Abstract
DNA-PKcs is the catalytic subunit of the DNA-dependent protein kinase (DNA-PK) complex that functions in the non-homologous end-joining of double-strand breaks, and it has been shown previously to have a role in telomere capping. In particular, DNA-PKcs deficiency leads to chromosome fusions involving telomeres produced by leading-strand synthesis. Here, by generating mice doubly deficient in DNA-PKcs and telomerase (Terc–/–/DNA-PKcs–/–), we demonstrate that DNA-PKcs also has a fundamental role in telomere length maintenance. In particular, Terc–/–/DNA-PKcs–/– mice displayed an accelerated rate of telomere shortening when compared with Terc–/– controls, suggesting a functional interaction between both activities in maintaining telomere length. In addition, we also provide direct demonstration that DNA-PKcs is essential for both end-to-end fusions and apoptosis triggered by critically short telomeres. Our data predict that, in telomerase-deficient cells, i.e. human somatic cells, DNA-PKcs abrogation may lead to a faster rate of telomere degradation and cell cycle arrest in the absence of increased apoptosis and/or fusion of telomere-exhausted chromosomes. These results suggest a critical role of DNA-PKcs in both cancer and aging.
Keywords: apoptosis/chromosome fusions/DNA-PKcs/telomerase/telomere length
Introduction
Telomeres cap the ends of eukaryotic chromosomes preventing them from being recognized as DNA breaks and joined by DNA repair activities (reviewed in Blackburn, 2001; Chan and Blackburn, 2002). Vertebrate telomeres are composed of tandem repeats of the TTAGGG sequence and an array of associated proteins (reviewed in de Lange, 2002). In addition, telomeres end in a 3′ overhang, known as the G-strand overhang (de Lange, 2002). The 3′ G-strand overhang is able to fold back and invade the double-stranded region of the telomere forming a loop, the so-called T-loop, which is stabilized by a set of specialized proteins (Griffith et al., 1999). T-loops facilitate the formation of a higher order structure that has been proposed to mediate end-capping by masking telomeric DNA ends from recognition by the DNA repair system, as well as regulating the access of telomerase to the telomere (Griffith et al., 1999). Loss of telomeric capping leads to chromosome end-to-end fusions and/or triggers cell cycle arrest and apoptosis, suggesting that dysfunctional telomeres are detected as damaged DNA (reviewed in de Lange, 2002; Goytisolo and Blasco, 2002).
One of the causes of telomere dysfunction in human cells is loss of TTAGGG repeats coupled to cell division (Harley et al., 1990). Eventual TTAGGG exhaustion from telomeres results in chromosomal fusions and/or proliferative defects and increased apoptosis, which are thought to contribute to the aging process (Collins and Mitchell, 2002). This progressive telomere loss can be prevented if cells have sufficiently high levels of telomerase, a cellular reverse transcriptase that adds TTAGGG repeats onto the chromosome ends (reviewed in Collins and Mitchell, 2002). Telomerase has two essential components, an RNA molecule or Terc (telomerase RNA component), and a catalytic subunit or Tert (telomerase reverse transcriptase). A mouse model for telomere dysfunction due to absence of telomerase has been generated in the past (Blasco et al., 1997). These mice lack the Terc component of the telomerase complex (Terc–/–) and display a progressive loss of telomeric DNA that eventually results in loss of fertility and in a high frequency of end-to-end fusion, as well as in premature aging phenotypes (Blasco et al., 1997; Lee et al., 1998; Herrera et al., 1999; Rudolph et al., 1999). Short telomeres are responsible for these outcomes of telomerase deficiency since re-introduction of telomerase is sufficient to specifically elongate short telomeres and to prevent end-to-end chromosomal fusions (Hemann et al., 2001b; Samper et al., 2001), as well as premature aging phenotypes in these mice (Samper et al., 2001).
