in pso4-1 using RT–PCR. ECM33 was chosen due to its nearly constitutive expression during the cell cycle, the hypersensitivity of a ecm33 mutant strain to calcofluor white (25) and the reasonable size of its sole intron (329 bp), which allows easy distinction between processed and non-processed gene products. The differential efficiency of pre-mRNA processing relative to growth temperature is shown in Figure 2B. Confirming our data obtained with the ACT1-lacZ reporter, the amount of processed mRNA is significantly lower at 33°C as compared with ECM33 mRNA obtained from cells grown at 23°C. This supports the hypothesis of a thermoconditional splicing-related pleiotropic sensitivity phenotype for pso4-1. Similar influence by temperature variation changes was observed for the UVC-sensitivity phenotype of pso4-1 (described in detail below).
Is pso4-1-mediated UV sensitivity due to non-processed pre-mRNA of DNA repair genes?
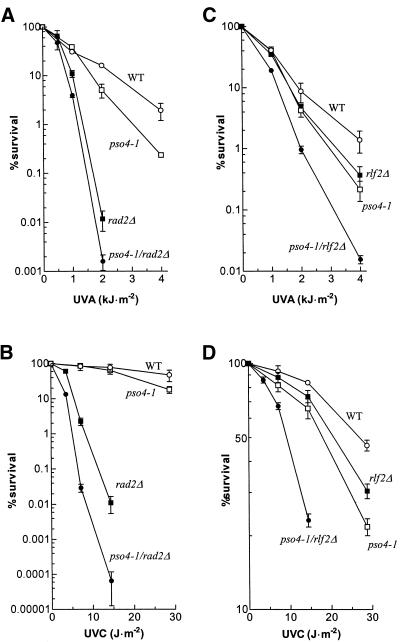
RAD14 is the only intron-containing DNA repair gene indispensable for the recognition/incision step of the nucleotide excision repair (NER) pathway in yeast (29). To exclude the possibility that most of the DNA repair-associated phenotypes of the pso4-1 mutant could result solely from faulty processing of RAD14-transcribed pre-mRNA, we tested rad14/pso4-1 double mutants for survival after UV irradiation at permissive and restrictive temperatures. Placing a rad14Δ mutant allele into a pso4-1 background yielded double mutants already with higher UVC sensitivity at 23°C than rad14Δ alone (synergistic interaction; Fig. 3, upper panel). The pso4-1 single mutant strain showed a temperature-dependent UV sensitivity phenotype with nearly wild-type resistance at 23°C and increased sensitivity at 30 and 33°C, respectively (Fig. 3, lower panel). This suggests a further indirect contribution of Pso4p/Prp19p in the repair of UVC damage, independent of RAD14 pre-mRNA processing. An overlapping effect of additional intron-containing genes involved in DNA repair (e.g. MMS2, RFA2 and KIN28), due to reduced splicing, cannot be excluded (see Discussion).
Figure 3.
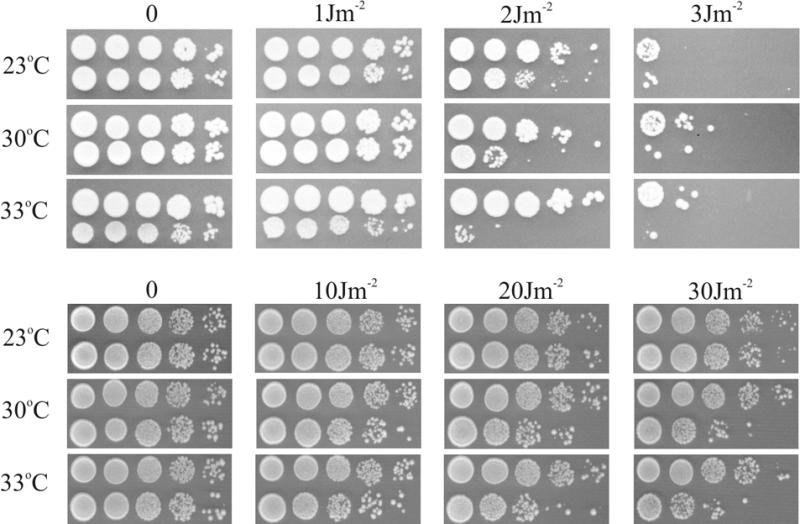
Thermoconditional UVC-sensitivity of single (pso4-1 or rad14Δ) and double mutant (pso4-1/rad14Δ) strains at given doses (J·m–2) and temperatures. Upper panel shows survival of rad14Δ (upper lines) and the double mutant strain pso4-1/rad14Δ (lower lines). Lower panel shows survival of pso4-1 strain (lower lines) and wild-type strain (upper lines).
Thermoconditional survival, mutability and sporulation in pso4-1 mutants
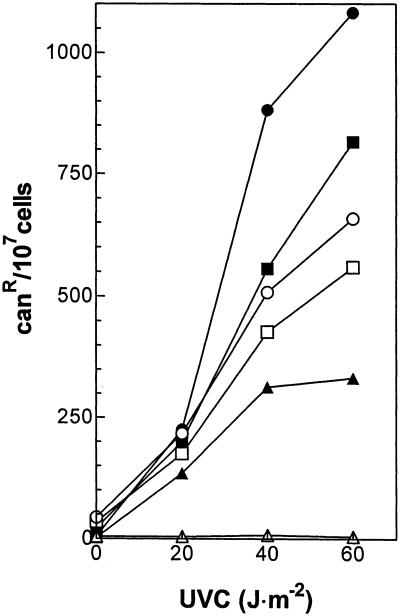
The association of defects in DNA repair with impaired recombination, induced mutagenesis and sporulation (3) in pso4-1 mutant strains prompted us to investigate further the influence of temperature on induced mutagenesis and sporulation efficiency. Table 2 summarizes the influence of temperature on sporulation efficiency of homoallelic (pso4-1/pso4-1) and heteroallelic (pso4-1/pso4::HIS3) diploid strains compared with the corresponding wild-type. Efficiency of sporulation decreases with increasing growth temperature, and no asci are detectable at 33°C in the mutant strains. A significant further decrease in sporulation efficiency can be observed in the heteroallelic pso4 mutant where the presence of only one pso4-1 mutant allele leads to a dramatic decrease of sporulation already at 23°C. When examining the temperature influence on induced mutagenesis in the canavanine forward mutagenesis assay, a similar response is found in the haploid pso4-1 mutant. Transformation of pso4-1 with a single copy plasmid containing the intact PRP19 gene can restore forward mutability and viability of the cells at 33°C to a significant extent (Fig. 4).
