Abstract
With the increasing demand for higher throughput single nucleotide polymorphism (SNP) genotyping, the quantity of genomic DNA often falls short of the number of assays required. We investigated the use of degenerate oligonucleotide primed polymerase chain reaction (DOP-PCR) to generate a template for our SNP genotyping methodology of fluorescence polarization template-directed dye-terminator incorporation detection. DOP-PCR employs a degenerate primer (5′-CCGACTCGAGNNNNNNATGTGG-3′) to produce non-specific uniform amplification of DNA. This approach has been successfully applied to microsatellite genotyping. We compared genotyping of DOP-PCR-amplified genomic DNA to genomic DNA as a template. Results were analyzed with respect to feasibility, allele loss of alleles, genotyping accuracy and storage conditions in a high-throughput genotyping environment. DOP-PCR yielded overall satisfactory results, with a certain loss in accuracy and quality of the genotype assignments. Accuracy and quality of genotypes generated from the DOP-PCR template also depended on storage conditions. Adding carrier DNA to a final concentration of 10 ng/µl improved results. In conclusion, we have successfully used DOP-PCR to amplify our genomic DNA collection for subsequent SNP genotyping as a standard process.
INTRODUCTION
The role of single nucleotide polymorphisms (SNPs) has been implicated in a number of complex diseases including Alzheimer’s disease (1), osteoporosis (2,3), Crohn’s disease (4) and obesity (5). More recently, they have played a fundamental role in the emerging field of pharmacogenomics and adverse drug reactions (6,7). As such, it is becoming clear that SNPs have the potential to act as informative markers for the discovery and characterization of genes in complex disease (8). This will be made possible, in part, due to greater knowledge of the human genome sequence (9,10) and the increasing number of SNPs in databases (11). The estimated number of SNPs required to be screened genome-wide in order to detect a disease-causing gene could be as high as 500 000 (12) but this number may be lowered with the ongoing ascertainment of the extent of linkage disequilibrium (13–15).
With this increase in knowledge, there has been an associated explosion in the number of technologies that offer higher and higher throughput SNP genotyping on an industrial scale (16). These assays offer up to 500 000 genotypes a day at reagent costs as low as 1 c/SNP. However, with this throughput comes an increasing demand for DNA template on which to carry out these large numbers of reactions. For example, if 10 ng of DNA is required per reaction and 500 000 SNPs were to be screened then 5 mg of DNA is required. This would heavily deplete a valuable genomic DNA resource.
Two similar approaches have employed total genomic amplification to produce non-specific uniform amplification of DNA. By using a degenerate primer, a representation of the genome can be produced by means of the polymerase chain reaction (PCR) to act as a template for subsequent typing efforts. First, primer extension preamplification (PEP), which was originally developed to amplify DNA from a single cell (17), has been applied to HLA typing from mouth swabs (18) by generating a mixture of PEP reactions carried out in quadruplet to gain sufficient genome coverage. The primer for this approach is totally degenerate, made up simply of 5′-NNNNNNNNNNNNNNN-3′.
A more recognized approach for complete genome coverage in one reaction is the use of degenerate oligonucleotide primed PCR (DOP-PCR) which employs a more specific primer: 5′-CCGACTCGAGNNNNNNATGTGG-3′ (19). This approach has been successfully modified and applied to genomic DNA in order to carry out microsatellite genotyping (20). Although there was 100% success in amplification of each marker and correct assignment of genotypes, it was noticed that there was some preferential amplification of the shorter allele. A similar observation was made with microsatellites, Alu insertions and variable-length segments of the lipoprotein lipase gene (LPL) when using long DOP-PCR on rare archival anthropological samples (21). As microsatellite typing is based on the length of a repeat, the issue of band intensity is not of great importance. However, if the same approach is to be employed for SNP typing, it is vital that uniform amplification of alleles occurs for current technologies to call the genotype with confidence. The use of a DOP-PCR-amplified template has been applied previously to a small cohort and the typing of two SNPs (22). Another emerging application of this procedure in the context of SNP typing is the use of DOP-PCR for the reduction of genome complexity (23). In addition, the first paper published on a non-PCR-based approach was published only a few months ago using multiple displacement amplification (24).