Mutation of telomere binding proteins has also been shown to disrupt telomeric capping in the absence of telomere shortening (reviewed in de Lange, 2002; Goytisolo and Blasco, 2002). Impairing the binding of TRF2 to the telomere using a dominant mutant results in telomere fusions with long telomeres at the fusion point (van Steensel et al., 1998). Proteins associated with double-strand break (DSB) repair pathways are also key components of the telomere (reviewed in de Lange, 2002). In this regard, mice deficient for either Ku86 or DNA-PKcs activity, both of which are essential components of the non-homologous end-joining (NHEJ) machinery for repairing DSBs (Smith and Jackson, 1999), also result in end-to-end telomere fusions in the absence of significant telomere shortening (Bailey et al., 1999; Samper et al., 2000; Gilley et al., 2001; Goytisolo et al., 2001; Espejel and Blasco, 2002; Espejel et al., 2002). These findings indicate that telomeres can be dysfunctional in the absence of significant telomere loss, furthermore, they suggest an interplay between DNA repair activities and telomere function. In particular, TRF2 and DNA-PKcs mutations share the outcome that the resulting end-to-end fusions preferentially involve telomeres produced via leading-strand synthesis. This scenario suggests that these proteins are required for the post-replicative processing of this strand, possibly to generate the 3′ G-strand overhang, thus contributing to telomere capping (Bailey et al., 2001).
To study further the role of DNA-PKcs at the telomere, we have generated mice that are deficient for both telomerase and DNA-PKcs activities, Terc–/–/DNA-PKcs–/– mice. The results presented here show that cells from Terc–/–/DNA-PKcs–/– mice show a faster rate of telomere loss than those from the corresponding Terc–/–/DNA-PKcs+/+ littermates. Therefore, if telomerase activity is abrogated, such as in Terc–/– mice, DNA-PKcs deficiency results in a faster rate of telomere degradation. The role of DNA-PKcs in telomere length maintenance is specific, since a similar role for Ku86 was not found when Terc–/–/Ku86–/– double mutant mice were studied, which showed a similar rate of telomere shortening to the corresponding Terc–/–/Ku86+/+ controls (Espejel et al., 2002). Indeed, the study of Terc–/–/Ku86–/– mice demonstrated that Ku86 specifically serves as a negative regulator of telomerase at the telomere, blocking access of telomerase to the telomere (Espejel et al., 2002). Therefore, DNA-PKcs activity, but not Ku86, functionally interacts with telomerase in maintaining telomere length. Concomitant with a faster rate of telomere loss, Terc–/–/DNA-PKcs–/– mice also show an earlier loss of fertility and viability than the corresponding telomerase-deficient controls; however, this loss of viability was not mediated by a further increase in end-to-end fusions or in apoptosis, demonstrating that DNA-PKcs is essential for the signaling of apoptosis and for the fusion of short telomeres. In the particular case of human adult somatic cells, which lack telomerase activity, DNA-PKcs mutation may lead to a faster rate of telomere loss and to telomere dysfunction, predicting an important role for DNA-PKcs activity in organismal aging and cancer progression.
Results
Severe germ cell depletion and telomere loss in first generation (G1) Terc–/–/DNA-PKcs–/– testis
We generated mutant mice simultaneously lacking both telomerase and DNA-PKcs in a defined C57BL6 genetic background (Materials and methods). Strikingly, double Terc–/–/DNA-PKcs–/– mice showed reduced fertility at the first generation (G1), and were completely infertile at the second generation (G2) as indicated by failure to obtain litters from G2 Terc–/–/DNA-PKcs–/– intercrosses (data not shown).
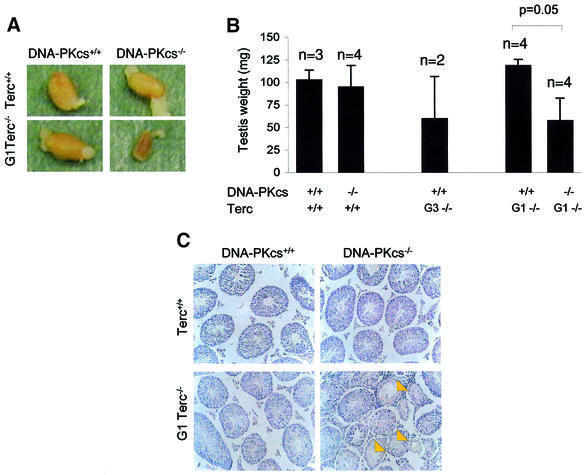
One of the most characteristic phenotypes associated with critical telomere shortening in single Terc–/– mice is severe testicular atrophy (Lee et al., 1998; Hemann et al., 2001a; this paper). In Terc–/– animals in a C57BL6 genetic background testicular atrophy is apparent after three or four generations (Herrera et al., 1999; Figure 1B; Table I). In contrast, first generation (G1) Terc–/– mice, which still have long telomeres, have been described as showing normal size and histology of the testis, as we also demonstrate here for the G1 Terc–/–/DNA-PKcs+/+ testis (Figure 1A–C; Table I). In the case of Terc+/+/DNA-PKcs–/– mice, that lack DNA-PKcs but have telomerase activity, they also showed normal testis size and histology (Figure 1A–C; Table I). Strikingly, G1 Terc–/–/DNA-PKcs–/– mice, which lack both telomerase and DNA-PKcs activity, showed a severe reduction in testis size that was statistically significant (Student’s t-test; P = 0.05) (Figure 1A and B; Table I), coincidental with a marked depletion of germ cells in the seminiferous tubules (see yellow arrows in Figure 1C; Table I).