Table 2. Sporulation efficiency (%) of diploid strains containing different pso4-1 alleles (pso4-1/pso4-1, pso4-1/pso4::HIS3 and PSO4/PSO4), at 23, 30 and 33°C.
| Strains | 23°C | 30°C | 33°C |
|---|---|---|---|
| W303 WT(PSO4/PSO4) | 43.8 ± 2.3 | 50.0 ± 3.0 | 29.8 ± 4.7 |
| MG5128(pso4-1/pso4-1) | 13.2 ± 0.9 | 0.5 ± 0.7 | 0 |
| MG5101(pso4-1/pso4::HIS3) | 1.1 ± 0.3 | 0.1 ± 0.2 | 0 |
Figure 4.
Thermoconditional mutability in pso4-1 mutant strains transformed with the single-copy pMG470 (5), containing a functional PRP19 gene. Open symbols show pso4-1 mutant strains harboring the empty plasmid tested at 23°C (circles), 28°C (squares) and 33°C (triangles). Closed symbols show pso4-1 mutant strains harboring single copy pMG470, tested at 23°C (circles), 28°C (squares) and 33°C (triangles).
A single amino acid change in Pso4-1p leads to the thermoconditional phenotype and affects Pso4-1p self-interaction
With the aim to investigate why the pso4-1 mutation confers a pleiotropic phenotype, the complete sequence of the mutant allele was determined. The comparison of obtained sequences from five different clones with the databank-deposited wild-type sequence revealed a single amino acid substitution in triplet 45 (T→C transition, changing TTA to TCA), replacing leucine with serine [L45S] and thus changing this amino acid position from apolar to polar. The Pso4-[L45S] mutation affects a highly conserved amino acid residue located within the 130 amino acids encompassing the N-terminal region similar in all PSO4/PRP19 homologs. This region has a minimal similarity of 50% amongst potential orthologs of PSO4/PRP19 in various organisms (Fig. 5), a fact that suggests a high functional relevance of this protein domain.
Figure 5.
Amino acid alignment of Prp19p N-terminal domain and potential orthologs from various organisms. The dotted line denotes the first 130 amino acids, which display the highest degree of conservation. The L45S substitution position in Pso4-1p is indicated by an arrow. Identical residues are shown in red and conserved amino acids in yellow and gray. The accession numbers are PRP19At (Arabidopsis thaliana AAB80652), Q9AV81Os (Oryza sativa AAK27816), NMP200Hs (Homo sapiens AJ131186), SNEVMm (Mus musculus AAK49039), Q0JMJ4Rn (Rattus novergicus BAA95215), PRP19Dm (Drosophila melanogaster AAD46846), T10F2.4Ce (Caenorhabiditis elegans Q10051), CWF8Sp (S.pombe O14011), O77325Pf (Plasmodium falciparum CAB11109) and PRP19Sc (S.cerevisiae P32523).
Prp19p interacts with itself and is thought to exist in an oligomeric form in the PRP19-AC (9). Although not required for yeast growth, Snt309p interacts directly with Prp19p, playing an important role in modulating interactions of Prp19p with other associated components in stabilizing Prp19-AC (12,13). Therefore, the two hybrid assay (THA) was employed to evaluate the effect of the L45S mutation on the interaction of Pso4p with itself and with Snt309p. The LexAPso4-1 mutant bait was co-expressed with the activation domain fusion plasmids pACT2PRP19 and pACT2SNT309, expressing the Pso4p wild-type and Snt309p activation domain fusion proteins, respectively. Table 3 shows that Pso4-[L45S] considerably reduces the interaction of this altered Prp19p with itself but does not affect the interaction with the Prp19-AC-stabilizing protein Snt309p. This implies that the L45S mutation specifically affects the interaction of Pso4-1p with itself.
Table 3. Effect of the L45S mutation on the Prp19p/Pso4p self interaction and on interaction with Snt309pa.
| DNA binding domain plasmid (pEG202) |
Activation domain plasmid (pACT2) |
β-Galactosidase activityb |
| PLexA PRP19 | pACT2 PRP19 | 34.1 ± 4.9 |
| PLexA PRP19 | pACT2 SNT309 | 37.9 ± 6.2 |
| PLexA pso4-1 | pACT2 PRP19 | 12.4 ± 3.8 |
| PLexA pso4-1 | pACT2 SNT309 | 48.4 ± 16.8 |
| PLexA SNT309c | pACT2 SNT309 | 1.22 ± 0.24 |
aPlasmids that directed the synthesis of the DNA-binding domain (pEG202) and activation domain (pACT2) fusion proteins were introduced into EGY48. The strain also harbored pSH18–34, a very sensitive LexAop-lacZ reporter. All constructions expressed full length fusion proteins.
bExpressed in Miller Units. The values are averages from three to four independent transformants each assayed in duplicate.