In this work, we analyze the application of a DOP-PCR-amplified template in a high-throughput setting and the impact on quality. In addition, the role of DOP-PCR storage conditions is assessed.
MATERIALS AND METHODS
DOP-PCR amplification
Genomic DOP-PCR amplification was performed in a final volume of 10 µl on an ABI 9700 thermal cycler (PE Applied Biosystems, Foster City, CA). The PCRs contained 2 µM DOP primer (5′-CCGACTCGAGNNNNNNATGTGG-3′) and phenol/chloroform purified genomic DNA template with amounts ranging from 1 to 40 ng. In the PCR mix, using components of the DOP Master kit (Roche, Basel, Switzerland), were 200 µM dNTPs, 1.5 mM MgCl2, 0.01% (v/v) Brj35, 10 mM Tris–HCl (pH 8.3), 50 mM KCl and 0.25 U Taq DNA polymerase. An initial denaturation of 95°C for 5 min was followed by five cycles of 94°C for 1 min, 30°C for 90 s, ramping to 72°C over a 3 min period (3.5°C/15 s) and 72°C for 3 min, then 35 cycles of 94°C for 1 min, 62°C for 1 min and 72°C for 2 min (and increasing by 14 s each subsequent cycle) and completed by a final extension step of 72°C for 7 min.
Storage
DOP-PCR products were either used directly or stored at –20 or –70°C. In a subsequent experiment, column-purified pUC19 plasmid DNA was added to the DOP-PCR product to a final concentration of 10 ng/µl before freezing.
PCR and SNP genotyping
The DOP-PCR product was diluted to 10% in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). A 0.25 µl aliquot of this dilution was transferred as template to the provided PCR mixture by stamping twice using a 96 Long Pin Replicator (Incyte Genomics, St Louis, MO).
Touchdown PCR (25) was performed in a reduced primer and dNTP environment of 5 µl. This environment consisted of 0.2 µM primer mix, 50 µM dNTPs, 1.5 mM MgCl2, 20 mM Tris–HCl (pH 8.4), 50 mM KCl and 0.5 U hot start Taq DNA polymerase (Invitrogen, Carlsbad, CA). SNP accession numbers and PCR primers are reported in Table 1. An initial denaturation of 5 min at 95°C was followed by five cycles of 94°C for 30 s, 63°C for 20 s and 72°C for 30 s, then similarly five cycles each of touchdown (25) with annealing temperature from 60.5 to 55.5°C by gradation of 2.5°C and then 20 cycles of 94°C for 30 s, 53°C for 20 s and 72°C for 30 s and a final extension step of 72°C for 5 min. To remove excess primers and nucleotides, the resultant PCR product was incubated with 2 µl of 50% concentration Exo-SAP-IT (Amersham Biosciences, Freiburg, Germany) at 37°C for 45 min and subsequently denatured at 80°C for 15 min.
Table 1. Primer sequencesa used for PCR and subsequent FP-TDI SNP genotyping.