Fig. 1. Early infertility phenotype in G1 Terc–/–/DNA-PKcs–/– mice. (A) Representative images of testis derived from the indicated genotypes. Note the smaller testis size in the G1 Terc–/–/DNA-PKcs–/– mice. (B) Quantification of testis weight in the number of mice indicated from each genotype. Standard deviation bars are also shown. There was a statistically significant difference in testis weight between G1 Terc–/–/DNA-PKcs–/– and G1 Terc–/–/DNA-PKcs+/+ mice (P = 0.05). n = number of mice of the genotype indicated used in the study. (C) Representative images of testis sections from mice of the indicated genotypes. Arrows point to seminiferous tubules lacking meiotic cells in the G1 Terc–/–/DNA-PKcs–/– mice. (D) Quantification of BrdU-positive cells per meiotic tubule. For each mouse, >100 meiotic tubules were counted. n = number of mice of the genotype indicated used in the study. Bars represent average values for the number of mice indicated from each genotype. Standard deviation bars of the average values are shown. Statistical calculations are done using >100 values of BrdU+ cells/tubule for each individual mouse. G2 Terc–/–/DNA-PKcs+/+ mice showed significantly decreased BrdU incorporation compared with wild-type mice (P = 0.01). Similarly, G1 Terc–/–/DNA-PKcs–/– mice showed significantly decreased BrdU incorporation compared with the G1 Terc–/–/DNA-PKcs+/+ controls (P = 0.003). (E) Representative images of anti-BrdU-stained meiotic tubules from mice of the genotypes indicated. BrdU-positive cells are the ones with dark brown staining. (F) Quantification of TUNEL+ cells per 100 meiotic tubules. The total number of meiotic tubules analyzed from each mouse are shown in Table I (numbers in parenthesis). n = number of mice of the indicated genotype used in the study. Bars represent average values for the number of mice indicated from each genotype. Standard deviation bars of the average values are shown. Statistical calculations are done using between 339 and 2243 values of TUNEL+ cells/tubule for each genotype (Table I). G3 Terc–/–/DNA-PKcs+/+ mice showed significantly increased apoptosis compared with wild-type mice (P = 0.006). In contrast, G1 Terc–/–/DNA-PKcs–/– testis did not show increased apoptosis compared with the G1 Terc–/–/DNA-PKcs+/+ controls (P = 0.76). (G) Representative images of TUNEL+ cells in testis sections from mice of the genotypes indicated. Arrows indicate TUNEL+ cells that appear with dark brown staining. Note the low number of TUNEL+ cells in G1 Terc–/–/DNA-PKcs–/– testis.
Table I. Detailed analysis of the testis of different genotype mice.