Isolation of potential molecular partners of Pso4p/Prp19p using the two hybrid system
A two hybrid screen (THS) was used to identify proteins that may interact physically with Pso4p/Prp19p. The in-frame fusion of the entire PSO4/PRP19 ORF to the LexA DNA binding domain (pLexA-PSO4) displayed no intrinsic transcriptional activation when transformed into strains containing reporter constructs regulated by LexA DNA-binding sites (data not shown). A population of 1.1 × 106 independent transformants obtained after transformation with the prey library was pooled and aliquots were plated on selective medium containing galactose for induced expression of the activation domain fusion library. Some 224 clones were identified that allowed growth on SD-LEU as a result of the activation of the LexAop::LEU2 reporter construct. These clones were colony-purified, molecularly characterized by restriction mapping and re-tested for their ability to activate Gal-inducible transcription of two reporter constructs (LexAop:: LEU2 and pSH18–34; 22). Twenty-four transformants containing putative interactors, able to activate transcription of the two independent reporter constructs in the presence of LexA-PSO4/PRP19, were isolated. These same clones were unable to induce transcription when co-expressed with unrelated LexA DNA-binding domain fusions (pRFHM1; 22, LexAPSO5—yeast RAD16 allele and LexAHDF1—yeast Ku70 protein), indicating that the interaction with LexA-PSO4/PRP19 was specific. Further sequencing of the library plasmids revealed 13 different fusion protein products (Table 4). Five of these putative Pso4p/Prp19p interacting proteins are encoded by ORFs with as yet unknown function (with one being essential). The remaining eight interacting proteins can be roughly grouped into four functional classes: (i) DNA repair (one ORF), (ii) meiosis and cell cycle regulation (four ORFs, with one being essential), (iii) pre-mRNA splicing (one ORF) and (iv) chromatin structure and chromosome dynamics (two ORFs).
Table 4. Genes identified in a two-hybrid screen using pLEXAPRP19/PSO4 as bait.
| Gene |
Gene informationc |
Extent of fusiond |
|---|---|---|
| RAD2 (5)a | DNA repair; DNA endonuclease; repairosome | C-terminal second half of the protein |
| SWM1 (4) | Nuclear; required for the last phase of sporulation | Contains the whole ORF |
| YNL018c (3)b | Unknown function | Starts at amino acid 10 |
| HSL1 (2) | Cell cycle control, C156 and C111 | C-terminal two-thirds of the protein |
| YLR132c (2) | Unknown function, essential, induced during meiosis | Contains the whole ORF; starts at amino acid 2 |
| DBR1/PRP26 (1) | Lariat RNA splicing debranching enzyme | C-terminal third of the protein |
| MDS3 (1) | Meiosis; regulator of early meiotic genes, C16, C46 and C106 | C-terminal quarter of the protein |
| NUD1 (1) | Cell cycle regulatory protein, essential, C106 | C-terminal two-thirds of the protein |
| PLO1 (1) | Unknown function, weak similarity to Nip80p, C152 (the same as RAD16 and RAD52) and C185 | C-terminal half of the protein |
| RLF2 (1) | Chromatin assembly complex, subunit p90 | C-terminal two-thirds of the protein |
| YAL028w (1) | Interacts with EST1, a telomere elongation protein | C-terminal; starts at amino acid 494 |
| YDR124w (1) | Unknown function | C-terminal third of the protein |
| YML083c (1) | Unknown function | C-terminal half of the protein |
aFrequency of independent isolation is shown in parentheses.
bC indicates association of the respective isolate with the protein complex from the Yeast protein complex database (http://yeast.cellzome.com/).
cAccording to Mewes et al. (28) and Costanzo et al. (44), SGD (http://genome-www.stanford.edu/Saccharomyces/), and the Yeast protein complex database (http://yeast.cellzome.com/).
dSegment of each ORF fused to the transcriptional activation domain constructs.
Out of the 13 different fusion proteins, we focused our efforts on the study of Pso4p/Prp19p interaction with the three most relevant candidates, corresponding to the genes RAD2, RLF2 and DBR1. Encoding a protein homologous to the human xeroderma pigmentosum group G (XPG) and indispensable for the 3′ recognition/incision step of the NER (30,31), Rad2p is a component of the nucleotide excision factor 3 (32). RLF2 has been isolated as a gene required for normal distribution of the telomere-binding Rap1p protein within the nucleus (33) and is also involved in assembling of nucleosomes on newly replicated DNA, in DNA repair and telomere silencing (33–35). Deletion of RLF2 increases the UVC sensitivity of yeast mutants (36) blocked in any of the three known DNA repair groups (NER, RAD52-mediated recombinational DNA repair and RAD6-mediated postreplicative DNA repair) (37). DBR1 was identified and first cloned in yeast using a genetic screen aimed at identifying cellular factors involved in Ty1 retro-transposition (38). The dbr1 mutation in S.cerevisiae reduces the Ty1 transposition frequency about 10-fold and leads to a severe defect in intron degradation, i.e. high levels of excised introns accumulate as lariat structures lacking their 3′ tail (38). The C-terminal second half of the RAD2 gene-coding region was independently isolated five times via THS whereas RLF2 and DBR1 were isolated only once. Table 5 shows the induced specific expression of lacZ reporter gene in the presence of the LexAPSO4/PRP19 bait and Rad2, Dbr1 and Rlf2 prey proteins. The interaction study of Rad2, Rlf2 and Dbr1 prey proteins was further investigated by testing their ability to interact with the LexApso4-1 mutant bait (harboring the L45S mutation). Comparing the induced β-galactosidase activity obtained with wild-type and mutated LexA fusion baits of Prp19p/Pso4p, we show that the L45S single amino acid substitution drastically reduces the level of interaction of Pso4-1p with itself and with these three potential interactors.