| SNPb | PCR sense | PCR antisense | TDI-primer extension |
|---|---|---|---|
| rs1457042 | AAGAGAGTTATCCTTATGTGG | CCTGTCTCAGTAGAGCATTTA | TTTCTAAATGATTATTACTAAGAAGCTA |
| rs943430 | CTTGCCTGCCACTCTCACTCT | GGCTGGGGGCTCATCTT | TCAGCGGGGGCAAGTACATCAGCCAA |
| rs877954 | CACTGCGCTCCCCTACC | ACCGCCTATTCCGTACACACT | GGCCATCCATTCTACACATGCTTG |
| rs1574570 | GCTGGATTGCAGGAAGGAT | TGCTGCCCTCAGGTTGG | AGCCTCTGCTGCTGTTTCCCCCA |
| rs731703 | GGTATGTGGTAGCCAGAGTCC | CCACAGCGGTAATAAGACAAG | GCACAGACCTGTTCTTTGCTGTCCTG |
| rs1800775 | CAGTTTCTCCCGGAGGCAGCCAATG | ACCCCAGAAACAGTCCTCTATGTAG | GCCAATGATCTCAGAGGCTGTATACCC |
| rs35735 | CACCTTCCTTTATTATTTACCCA | TCTGAATCTTGATCCAACACA | CAGGCAGCTGTAATCATGGGTAA |
| rs688 | AACTTCACTCCATCTCAAGCATC | CGTAAGCCACACCTCAAAGAC | CACTCCATCTCAAGCATCGATGTCAA |
| rs2076167 | GCTGAGAAGAGGAAGCTGGTG | GGCATTGTAGATGTGCTTGGAG | ACCTGTGGGTTGTACTGGCTCCCCTC |
| rs1846223 | CATGCCTAATATAATGGTGGAA | CAGCAATTGTTTCAAGGTAACT | CTTGCCTGTTCGATGAATATTCTGCATA |
aAll sequences in 5′→3′ orientation.
bNCBI dbSNP accession number.
An additional volume of 13 µl of primer extension mix was added containing 0.38 µM primer (Table 1) and the AcycloPrime-FP components of reaction buffer, AcycloPol and AcycloTerminator Mix (PerkinElmer Life Sciences, Boston, MA). An initial denaturation of 2 min at 95°C was followed by 10–40 cycles (based on optimization tests) of 94°C for 15 s and 55°C for 30 s. The resultant 30 µl assay was read in a 96-well black propylene plate (ABgene, Epsom, UK) at 1 s/well in the VICTOR2 V multilabel counter (PerkinElmer Life Sciences) using 485 nm excitation and 520 nm emission filters for the R110 dye and 544 nm excitation and 580 nm emission filters for the TAMRA label. The subsequent data were corrected using the G-factors 1.17 and 1.51 for R110 and TAMRA, respectively. The G-factor was calculated based on the average of the raw values from 10 water controls that corrected the negative reading to 50 mP for both dyes.
The obtained mP readings were clustered by means of an in-house software program using the K-means clustering algorithm. The software automatically assigns each data point to one of four clusters representing the three respective genotypes or non-callable data points (M.Kschischo, R.Kern, C.Gieger, M.Steinhauser and R.Tolle, manuscript in preparation). Automatically assigned genotypes were visually inspected and genotype calls were deleted for those data points that were not unequivocally assignable to a single cluster. The fluorescence polarization template-directed dye-terminator incorporation (FP-TDI) assay conditions were pre-tested for all SNPs on genomic and DOP-PCR-amplified template before starting the microtiter plate screen.
Sequencing
Individuals that gave differing genotypes between DOP and genomic templates were sequenced to resolve the correct genotype. In the 10 µl PCR mix was 200 µM dNTPs, 1.5 mM MgCl2, 20 mM Tris–HCl (pH 8.4), 50 mM KCl and 2.5 U hot start Taq DNA polymerase (Invitrogen). The cycling procedure was the same touchdown protocol as for the FP-TDI PCR. A 1 µl PCR was sequenced in a total volume of 10 µl with 1 µM primer and 4 µl of Big Dye terminator ready reaction mix (PE Applied Biosystems) with an initial denaturation of 95°C for 2 min 30 s followed by 25–40 cycles of 95°C for 10 s, 50°C for 10 s and 60°C for 4 min. The sequencing reactions were then vacuum dried and resuspended in 3 µl of dye and formamide. A 1 µl aliquot was loaded on an ABI 377 and run for 4 h. The data generated were processed in GeneScan (PE Applied Biosystems) and analyzed with Polyphred software (26).