| Mouse ID | Genotype | Testis weight, mg | Meiotic tubules, % (total number of tubules scored) | Germ cell proliferation, BrdU+ cells/tubule | Germ cell apoptosis, TUNEL+/100 tubules (total number of tubules scored) | Germ cell telomere length, a.u.f. |
|---|---|---|---|---|---|---|
| TB11 | Terc+/+/DNA-PKcs+/+ | 104.0 | 100.0 (319) | 12.9 ± 15.1 | 2.8 (319) | 1378.2 ± 497.4 |
| TB30 | Terc+/+/DNA-PKcs+/+ | 113.0 | 100.0 (439) | 13.3 ± 15.8 | 6.2 (439) | 1571.8 ± 389.0 |
| TB85 | Terc+/+/DNA-PKcs+/+ | 91.3 | 100.0 (837) | n.d. | 5.6 (837) | 1277.7 ± 461.9 |
| Average | 102.8 ± 10.9 | 100.0 ± 0.0 | 13.1 ± 0.3 | 4.9 ± 1.8 | 1408.7 ± 149.4 | |
| TB17 | Terc+/+/DNA-PKcs–/– | 103.0 | 100.0 (618) | 12.0 ± 14.2 | 11 (618) | 916.8 ± 241.2 |
| TB28 | Terc+/+/DNA-PKcs–/– | 65.0 | 100.0 (246) | 13.5 ± 16.3 | 8.1 (246) | 1950.4 ± 489.0 |
| TB66 | Terc+/+/DNA-PKcs–/– | 88.0 | 100.0 (349) | 11.8 ± 14.7 | 0.0 (349) | 1199.2 ± 404.3 |
| TB84 | Terc+/+/DNA-PKcs–/– | 122.5 | 100.0 (785) | n.d. | 3.9 (785) | 1186.0 ± 256.2 |
| Average | 94.6 ± 24.3 | 100.0 ± 0.0 | 12.4 ± 0.9 | 5.8 ± 4.8 | 1313.2 ± 444.3 | |
| TB74 | G3 Terc–/–/DNA-PKcs+/+ | 92.8 | 97.6 (338) | n.d. | 23.0 (330) | 860.9 ± 241.1 |
| TB76 | G3 Terc–/–/DNA-PKcs+/+ | 26.7 | 7.8 (116) | n.d. | 222.2 (9) | 742.0 ± 203.4 |
| Average | 59.8 ± 46.7 | 52.7 ± 63.6 | – | 122.6 ± 140.8 | 801.4 ± 84.1 | |
| TB16 | G1 Terc–/–/DNA-PKcs+/+ | 128.0 | 100.0 (449) | 14.3 ± 17.1 | 11.4 (449) | 1411.1 ± 384.3 |
| TB10 | G1 Terc–/–/DNA-PKcs+/+ | 117.0 | 100.0 (548) | 12.6 ± 15.9 | 7.1 (548) | 1254.8 ± 477.4 |
| TB18 | G1 Terc–/–/DNA-PKcs+/+ | 118.0 | 100 (998) | 9.8 ± 14.5 | 4.7 (998) | 1381.6 ± 437.3 |
| TA35 | G1 Terc–/–/DNA-PKcs+/+ | 111.6 | 100.0 (248) | n.d. | 103.6 (248) | 1418.9 ± 435.9 |
| Average | 118.7 ± 6.8 | 100.0 ± 0.0 | 12.2 ± 2.3 | 31.7 ± 48.0a | 1366.5 ± 76.2 | |
| TB29 | G1 Terc–/–/DNA-PKcs–/– | 71.0 | 100.0 (315) | 9.3 ± 13.7 | 20.6 (315) | 761.3 ± 300.3 |
| TB13 | G1 Terc–/–/DNA-PKcs–/– | 36.0 | 78.5 (325) | 7.1 ± 13.1 | 7.8 (255) | 985.4 ± 331.0 |
| TA38 | G1 Terc–/–/DNA-PKcs–/– | 36.2 | 48.7 (546) | n.d. | 11.7 (266) | 1075.1 ± 368.8 |
| TB69 | G1 Terc–/–/DNA-PKcs–/– | 86.0 | 100.0 (366) | 9.8 ± 14.3 | 12.3 (366) | 722.7 ± 151.2 |
| Average | 57.3 ± 25.2 | 81.8 ± 24.3 | 8.7 ± 1.4 | 13.1 ± 5.4 | 886.1 ± 171.1 |
Average values of all mice from the same genotype are indicated in bold.
n.d., not determined.
For germ cell telomere length determination pachytene meiocytes were scored.
aStandard deviation is so high in this group of mice due to the fact that only one (TA35) of four mice showed increased apoptosis in the testis. This was not observed in any of the G1 Terc–/–/DNA-PKcs–/– testes analyzed.