Table 5. Effect of the L45S mutation on the interaction between Prp19p/Pso4p and its potential interacting proteins isolated in the THSa.
| DNA binding domain plasmid (pEG202) | Activation domain plasmid (pJG4-5) | Galactose | |
|---|---|---|---|
| Leu | β-Galactosidase activityb | ||
| PLexA GAL4 | pJG4-5 (HA-tagged) | +++ | 1075 ± 210.4 |
| PLexA Bicoid | pJG4-5 (HA-tagged) | – | 0.3 ± 0.1 |
| PLexA PRP19/PSO4 | pJG4-5-RAD2 (amino acid 481–1031) | +++ | 140.0 ± 49.1 |
| PLexA PRP19/PSO4 | pJG4-5-RLF2 (amino acid 81–606) | +++ | 188.4 ± 64.2 |
| PLexA PRP19/PSO4 | pJG4-5-DBR1 (amino acid 239–405) | +++ | 54.0 ± 27.2 |
| PLexA pso4-1 | pJG4-5-RAD2 (amino acid 507–1031) | – | 19.3 ± 5.6 |
| PLexA pso4-1 | pJG4-5-RLF2 (amino acid 81–606) | – | 13.3 ± 7.5 |
| PLexA pso4-1 | pJG4-5-DBR1 (amino acid 239–405) | – | 14.9 ± 1.9 |
aPlasmids that directed the synthesis of the DNA-binding domain (pEG202) and activation domain (pJG4-5) fusion proteins were introduced into EGY48. In addition to the LexAop(6)-LEU2, the strain also harbored pSH18-34, a very sensitive LexAop-lacZ reporter. All constructions expressed full length fusion proteins except when indicated. pLexA GAL4 is a transcription activator and was used as positive control; pLexA Bicoid contains residues 2–160 of the Drosophila bicoid gene product and was used as negative control.
bExpressed in Miller Units. The values are averages from three to four independent transformants each assayed in duplicate.
Genetic interaction between PSO4, RAD2 and RLF2 after induced DNA damage
The pso4-1 mutant is sensitive to UVC and 8-MOP + UVA (2). In order to study the nature of the interactions found between PSO4, RAD2 and RLF2 with respect to DNA repair, survival was determined in single and double mutants after UVC and 8-MOP + UVA. The effect of temperature on pre-mRNA processing in pso4-1 strains was minimized by incubating at 25°C. Survival data in Figure 6 show that the combination of the two mutant alleles always increased the sensitivity to UVC (synergistic interaction) or to 8-MOP + UVA (additivity). Although the encoded proteins are needed for different cellular functions, their interference suggests an indirect or direct physiological interaction between them (via Prp19-AC, see Discussion) as both are needed for a complete wild-type-like DNA repair.
Figure 6.
Survival of haploid wild-type, pso4-1, rlf2Δ and rad2Δ mutants after UVC and 8-MOP + UVA. (A) and (B) show survival of pso4-1 in combination with rad2Δ for 8-MOP + UVA and UVC, respectively. (C) and (D) show survival of pso4-1 in combination with rlf2Δ for 8-MOP + UVA and UVC, respectively.
DISCUSSION
We aimed to test whether all repair-related phenotypes of the thermoconditional pso4-1 mutant, i.e. mutagen sensitivity, low induced mutability and impaired sporulation, can be explained solely by splicing defects, or if the PSO4/PRP19-encoded protein may also play a direct role in DNA repair. Thus, we first established a high evidence of correlation between growth temperature and phenotypes related to intron-containing genes and to the respective processing of the encoded pre-mRNAs. By using splicing reporter systems and RT–PCR we could confirm that splicing efficiency in pso4-1 was inversely correlated to growth temperature and that this mutant accumulates pre-mRNA at non-permissive temperatures, confirming results obtained with a conditional prp19 mutant (6,39). The differential influence of splicing efficiency on the phenotypes (Fig. 1) might be explained by the number of intron-containing genes contributing to its expression. Osmosensitivity can be associated with eight known intron-containing genes (28), three of which are essential [ARP2/ACT2 (40), GLC7 (41) and ACT1 (42)], while benomyl resistance depends on four known intron-containing genes (28), one of them the essential TUB1 (43). Calcofluor white sensitivity depends on three intron-containing genes, including the essential SPT14 (28), while caffeine sensitivity associates with two non-essential intron-containing genes (28).
Of over 6200 genes of the S.cerevisiae genome, 238 are thought to contain introns (28). These genes encode proteins with functions in diverse physiological tasks and thus their lower or non-expression by sub-optimal splicing will result in a pleiotropic phenotype (28). The high number of genes, and the existence of ORFs of yet unknown function, complicate analysis of the repair phenotype of the pso4-1 mutant. The intron-containing repair gene RAD14 (29) could be responsible for most of the thermoconditional UVC sensitivity phenotype of pso4-1 as reduced processing of its pre-mRNA at 33°C would lead to a failure of the DNA damage recognition/incision step of the very effective NER pathway (37). Comparison of single and double mutants of rad14Δ and pso4-1 alleles for their UVC sensitivity suggested an indirect contribution of Pso4p independently of RAD14 pre-mRNA processing. At least another three intron-containing genes, MMS2, KIN28 and RFA2, function in DNA repair (28,44). Thus, an overlapping synergistic effect due to reduced splicing of their pre-mRNAs in pso4-1 cannot be excluded. MMS2 is a member of the error-free postreplication repair pathway and encodes an ubiquitin-conjugating enzyme, whose deletion renders mutants only slightly UVC-sensitive (45). Both KIN28 and RFA2 are essential genes and encode proteins associated with the NER pathway (46–48). Therefore, reduced splicing activity in a pso4-1 mutant and hence impaired functionality of these and other unknown repair genes could negatively affect the NER and postreplication repair pathways, resulting in the observed pleiotropic mutagen sensitivity. However, at 23°C, the pso4-1 single mutant strain shows a phenotype with nearly wild-type resistance when compared with the double mutant at the same temperature (Fig. 3, lower panel), suggesting that Prp19p/Pso4p may contribute to DNA repair apart from its function in pre-mRNA processing. Interestingly, over-expressed RecA protein of E.coli can complement most of the UVC sensitivity of the pso4-1 mutant, a function that can be correlated with the recombination involvement of Rfa2p (4,49,50).
Since several intron-containing genes, e.g. MEI4, CIN2 and HOP2 and the recA-like DMC1, REC107/MER2 and REC114 are involved in specific steps in meiosis (51–59, respectively), sporulation should be a thermoconditional biological endpoint in pso4-1. Indeed, sporulation efficiency of homoallelic pso4-1 and heteroallelic pso4-1/pso4-1::HIS3 diploid strains was temperature-dependent (Table 2) and thus correlated to efficiency of pre-mRNA processing (Fig. 2). The residual splicing activity contributed by the single pso4-1 mutant allele in the heteroallelic diploid led to a significantly lower sporulation already at permissive temperature, thus indicating co-dominance of PSO4/PRP19 alleles in splicing activity in diploid yeast, as suggested previously for DNA repair (2,5).