RESULTS AND DISCUSSION
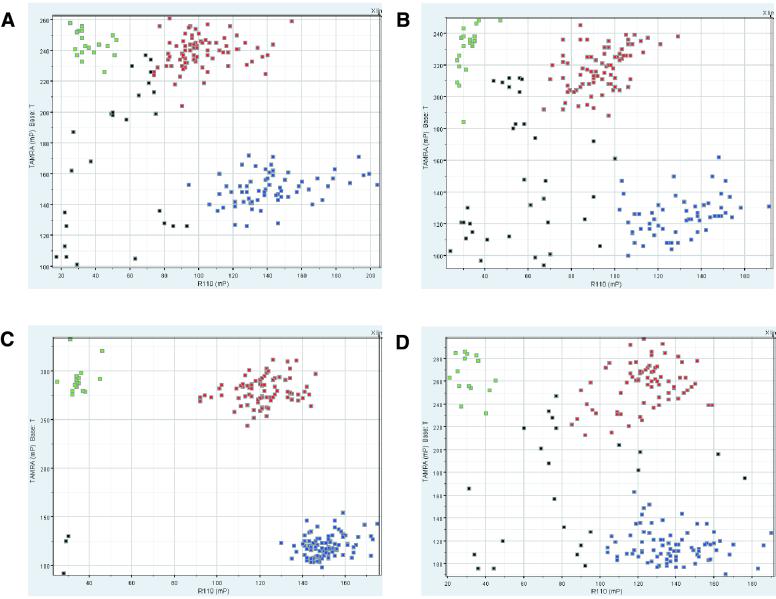
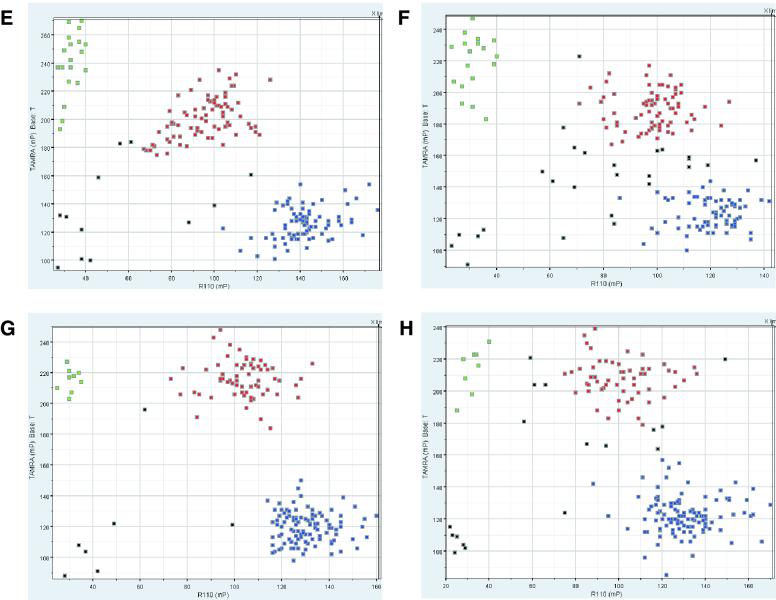
SNP genotyping was carried out on 10 SNPs from 10 different genes selected from the SNP database at the National Center for Biotechnology Information (dbSNP) (27). For six SNPs, two 96-well plates each and for four SNPs single 96-well plates were typed. Five wells on each plate were reserved for control reactions: reagent blank, commercially obtained CEPH-DNA (Applied Biosystems, Weiterstadt Germany), and three DNA samples to represent all SNP genotypes. When comparing FP-TDI results from genomic and DOP-PCR-amplified template, it is clear that different qualities of results are observed (Fig. 1). Data clusters are not as tight for the DOP-PCR-amplified template as for the genomic template independent of the SNP and the neighboring sequence. Although discrete clusters can be derived, the number of data points which cannot be assigned unequivocally to a single cluster increases when using the DOP-PCR-amplified template. The increase is reflected in the higher number of DNAs for which no genotype call can be made (Table 2). Samples without a genotype call can be differentiated into those which do not produce a signal (‘no call’) and those that fall into the overlap area between two genotype clusters (‘ambiguous call’). The DOP-PCR template and the genomic template were very similar with respect to no calls (1.3 versus 1.6% based on 1462 measured samples). A clear difference can be observed for ambiguous calls, where only 2.0% of genomic templates fall under this category compared to 5.8% from the DOP-PCR template. Discrepancies in unequivocal genotype calls were resolved by sequencing the genomic template, a technique considered the gold standard method for SNP genotype determination. Genotyping errors were discovered for both the genomic template and the DOP-PCR template. Whole genome amplification resulted in a slightly higher overall error rate of 0.7%. There is no specific trend in the type of errors obtained when genotyping the DOP-PCR-amplified template. One does not only observe the loss of alleles, i.e. the conversion of a true heterozygous sample into an apparent homozygous sample but also the creation of additional alleles or the complete conversion of alleles at a low rate. We investigated the role of the amount of DOP-PCR template by repeating an assay with 50% of our standard concentration (Table 2). This reduction resulted in an increase of no calls, ambiguous calls and genotyping errors. A visual inspection of PCR products from DOP-PCR templates on agarose gels demonstrated that samples without calls were normally characterized by absent bands. Samples that produced ambiguous or erroneous calls, however, did not display visible differences from normal samples (data not shown). Furthermore, the introduction of errors or ambiguous calls could not be attributed to differences in quality of the original DNA samples, as a repetition of the same experiment does produce erroneous and ambiguous calls for DNAs which are different from those of the first experiment (data not shown).