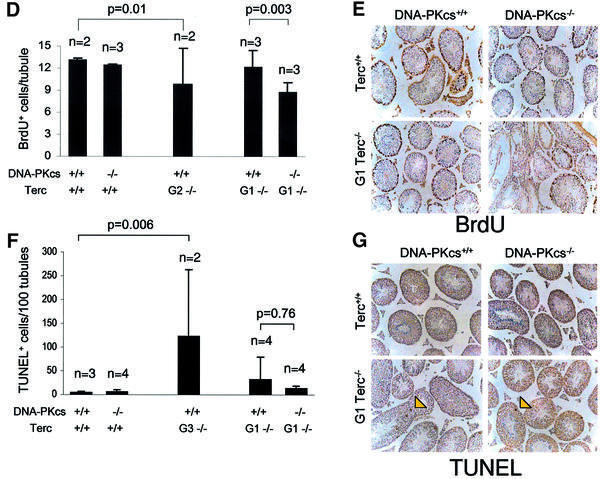
The severity of the testis phenotype in the G1 Terc–/–/DNA-PKcs–/– mice was similar to that of the single G3 Terc–/–/DNA-PKcs+/+ mice (Figure 1A–C; Table I). This fact suggested that the G1 Terc–/–/DNA-PKcs–/– mice could suffer a faster rate of telomere loss than the G1 Terc–/–/DNA-PKcs+/+ controls. To demonstrate this, we performed quantitative fluorescence in situ hybridization using a PNA-telomeric probe [telomeric quantitative fluorescence in situ hybridization (Q-FISH)] on testis sections, which allowed us to measure average telomere length in meiotic cells from the different genotypes (González-Suárez et al., 2000). Approximately 100 meiotic cells were analyzed from each mouse, and at least two mice from each genotype were studied (Figure 2A shows average fluorescence of 100 meiotic tubules from individual mice, and Figure 2B that of mice grouped by genotype; Table I). Consistent with published data (Gilley et al., 2001; Goytisolo et al., 2001), average telomere length in Terc+/+/DNA-PKcs–/– meiocytes was similar to that of the Terc+/+/DNA-PKcs+/+ controls, and there were no statistically significant differences between both groups of mice (Figure 2A and B; Table I). Similarly, average telomere length in G1 Terc–/–/DNA-PKcs+/+ testis was not significantly different from that of the Terc+/+/DNA-PKcs+/+ controls, in agreement with previous reports on single G1 Terc–/– mice in a C57Bl6 background (Figure 2A and B) (Herrera et al., 1999). Strikingly, all G1 Terc–/–/DNA-PKcs–/– mice studied showed germ cells with a significant decrease in telomere fluorescence compared with those of the corresponding G1 Terc–/–/DNA-PKcs+/+ controls (Figure 2A and B; Student’s t-test, P < 0.001) (Table I), suggesting that DNA-PKcs and telomerase functionally interact in maintaining telomere length in germ cells. The severity of the telomere shortening phenotype in G1 Terc–/–/DNA-PKcs–/– mice was equivalent to that of the late generation G3 Terc–/–/DNA-PKcs+/+ mice, which also showed significantly shorter telomeres than wild-type controls (Figure 2A and B; Student’s t-test, P < 0.001) (Table I).
Fig. 2. Telomere length determination using Q-FISH on testis sections. (A) Average telomere fluorescence in arbitrary units (a.u.f.) of 100 meiotic cells from each mouse of the indicated genotype. Standard deviation, as well as the total number of meiocytes analyzed per mouse are indicated. (B) Average telomere fluorescence in arbitrary units (a.u.f.) of three or four mice grouped per genotype. Standard deviation, as well as the total number of mice of each genotype, are indicated. There was a significant decrease in average telomere fluorescence in G1 Terc–/–/DNA-PKcs–/– meiotic cells compared with G1 Terc–/–/DNA-PKcs+/+ controls (P < 0.001).
Collectively, these results suggest that the accelerated rate of telomere loss in germ cells derived from G1 Terc–/–/DNA-PKcs–/– mice compared with single mutant G1 Terc–/–/DNA-PKcs+/+ controls is responsible for the early onset of infertility, and thus testicular atrophy. Indeed, telomere shortening in G1 Terc–/–/DNA-PKcs–/– germ cells was comparable to that of late generation G3 Terc–/–/DNA-PKcs+/+ mice, which also show a severe testis atrophy phenotype (Herrera et al., 1999; this paper).