UVC-induced mutagenesis in pso4-1 in the canavanine resistance forward mutational test was strictly temperature-dependent, and no mutation induction could be observed at 33°C (Fig. 4). However, this temperature was also prohibitive for cell survival, so that non-viability and mutability are closely correlated on SynCo media, whereas YEPD still allowed for some survival at this condition (Figs 1, 3 and 6). Mutability could be restored in pso4-1 by transforming with a single-copy PRP19/PSO4-containing plasmid (Fig. 4) so that reintroducing a functional Ppr19p/Pso4p suffices for nearly normal mutability in a pso4-1 background. It should be noted that mutation induction is shown as mutants/treated cells; since the transformants had higher UVC sensitivity than wild-type, induced mutants/surviving cells would be considerably higher and similar to values obtained for pso4-1 at 23°C.
The similarities existing between Prp19p-related proteins of several species (Fig. 5) (60) suggest an important biological function since early evolution. Ranging from 50 to >90% similarity, the region of the first 130 N-terminal amino acid residues is highly conserved, and the first 140 amino acids are 100% identical in rat, mouse and humans (60). Because of this conserved region, Prp19p has been proposed recently as a member of the U-box protein family (61). Amongst the known homologs, Prp19p/Pso4p of S.cerevisiae represents the only thoroughly characterized protein. The N-terminal region of yeast Prp19p is responsible for interaction with itself (homo-oligomerization) and with other proteins of the Prp19-AC (9). The L45S single amino acid substitution of Pso4-1p is located within this region, and this amino acid is conserved in all but one (Schizosaccharomyces pombe) of the Prp19p-related proteins (Fig. 5). A computer-based analysis (62) showed a 20% reduced predicted probability of a coil or turn in the secondary structure in the N-terminal domain of the altered Pso4-1p. Indeed the THA confirmed that the Pso4-[L45S] mutation strongly reduces the interaction of Pso4-1p with itself but not with the Prp19-AC stabilizing protein Snt309p (Table 3).
According to Tarn et al. (9) homo-oligomerization of Prp19p is a pre-requisite for Prp19-AC assembly. It is now believed that formation of the Prp19p-AC involves sequential interactions of its members (11,14). First, the stabilizing protein Snt309p binds to the Prp19p homotetramer, followed by binding of Cef1p; then Ntc30p, Ntc20p, Ntc31p, Ntc77p and Ntc90p interact with each other, building a sub-complex, which binds to Cef1p to form the functional Prp19-AC. By affecting the interaction of Prp19p with itself, the L45S mutation, therefore, confirms the importance of the first 130 N-terminal amino acid residues of Prp19p for homo-oligomerization. Failing in this process it may also indirectly lower the interaction of the complex with other nuclear proteins, thus leading to the observed pleiotropic phenotype of the pso4-1 mutant. Given its thermoconditional expressivity, one could assume that, at lower temperatures, Pso4-1p might still function in a near-wild-type fashion, while higher temperatures would prevent correct assembly of the Prp19-AC. This hypothesis is supported by the fact that over-expression of the pso4-1 coding ORF from an ADH1-directed expression vector was able to complement growth at restrictive temperatures in a pso4-1 mutant, an expected result for a temperature-sensitive protein (35–37°C, data not shown).
Using THS we could isolate a set of putative binding partners of Prp19p (Table 4), among them the structure-specific DNA endonuclease Rad2p (30,31), the major subunit (p90) of the yCAF-1, Rlf2p (34) and the lariat-debranching enzyme, Dbr1p (38), but none of the already characterized Prp19-AC components (11). Perhaps our prey library is not representative, or assembly of Prp19-AC relies on indirect interactions, where some of the yeast cell Prp19-AC components would serve as interacting bridges, as these native proteins could bind the bait fusion construct. This last explanation is consistent with the observation that the LexAPRP19/PSO4 bait was able to complement the phenotypes of DNA repair and temperature sensitivity of the pso4-1 mutant strain (data not shown).
Primarily, the putative Prp19p interactors were evaluated for their ability to bind to the Pso4-1p, where the L45S mutation substantially reduced the activity of the lacZ reporter gene. In the same manner, as discussed above, the L45S mutation may interfere with the direct or indirect interaction (via Prp19-AC components) between Pso4-1p and its putative binding partners.
Because rad2Δ and rlf2Δ mutant strains both display a DNA repair-deficiency phenotype, for example, are sensitive to UVC and to 8-MOP + UVA (31,36,63) (Fig. 6), we checked whether they belonged to the same epistasis group of repair as PSO4. The double mutants rad2Δ/pso4-1 and rlf2Δ/pso4-1 showed increased sensitivity over the single mutant strains, for both UVC and 8-MOP + UVA (Fig. 6). According to analysis of survival (64) the mutant alleles interacted synergistically for UVC sensitivity (formally indicating competition for the substrate in DNA repair) and showed additivity for 8-MOP + UVA (indicating non-overlap in removal of DNA damage controlled by the two proteins).
Our results are consistent with the proposed function for Prp19p as a spliceosome-associated protein in pre-mRNA processing (11). Regarding DNA repair, impaired processing of pre-mRNA of intron-containing genes, especially those involved in recombination (e.g. MEI4 and the recA-like MER2/REC107, REC114 and DMC1), repair (e.g. MMS2, RFA2, RAD14 and KIN28), cell cycle/chromosomal structure/segregation (e.g. UBC9, GLC7, HOP2, CIN2, MOB1) (28,44) could lead to the observed phenotypes. As the NER pathway is responsible for most of the UVC-induced repair, impairing its function by blocking expression of genes RFA2, RAD14 and KIN28 would lead to the observed UVC sensitivity.