Figure 1.
(Previous page and above) DNA template comparison for FP-TDI genotyping with four public SNPs—NCBI dbSNP accession numbers rs1457042 (A and B), rs943430 (C and D), rs877954 (E and F) and rs1574570 (G and H). The green and blue squares represent the two forms of homozygous genotype calls, heterozygous calls are indicated by red squares, no calls are marked by black squares. A genomic DNA template (A, C, E and G) generally yielded tighter clusters than its DOP-PCR-amplified counterpart (B, D, F and H).
Table 2. Number of samples lacking a genotype call (no call), yielding ambiguous genotype calls (ambiguous call) or yielding erroneous genotype calls by use of genomic and DOP-PCR-amplified template.
| SNP-IDa | Sample size | No call | Ambiguous call | Total errorsb | Homoz. > heteroz.c | Heteroz. > homoz.c | Homoz. > homoz.c | Typed bases | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomic | DOP | Genomic | DOP | Genomic | DOP | |||||||
| rs1457042 | abs. | 182 | 4 | 8 | 6 | 7 | 0 | 0 | c/t | |||
| % | 2.2 | 2.2 | 6.6 | 6.0 | 0.0 | 0.0 | ||||||
| rs943430 | abs. | 182 | 1 | 3 | 0 | 15 | 0 | 1 | 1 | c/t | ||
| % | 0.5 | 1.6 | 0.0 | 8.2 | 0.0 | 0.5 | 0.5 | |||||
| rs877954 | abs. | 182 | 3 | 4 | 3 | 21 | 0 | 3 | 3 | c/t | ||
| % | 1.6 | 2.2 | 1.6 | 11.5 | 0.0 | 1.6 | 1.6 | |||||
| rs1574570 | abs. | 182 | 2 | 4 | 0 | 17 | 0 | 1 | 1 | c/t | ||
| % | 1.1 | 2.2 | 0.0 | 9.3 | 0.0 | 0.5 | 0.5 | |||||
| rs731703 | abs. | 182 | 6 | 2 | 11 | 16 | 3 | 1 | 1 | a/g | ||
| % | 3.3 | 1.1 | 6.0 | 8.8 | 1.6 | 0.5 | 0.5 | |||||
| rs1800775 | abs. | 91 | 0 | 1 | 3 | 1 | 0 | 1 | 1 | a/c | ||
| % | 0.0 | 1.1 | 3.3 | 1.1 | 0 | 1.1 | 1.1 | |||||
| rs35735 | abs. | 91 | 2 | 1 | 0 | 0 | 2 | 1 | 1 | a/c | ||
| % | 2.2 | 1.1 | 0.0 | 0.0 | 2.2 | 1.1 | 1.1 | |||||
| rs688 | abs. | 91 | 1 | 0 | 3 | 4 | 0 | 1 | 1 | c/t | ||
| % | 1.1 | 0.0 | 3.3 | 4.4 | 0 | 1.1 | 1.1 | |||||
| rs2076167 | abs. | 91 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | a/g | ||
| % | 0.0 | 0.0 | 1.1 | 2.2 | 0 | 1.1 | 1.1 | |||||
| rs1846223 | abs. | 188 | 0 | 1 | 2 | 2 | 3 | 0 | a/t | |||
| % | 0.0 | 0.5 | 1.1 | 1.1 | 1.6 | 0.0 | ||||||
| rs1846223d | abs. | 188 | n.d. | 4 | n.d. | 6 | n.d. | 3 | 3 | a/t | ||
| 0.5× | % | 2.1 | 3.2 | 1.6 | 1.6 | |||||||
| Sum | abs. | 1462 | 19 | 24 | 29 | 85 | 8 | 10 | 6 | 3 | 1 | a/t |
| % | 1.3 | 1.6 | 2.0 | 5.8 | 0.5 | 0.7 | 0.41 | 0.20 | 0.07 | |||
abs., absolute; homoz., homozygous; heteroz, heterozygous; n.d., not determined.
aNCBI dbSNP accession number.
bDiscordant genotypes were resolved using sequencing as the gold standard.
cNature of allelic errors obtained with the DOP-PCR-amplified template denoted as true genotype > apparent genotype.