Short telomeres in G1 Terc–/–/DNA-PKcs–/– testis lead to decreased proliferation but not increased apoptosis of the male germ cells
Telomere loss to a critically short length in Terc–/– testis has been shown to be coincidental with both decreased proliferation and massive apoptosis of the germ cells (Lee et al., 1998; Hemann et al., 2001a; this paper). To address whether this was also the case for the G1 Terc–/–/DNA-PKcs–/– testis with short telomeres, we determined both germ cell proliferation [by measuring 5-bromo-2′-deoxyuridine (BrdU) incorporation] and apoptosis (by using the TUNEL assay) on testis sections from different genotypes (Materials and methods). The number of BrdU-positive cells per testis tubule was significantly decreased in G1 Terc–/–/DNA-PKcs–/– testis compared with G1 Terc–/–/DNA-PKcs+/+ testis (see Table I and Figure 1D for quantification; Figure 1E for examples) (>100 meiotic tubules were analyzed for each mouse; Student’s t-test, P = 0.003). This indicates that G1 Terc–/–/DNA-PKcs–/– germ cells have a reduced proliferative capacity compared with the G1 Terc–/–/DNA-PKcs+/+ controls. As a control, G2 Terc–/–/DNA-PKcs+/+ mice also showed a significantly reduced number of BrdU-positive cells per meiotic tubule compared with wild-type mice (Figure 1D; >100 meiotic tubules were analyzed for each mouse; Student’s t-test, P = 0.01). Interestingly, the number of TUNEL+ cells per 100 meiotic tubules was not significantly increased in the G1 Terc–/–/DNA-PKcs–/– testis compared with the G1 Terc–/–/DNA-PKcs+/+ controls, despite the fact that they had significantly shorter telomeres. The number of meiotic tubules analyzed for each mouse is shown in Table I (numbers in parenthesis). Comparison of 2243 values from a total of four G1 Terc–/–/DNA-PKcs+/+ mice with 1202 values from a total of four G1 Terc–/–/DNA-PKcs–/– mice, indicated that there were no significant differences in the number of TUNEL+ cells per tubule (Student’s t-test, P = 0.76) (see Table I and Figure 1F for quantification; Figure 1G for representative images showing no increased apoptosis). In contrast, late generation G3 Terc–/–/DNA-PKcs+/+ testis showed a significantly increased number of TUNEL+ cells per tubule compared with the wild-type controls, in agreement with previous reports (339 meiotic tubules were analyzed from a total of two G3 Terc–/–/DNA-PKcs+/+ mice and 1595 tubules from a total of three wild-type controls; Student’s t-test, P = 0.006) (Table I; Figure 1F; Herrera et al., 1999).
Therefore, in contrast to late generation single mutant G3–G4 Terc–/– mice, germ cell depletion in testis from G1 Terc–/–/DNA-PKcs–/– animals is the result of decreased germ cell proliferation but not of increased apoptosis. These findings suggest that DNA-PKcs activity is required to mediate germ cell apoptosis, but not cell cycle arrest, that occurs as a consequence of critical telomere shortening in Terc–/– testis (see Discussion).
Faster rate of telomere loss in different Terc–/–/DNA-PKcs–/– cell types compared with the corresponding Terc–/–/DNA-PKcs+/+ controls
To investigate whether the role of DNA-PKcs in telomere length maintenance was general for other cell types in the mouse, we measured telomere length by different techniques in mouse embryonic fibroblasts (MEFs), splenocytes, as well as hepatocytes from mice of the various genotypes.