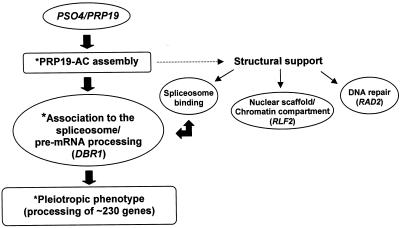
The recent characterization of the human hNMP200 ortholog of Pso4p/Prp19p as a component of the nuclear matrix (15) allows us, however, to discuss an additional function for Prp19p. There is mounting evidence that the nuclear matrix is involved in a variety of nuclear processes and in genome maintenance, e.g. DNA replication, DNA repair, transcription and RNA processing (65–76). Like hNMP200, Prp19p could thus be a yeast nuclear matrix protein and, as a member of the nuclear non-chromatin scaffold, it might provide structural support for the RNA processing machinery and for protein complexes engaged in DNA replication, transcription and repair. Thus, the isolation (via THS) of Rad2p, Rlf2p and Dbr1p interacting with Prp19p would be meaningful (see model in Fig. 7). By adopting a biochemical procedure originally developed to identify proteins associated with the nuclear matrix, Park et al. (77) could show tight binding of human XPG protein (the yeast Rad2p equivalent) to this nuclear fraction. As part of yCAF-1 major complex, Rfl2p fulfils its dual role in replication- and repair-coupled chromatin assembly (78,79) and its malfunction renders rfl2 mutants UVC and 8-MOP + UVA sensitive (Fig. 6). Finally, our isolation of DBR1 is fully consistent with the functional localization of its encoded protein, a lariat debranching enzyme (38) at splicing sites. Its localization at splicing sites via interaction with Prp19pAC would agree with the proposed dynamics of splicing reactions (11). In view of this, and the results obtained with the pso4-1 mutant, a possible role of Prp19p/Pso4p as a member of the nuclear scaffold cannot be excluded.
Figure 7.
Hypothetical model for Prp19p/Pso4p functions in the cell. The asterisk indicates the negative effect of Pso4-L45S mutation on PRP19-AC assembly and on pre-mRNA processing. The dotted arrow points to alternative roles with respect to proteins encoded by yeast genes RAD2, DBR1 and RLF2.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs R. Brent, E. C. Friedberg and C. Guthrie for kindly providing plasmids and strains. L.F.R., J.M.C. and D.B. held fellowships from CNPq. Collaborative research was sponsored by DAAD-CNPq and DAAD-CAPES travel grants to L.F.R., J.S., M.G., H.F., M.B. and J.A.P.H. This work was supported by CNPq, FAPERGS and GENOTOX (Laboratório de Genotoxicidade – UFRGS).
DDBJ/EMBL/GenBank accession no. AF500479
REFERENCES
- 1.Benathen A. and Beam,C.A. (1977) The genetic control of X-ray resistance of budding yeast cells. Radiat. Res., 69, 91–116. [PubMed] [Google Scholar]
- 2.Henriques J.A.P., Vicente,E.J., Silva,K.V.C.L. and Schenberg,A.C.G. (1989) PSO4: a novel gene involved in error-prone repair in Saccharomyces cerevisiae. Mutat. Res., 218, 111–124. [DOI] [PubMed] [Google Scholar]
- 3.Henriques J.A.P., Brozmanová,J. and Brendel,M. (1997) Role of PSO genes in the repair of photoinduced interstrand cross-links and photooxidative damage in the DNA of the yeast Saccharomyces cerevisiae. J. Photochem. Photobiol. B, 39, 185–196. [DOI] [PubMed] [Google Scholar]
- 4.Morais M.A. Jr, Brozmanová,J., Benfato,M., Duraj,J., Vlcková,V. and Henriques,J.A.P. (1994) The E.coli recA gene can restore the defect in mutagenesis of the pso4-1 mutant of S.cerevisiae. Mutat. Res., 314, 209–220. [DOI] [PubMed] [Google Scholar]
- 5.Grey M., Düsterhöft,A., Henriques,J.A.P. and Brendel,M. (1996) Allelism of PSO4 and PRP19 links pre-mRNA processing with recombination and error-prone DNA-repair in Saccharomyces cerevisiae. Nucleic Acids Res., 24, 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarn W.-Y., Lee,K.-R. and Cheng S.-C. (1993) PRP19: a novel spliceosomal component. Mol. Cell. Biol., 13, 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarn W.-Y., Lee,K.-R. and Cheng S.-C. (1993) Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl Acad. Sci. USA, 90, 10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarn W.-Y., Lee,K.R. and Cheng,S.C. (1993) The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol., 13, 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarn W.-Y., Hsu,C.-H., Huang,K.-T., Chen,H.-R., Kao,H.-Y., Lee,K.-R. and Cheng,S.-C. (1994) Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J., 13, 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai W.-Y., Chow,Y.-T., Chen,H.-R., Huang,K.-T., Hong,R.-I., Jan,S.-P., Kuo,N.-Y., Tsao,T.Y., Chen,C.-H. and Cheng,S.-C. (1999) Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem., 274, 9455–9462. [DOI] [PubMed] [Google Scholar]
- 11.Chen C.-H., Yu,W.-C., Tsao,T.Y., Wang,L.-Y., Chen,H.-R., Lin,J.-Y., Tsai,W.-Y. and Cheng,S.-C. (2002) Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res., 30, 1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H.-R., Jan,S.-P., Tsao,T.Y., Sheu,Y.-J., Banroques,J. and Cheng,S.-C. (1998) Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19. Mol. Cell. Biol., 18, 2196–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H.-R., Tsao,T.Y., Chen,C.-H., Tsai,W.-Y., Her,L.-S., Hsu,M.M.-T. and Cheng,S.-C. (1999) Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA processing. Proc. Natl Acad. Sci. USA, 96, 5406–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C.-H., Tsai,W.-Y., Chen,H.-R., Wang,C.-H. and Cheng,S.-C. (2001) Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J. Biol. Chem., 276, 488–494. [DOI] [PubMed] [Google Scholar]
- 15.Gotzmann J., Gerner,C., Meissner,M., Holzmann,K., Grimm,R., Mikulits,W. and Sauermann,G. (2000) hNMP200: a novel human common nuclear matrix protein combining structural and regulatory functions. Exp. Cell Res., 261, 166–179. [DOI] [PubMed] [Google Scholar]
- 16.Fields S. and Song,O.K. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 17.Umen J.G. and Guthrie,C. (1995) A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev., 9, 855–868. [DOI] [PubMed] [Google Scholar]
- 18.Lesser C.F. and Guthrie,C. (1993) Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics, 133, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose M.D., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Ausubel F., Brent,R., Kingston,R.E., More,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1996) Current Protocols in Molecular Biology, 3rd Edn. Wiley, USA.