dSNP rs1846223 was retyped with half-maximal concentration. Data not used in the sums below.
Whereas unequal amplification which has previously been described for microsatellite genotyping and DOP-PCR (20) could be the reason for the observed loss of heterozygous alleles, this effect cannot explain the observed generation of additional alleles at a low rate. Despite safety measures to avoid cross-contamination of samples, the extremely high rate of amplification makes it most likely that erroneous genotypes with additional alleles were generated by cross-contamination at a low rate. This assumption is supported by an analysis of the reagent blanks. Seven out of 44 wells with reagent blanks resulted in an erroneous genotype call. A simultaneous contamination of both negative control reactions per 192 samples was however never observed.
In summary, we do see a reduction in data quality when using a DOP-PCR-amplified template with FP-TDI genotyping that is mainly attributable to ambiguous genotype assignments. This is in contrast to the observations of Barbaux et al. (22) reporting a 100% concordance of genotypes derived from a DOP-PCR-amplified template. This higher success rate could be due to the much lower number of experiments and samples analyzed by these authors and by their use of a different SNP typing protocol. Their approach of using radiolabeled allele-specific oligos is however not suitable for automation and higher throughput SNP analysis.
How the decrease in accuracy impacts on the overall quality of a genetic study depends on the study design. In general, a certain degree of missing genotype information is less of a problem as these samples can be easily identified and data can be complemented. Thus, a conservative approach to genotype calling is recommended with a DOP-amplified template. The generation of erroneous genotypes poses more of a challenge. Error detection is very limited for case–control designs. In nuclear families, Mendelian inconsistencies will disclose in most cases ∼30% of the true genotyping errors (28,29) and even extended pedigrees cannot be considered to be free of errors. As this problem becomes more apparent with the increasing number of SNP studies, statistical methods have been developed to better accommodate for errors present in the genotype data set for both case–control design and transmission disequilibrium testing (30,31). Genotyping result quality can be improved both by analyzing samples in duplicate or by increasing the concentration of the DOP-PCR template in the subsequent PCR. However, both approaches will increase template consumption thwarting the initial intention of saving precious DNA. It remains to be seen if the recently reported advantages of multiple displacement amplification can be translated into a daily high-throughput routine, once this technology is available to the scientific community (24).