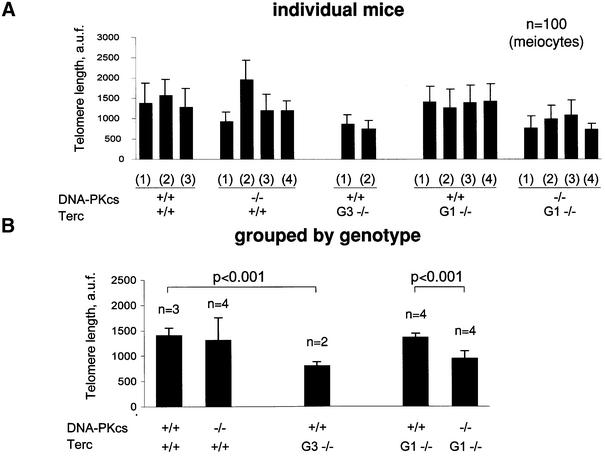
First, we performed Q-FISH on metaphases derived from primary (passage 1) MEFs generated by intercrossing wild-type, G1 Terc–/–/DNA-PKcs+/+ or G1 Terc–/–/DNA-PKcs–/– mice. Q-FISH on metaphasic chromosomes allows determination of telomere length frequencies, as well as of the percentage of undetectable telomeres per individual nuclei (the detection limit of the Q-FISH technique is 200–500 bp of TTAGGG repeats) (Zijlmans et al., 1997). Average telomere length (average of p- and q-arms) was decreased in two independent G2 Terc–/–/DNA-PKcs–/– MEFs compared with two G2 Terc–/–/DNA-PKcs+/+ MEFs, 24.2 ± 16.4 kb and 27.9 ± 17.5 kb compared with 33.2 ± 18.7 kb and 30.1 ± 17.4 kb, respectively (Table II). This difference was highly significant, as indicated by a Student’s t-test value of P = 6.6 × 10–36, which was calculated by comparing >3000 telomere values from each genotype. Histograms showing telomeric frequencies also illustrate that G2 Terc–/–/DNA-PKcs–/– MEFs showed an increased frequency of shorter telomeres compared with the corresponding G2 Terc–/–/DNA-PKcs+/+ MEFs, as indicated by a shift of the population towards shorter telomere length values (Figure 3A). As expected, G2 Terc–/–/DNA-PKcs+/+ MEFs showed significantly shorter telomeres than those of Terc+/+/DNA-PKcs+/+ control MEFs, 33.2 ± 18.7 kb and 30.1 ± 17.4 kb compared with 35.5 ± 15.2 kb and 36.0 ± 16.2 kb, respectively, in agreement with telomere shortening in these mice as a consequence of telomerase deficiency through two mouse generations (Student’s t-test, P = 9.4 × 10–20) (see Table II for telomere length values and Figure 3A for histograms of telomere length frequencies). From these Q-FISH data, we have estimated that the rate of telomere shortening per mouse generation in Terc–/–/DNA-PKcs–/– MEFs was 4.85 kb compared with 2.1 kb in the case of Terc–/–/DNA-PKcs+/+ MEFs.
Table II. Q-FISH determination of telomere length in passage 1 primary MEFs.
| Group | Genotype (MEF) | Metaphase number | Telomere number | p-arm (kb) | q-arm | Average p+q | No TTAGGG (%) |
|---|---|---|---|---|---|---|---|
| A | Wild-type (6) | 10 | 1592 | 33.0 ± 13.8 | 37.9 ± 16.7 | 35.5 ± 15.2 | 0 |
| Wild-type (7) | 10 | 1420 | 31.4 ± 14.2 | 40.8 ± 18.2 | 36.0 ± 16.2 | 0.9 | |
| B | G2 Terc–/–/DNA-PKcs+/+ (4) | 10 | 1596 | 32.3 ± 18.3 | 34.2 ± 18.8 | 33.2 ± 18.7 | 2.0 |
| G2 Terc–/–/DNA-PKcs+/+ (5) | 10 | 1576 | 28.3 ± 15.7 | 31.9 ± 19.1 | 30.1 ± 17.4 | 2.0 | |
| C | G2 Terc–/–/DNA-PKcs–/– (3) | 10 | 1580 | 22.6 ± 13.5 | 25.8 ± 19.3 | 24.2 ± 16.4 | 3.0 |
| G2 Terc–/–/DNA-PKcs–/– (4) | 10 | 1604 | 25.3 ± 14.8 | 30.5 ± 20.2 | 27.9 ± 17.5 | 2.5 |
Student’s t-test comparing all telomeres analyzed for the different genotypes (>3000 telomere values for each genotype) was used for statistical significance: A/B, P = 9.4 × 10–20; A/C, P = 3.9 × 10–108; B/C, P = 6.67 × 10–36.
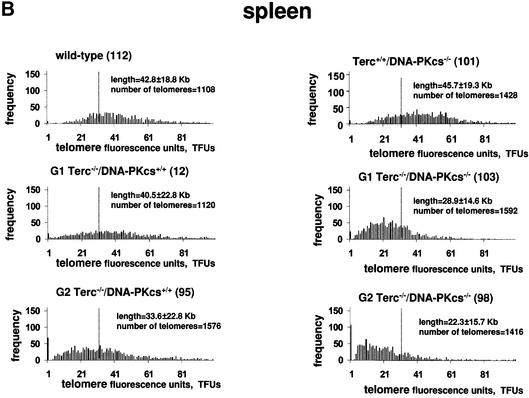
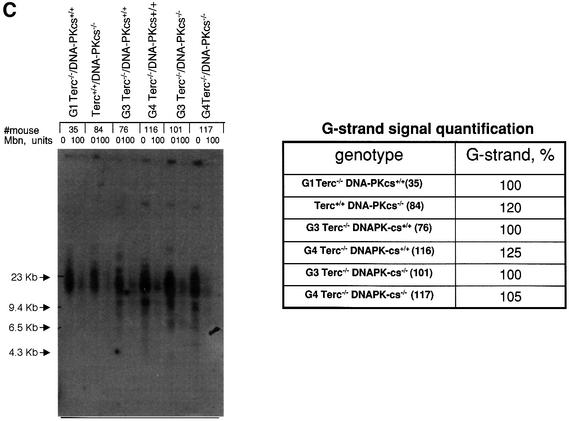