- 22.Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- 23.Chowdhury S., Smith,K.W. and Gustin,M.C. (1992) Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J. Cell Biol., 118, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neff N.F., Thomas,J.H., Grisafi,P. and Botstein,D. (1983) Isolation of the beta-tubulin gene from yeast and demonstration of its essential function in vivo. Cell, 33, 211–219. [DOI] [PubMed] [Google Scholar]
- 25.Lussier M., White,A.M., Sheraton,J., diPaolo,T., Treadwell,J., Southard,S.B., Horenstein,C.I., ChenWeiner,J., Ram,A.F.J., Kapteyn,J.C. et al. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics, 147, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriques J.A.P. and Moustacchi,E. (1980) Isolation and characterization of pso mutants sensitive to photoaddition of psoralen derivatives in Saccharomyces cerevisiae. Genetics, 95, 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watt P.M., Louis,E.J., Borts,R.H. and Hickson,I.D. (1995) Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell, 81, 253–260. [DOI] [PubMed] [Google Scholar]
- 28.Mewes H.W., Frishman,D., Gruber,C., Geier,B., Haase,D., Kaps,A., Lemcke,K., Mannhaupt,G., Pfeiffer,F., Schuller,C., Stocker,S. and Weil,B. (2000) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 28, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankmann M., Prakash,L. and Prakash,S. (1992) Yeast RAD14 and human xeroderma pigmentosum group A DNA-repair genes encode homologous proteins. Nature, 355, 555–558. [DOI] [PubMed] [Google Scholar]
- 30.Higgins D.R., Prakash,L., Reynolds,P. and Prakash,S. (1984) Isolation and characterization of the RAD2 gene of Saccharomyces cerevisiae. Gene, 30, 121–128. [DOI] [PubMed] [Google Scholar]
- 31.Araujo S.J. and Wood,R.D. (1999) Protein complexes in nucleotide excision repair. [Erratum (2000) Mutat. Res., 459, 171–172.] Mutat. Res., 435, 23–33. [DOI] [PubMed] [Google Scholar]
- 32.Habraken Y., Sung,P., Prakash,S. and Prakash,L. (1996) Transcription factor TFIIH and DNA endonuclease Rad2 constitute yeast nucleotide excision repair factor 3: implications for nucleotide excision repair and Cockayne syndrome. Proc. Natl Acad. Sci. USA, 93, 10718–10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enomoto S., McCune-Zierath,P.D., Gerami-Nejad,M., Sanders,M.A. and Berman,J. (1997) RLF2, a subunit of the yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev., 11, 358–370. [DOI] [PubMed] [Google Scholar]
- 34.Kaufman P.D., Kobayashi,R. and Stillman,B. (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev., 11, 345–357. [DOI] [PubMed] [Google Scholar]
- 35.Enomoto S. and Berman,J. (1998) Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev., 12, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Game J.C. and Kaufman,P.D. (1999) Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo.Genetics, 151, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 38.Chapman K.B. and Boeke,J.D. (1991) Isolation and characterization of the gene encoding yeast debranching enzyme. Cell, 65, 483–492. [DOI] [PubMed] [Google Scholar]
- 39.Vijayraghavan U., Company,M. and Abelson,J. (1989) Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev., 3, 1206–1216. [DOI] [PubMed] [Google Scholar]
- 40.Schwob E. and Martin,R.P. (1992) New yeast actin-like gene required late in the cell cycle. Nature, 355, 179–182. [DOI] [PubMed] [Google Scholar]
- 41.Feng Z.H., Wilson,S.E., Peng,Z.Y., Schlender,K.K., Reinmann,E.M. and Trumbly,R.J. (1991) The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J. Biol. Chem., 266, 23796–23801. [PubMed] [Google Scholar]
- 42.Ng R. and Abelson,J. (1980) Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 77, 3912–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schatz P.J., Solomon,F. and Botstein,D. (1986) Genetically essential and nonessential alpha-tubulin genes specify functionally interchangeable proteins. Mol. Cell. Biol., 6, 3722–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costanzo M.C., Crawford,M.E., Hirschman,J.E., Kranz,J.E., Olsen,P., Robertson,L.S., Skrzypek,M.S., Braun,B.R., Hopkins,K.L., Kondu,P. et al. (2001) YPD™, PombePD™ and WormPD™: model organism volumes of the BioKnowledge™ library, an integrated resource for protein information. Nucleic Acids Res., 29, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broomfield S., Chow,B.L. and Xiao,W. (1998) MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl Acad. Sci. USA, 95, 5678–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tijsterman M., Tasseron de Jong,J.G., Verhage,R.A. and Brouwer,J. (1998) Defective Kin28, a subunit of yeast TFIIH, impairs transcription-coupled but not global genome nucleotide excision repair. Mutat. Res., 409, 181–188. [DOI] [PubMed] [Google Scholar]
- 47.Guzder S.N., Habraken,Y., Sung,P., Prakash,L. and Prakash,S. (1995) Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem., 270, 12973–12976. [DOI] [PubMed] [Google Scholar]
- 48.He Z., Wong,J.M.S., Maniar,H.S., Brill,S.J. and Ingles,C.J. (1996) Assessing the requirements for nucleotide excision repair proteins of Saccharomyces cerevisiae in an in vitro system. J. Biol. Chem., 271, 28243–28249. [DOI] [PubMed] [Google Scholar]
- 49.Sugiyama T., Zaitseva,E.M. and Kowalczykowski,S.C. (1997) A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem., 272, 7940–7945. [DOI] [PubMed] [Google Scholar]