We also observed that upon storage of DOP-PCR products at –20°C subsequent SNP genotyping resulted in less compact data clusters. A visual inspection of DOP-PCR products fresh and after storage at –20°C for 2 and 4 weeks on agarose gels did not demonstrate any differences which would point to a sample degradation (data not shown). The effect could, however, be compensated for by adding additional plasmid DNA to the DOP-PCR template and at the same time by storing samples at –70°C.
In summary, we consider DOP-PCR amplification as a valuable means to substantially increase the amount of data that can be derived from precious clinical samples if researchers correctly deal with missing data and a modest increase in genotyping error.
REFERENCES
- 1.Martin E.R., Lai,E.H., Gilbert,J.R., Rogala,A.R., Afshari,A.J., Riley,J., Finch,K.L., Stevens,J.F., Livak,K.J., Slotterbeck,B.D. et al. (2000) SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am. J. Hum. Genet., 67, 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant S.F., Reid,D.M., Blake,G., Herd,R., Fogelman,I. and Ralston,S.H. (1996) Reduced bone density and osteoporosis associated with a polymorphic Sp1 binding site in the collagen type I alpha 1 gene. Nature Genet., 14, 203–205. [DOI] [PubMed] [Google Scholar]
- 3.Uitterlinden A.G., Burger,H., Huang,Q., Yue,F., McGuigan,F.E., Grant,S.F., Hofman,A., van Leeuwen,J.P., Pols,H.A. and Ralston,S.H. (1998) Relation of alleles of the collagen type Ialpha1 gene to bone density and the risk of osteoporotic fractures in postmenopausal women. N. Engl. J. Med., 338, 1016–1021. [DOI] [PubMed] [Google Scholar]
- 4.Hugot J.P., Chamaillard,M., Zouali,H., Lesage,S., Cezard,J.P., Belaiche,J., Almer,S., Tysk,C., O’Morain,C.A., Gassull,M. et al. (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature, 411, 599–603. [DOI] [PubMed] [Google Scholar]
- 5.Esterbauer H., Schneitler,C., Oberkofler,H., Ebenbichler,C., Paulweber,B., Sandhofer,F., Ladurner,G., Hell,E., Strosberg,A.D., Patsch,J.R. et al. (2001) A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans. Nature Genet., 28, 178–183. [DOI] [PubMed] [Google Scholar]
- 6.Evans W.E. and Relling,M.V. (1999) Pharmacogenomics: translating functional genomics into rational therapeutics. Science, 286, 487–491. [DOI] [PubMed] [Google Scholar]
- 7.Meyer U.A. (2000) Pharmacogenetics and adverse drug reactions. Lancet, 356, 1667–1671. [DOI] [PubMed] [Google Scholar]
- 8.Collins F.S., Guyer,M.S. and Charkravarti,A. (1997) Variations on a theme: cataloging human DNA sequence variation. Science, 278, 1580–1581. [DOI] [PubMed] [Google Scholar]
- 9.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 10.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 11.The International SNP Map Working Group (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 12.Kruglyak L. (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet., 22, 139–144. [DOI] [PubMed] [Google Scholar]
- 13.Taillon-Miller P., Bauer-Sardina,I., Saccone,N.L., Putzel,J., Laitinen,T., Cao,A., Kere,J., Pilia,G., Rice,J.P. and Kwok,P.Y. (2000) Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nature Genet., 25, 324–328. [DOI] [PubMed] [Google Scholar]
- 14.Reich D.E., Cargill,M., Bolk,S., Ireland,J., Sabeti,P.C., Richter,D.J., Lavery,T., Kouyoumjian,R., Farhadian,S.F., Ward,R. et al. (2001) Linkage disequilibrium in the human genome. Nature, 411, 199–204. [DOI] [PubMed] [Google Scholar]