- 50.Hays S.L., Firmenich,A.A., Massey,P., Banerjee,R. and Berg,P. (1998) Studies of the interaction between Rad52 protein and the yeast single stranded DNA binding protein RPA. Mol. Cell. Biol., 18, 4400–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menees T.M., Ross-MacDonald,P.B. and Roeder,G.S. (1992) MEI4, a meiosis-specific yeast gene required for chromosome synapsis. Mol. Cell. Biol., 12, 1340–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nag D.K., Scherthan,H., Rockmill,B., Bhargava,J. and Roeder,G.S. (1995) Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics, 141, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoyt M.A., Stearns,T. and Botstein,D. (1990) Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol. Cell. Biol., 10, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leu J.Y., Chua,P.R. and Roeder,G.S. (1998) The meiosis-specific Hop2 protein of S.cerevisiae ensures synapsis between homologous chromosomes. Cell, 94, 375–386. [DOI] [PubMed] [Google Scholar]
- 55.Leu J.Y. and Roeder,G.S. (1999) The pachytene checkpoint in S.cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell, 4, 805–814. [DOI] [PubMed] [Google Scholar]
- 56.Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 57.Rockmill B., Engebrecht,J.A., Scherthan,H., Loidl,J. and Roeder,G.S. (1995) The yeast MER2 gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics, 141, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malone R.E., Bullard,S., Hermiston,M., Rieger,R., Cool,M. and Galbraith,A. (1991) Isolation of mutants defective in early steps of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics, 128, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johzuka K. and Ogawa,H. (1995) Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics, 139, 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Rapp,B.A. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohi M.D. and Gould,K.L. (2002) Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA, 8, 798–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.King R.D. and Sternberg,M.J. (1996) Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci., 5, 2298–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meniel V., Magaña-Schwencke,N. and Averbeck,D. (1995) Preferential repair in Saccharomyces cerevisiae rad mutants after induction of interstrand cross-links by 8-methoxypsoralen plus UVA. Mutagenesis, 10, 543–548. [DOI] [PubMed] [Google Scholar]
- 64.Brendel M. and Haynes,R.H. (1973) Interactions among genes controlling sensitivity to radiation and alkylation in yeast. Mol. Gen. Genet., 125, 197–216. [DOI] [PubMed] [Google Scholar]
- 65.Pederson T. (1998) Thinking about a nuclear matrix. J. Mol. Biol., 277, 147–159. [DOI] [PubMed] [Google Scholar]
- 66.Martini E., Roche,D.M., Marheineke,K., Verreault,A. and Almouzni,G. (1998) Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol., 143, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ridgway P. and Almouzni,G. (2000) CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci., 113, 2647–2658. [DOI] [PubMed] [Google Scholar]
- 68.Bode J., Benham,C., Ernst,E., Knopp,A., Marschalek,R., Strick,R. and Strissel,P. (2000) Fatal connections: when DNA ends meet on the nuclear matrix. J. Cell. Biochem., 35 (suppl.), 3–22. [DOI] [PubMed] [Google Scholar]
- 69.Brown K. (1999) Nuclear structure, gene expression and development. Crit. Rev. Eukaryot. Gene Expr., 9, 203–212. [DOI] [PubMed] [Google Scholar]
- 70.Pederson T. (2000) Half a century of ‘the nuclear matrix’. Mol. Biol. Cell, 11, 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.vanDriel R., Wansink,D.G., van Steensel,B., Grande,M.A., Schul,W. and de Jong,L. (1995) Nuclear domains and the nuclear matrix. Int. Rev. Cytol., 162A, 151–189. [DOI] [PubMed] [Google Scholar]
- 72.Martelli A.M., Cocco,L., Riederer,B.M. and Neri,L.M. (1996) The nuclear matrix: a critical appraisal. Histol. Histopathol., 11, 1035–1048. [PubMed] [Google Scholar]
- 73.Kamiuchi S., Saijo,M., Citterio,E., de Jager,M., Hoeijmakers,J.H. and Tanaka,K. (2002) Translocation of Cockayne syndrome group A protein to the nuclear matrix: possible relevance to transcription-coupled DNA repair. Proc. Natl Acad. Sci. USA, 99, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Balajee A.S., Dianova,I. and Bohr,V.A. (1999) Oxidative damage-induced PCNA complex formation is efficient in xeroderma pigmentosum group A but reduced in Cockayne syndrome group B cells. Nucleic Acids Res., 27, 4476–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erdemir T., Bilican,B., Oncel,D., Goding,C.R. and Yavuzer,U. (2002) DNA damage-dependent interaction of the nuclear matrix protein C1D with translin-associated factor X (TRAX). J. Cell Sci., 115, 207–216. [DOI] [PubMed] [Google Scholar]
- 76.Lechner M.S., Levitan,I. and Dressler,G.R. (2000) PTIP, a novel BRCT domain-containing protein interacts with Pax2 and is associated with active chromatin. Nucleic Acids Res., 28, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park M.S., Knauf,J.A., Pendergrass,S.H., Coulon,C.H., Strniste,G.F., Marrone,B.L. and MacInnes,M.A. (1996) Ultraviolet-induced movement of the human DNA repair protein, Xeroderma pigmentosum type G, in the nucleus. Proc. Natl Acad. Sci. USA, 93, 8368–8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z., Shibahara,K. and Stillman,B. (2000) PCNA connects DNA replication to epigenetic inheritance in yeast. Nature, 408, 221–225. [DOI] [PubMed] [Google Scholar]
- 79.Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]