- 15.Stephens J.C., Schneider,J.A., Tanguay,D.A., Choi,J., Acharya,T., Stanley,S.E., Jiang,R., Messer,C.J., Chew,A., Han,J.-H. et al. (2001) Haplotype variation and linkage disequilibrium in 313 human genes. Science, 293, 489–493. [DOI] [PubMed] [Google Scholar]
- 16.Kwok P.Y. (2000) High-throughput genotyping assay approaches. Pharmacogenomics, 1, 95–100. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., Cui,X., Schmitt,K., Hubert,R., Navidi,W. and Arnheim,N. (1992) Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. USA, 89, 5847–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillespie K.M., Valovin,S.J., Saunby,J., Hunter,K.M., Savage,D.A., Middleton,D., Todd,J.A., Bingley,P.J. and Gale,E.A. (2000) HLA class II typing of whole genome amplified mouth swab DNA. Tissue Antigens, 56, 530–538. [DOI] [PubMed] [Google Scholar]
- 19.Telenius H., Carter,N.P., Bebb,C.E., Nordenskjold,M., Ponder,B.A. and Tunnacliffe,A. (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics, 13, 718–725. [DOI] [PubMed] [Google Scholar]
- 20.Cheung V.G. and Nelson,S.F. (1996) Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc. Natl Acad. Sci. USA, 93, 14676–14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchanan A.V., Risch,G.M., Robichaux,M., Sherry,S.T., Batzer,M.A. and Weiss,K.M. (2000) Long DOP-PCR of rare archival anthropological samples. Hum. Biol., 72, 911–925. [PubMed] [Google Scholar]
- 22.Barbaux S., Poirier,O. and Cambien,F. (2001) Use of degenerate oligonucleotide primed PCR (DOP-PCR) for the genotyping of low-concentration DNA samples. J. Mol. Med., 79, 329–332. [DOI] [PubMed] [Google Scholar]
- 23.Jordan B., Charest,A., Dowd,J.F., Blumenstiel,J.P., Yeh Rf,R.F., Osman,A., Housman,D.E. and Landers,J.E. (2002) Genome complexity reduction for SNP genotyping analysis. Proc. Natl Acad. Sci. USA, 99, 2942–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean F.B., Hosono,S., Fang,L., Wu,X., Faruqi,A.F., Bray-Ward,P., Sun,Z., Zong,Q., Du,Y., Du,J. et al. (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA, 99, 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Don R.H., Cox,P.T., Wainwright,B.J., Baker,K. and Mattick,J.S. (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res., 19, 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nickerson D.A., Tobe,V.O. and Taylor,S.L. (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res., 25, 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smigielski E.M., Sirotkin,K., Ward,M. and Sherry,S.T. (2000) dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res., 28, 352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douglas J.A., Skol,A.D. and Boehnke,M. (2002) Probability of detection of genotyping errors and mutations as inheritance inconsistencies in nuclear-family data. Am. J. Hum. Genet., 70, 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon D., Heath,S.C. and Ott,J. (1999) True pedigree errors more frequent than apparent errors for single nucleotide polymorphisms. Hum. Hered., 49, 65–70. [DOI] [PubMed] [Google Scholar]
- 30.Gordon D., Heath,S.C., Liu,X. and Ott,J. (2001) A transmission/disequilibrium test that allows for genotyping errors in the analysis of single-nucleotide polymorphism data. Am. J. Hum. Genet., 69, 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon D. and Ott,J. (2001) Assessment and management of single nucleotide polymorphism genotype errors in genetic association analysis. Pac. Symp. Biocomput., 6, 18–29. [DOI] [PubMed] [Google Scholar]