Fig. 3. (A) Telomere length distribution in primary MEFs from the indicated genotype. One telomere fluorescence unit (TFU) corresponds to 1 kb of TTAGGG repeats (Zijlmans et al., 1997). Average telomere length and the standard deviation, as well as the total number of telomeres analyzed, are indicated. The distribution of the telomere length frequencies for each MEF is an indication of the standard deviation. Notice the marked shift towards short telomeres in Terc–/–/DNA-PKcs–/– MEFs compared with the corresponding Terc–/–/DNA-PKcs+/+ controls. (B) Telomere length distribution in primary splenocytes from the indicated genotype. One TFU corresponds to 1 kb of TTAGGG repeats (Zijlmans et al., 1997). Average telomere length and the standard deviation, as well as the total number of telomeres analyzed, are indicated. The distribution of the telomere length frequencies for each splenocyte culture is an indication of the standard deviation. Notice the marked shift towards short telomeres in Terc–/–/DNA-PKcs–/– splenocytes compared with the corresponding Terc–/–/DNA-PKcs+/+ controls. (C) G-strand overhangs using liver nuclei visualized in native gel after hybridization with a (CCCTAA)4 probe. Notice that upon treatment with 100 U of mung bean nuclease (Mbn) the G-strand-specific signal decreases. Note that the G-strand overhang signal was present at lower molecular weights in the G3 and G4 Terc–/–/DNA-PKcs–/– liver cells compared with the corresponding G3 and G4 Terc–/–/DNA-PKcs+/+ controls, again indicating the presence of shorter telomeres in Terc–/–/DNA-PKcs–/– cells. Mice numbers 116 and 117 are littermates. Quantification of G-strand signals is also shown (see Materials and methods for details).
A similar result was obtained when we extended these studies to primary splenocytes derived from different generation Terc–/–/DNA-PKcs–/– and Terc–/–/DNA-PKcs+/+ mice. As shown in Table III, splenocytes isolated from G1 and G2 Terc–/–/DNA-PKcs–/– mice have shorter telomeres compared with splenocytes derived from G1 and G2 Terc–/–/DNA-PKcs+/+ controls, with average lengths of 28.9 ± 14.6 kb and 22.3 ± 15.7 kb, compared with 40.5 ± 22.8 kb and 33.6 ± 22.8 kb, respectively. These differences are highly significant, as shown by Student’s t-values of 1.8 × 10–81 and 3.6 × 10–52, respectively (see Table III for complete Student’s t-test values). The histograms of telomere length frequencies also show that Terc–/–/DNA-PKcs–/– splenocytes show a significantly higher proportion of shorter telomeres than the corresponding Terc–/–/DNA-PKcs+/+ splenocytes (Figure 3B). We have estimated that the rate of telomere shortening in splenocytes per mouse generation was increased from 3 kb for Terc–/–/DNA-PKcs+/+ to 6.8 kb for Terc–/–/DNA-PKcs–/– splenocytes, a similar 2-fold increase to that determined previously for MEFs (see above).
Table III. Q-FISH determination of telomere length in primary splenocytes.
| Group | Genotype (mice) | Metaphase number | Telomere number | p-arm (kb) | q-arm (kb) | Average p+q (kb) | No TTAGGG (%) |
|---|---|---|---|---|---|---|---|
| A | Wild-type (112) | 7 | 1108 | 36.6 ± 15.2 | 49.1 ± 22.7 | 42.8 ± 18.8 | 0.8 |
| B | Terc+/+/DNA-PKcs–/– (101) | 9 | 1428 | 39.9 ± 16.0 | 51.4 ± 22.4 | 45.7 ± 19.3 | 0.7 |
| C | G1 Terc–/–/DNA-PKcs+/+ (12) | 7 | 1120 | 33.3 ± 18.8 | 47.7 ± 25.8 | 40.5 ± 22.8 | 1.6 |
| D | G1 Terc–/–/DNA-PKcs–/– (103) | 10 | 1592 | 24.4 ± 12.2 | 29.3 ± 17.0 | 28.9 ± 14.6 | 1.4 |
| E | G2 Terc–/–/DNA-PKcs+/+ (95) | 10 | 1576 | 26.5 ± 21.4 | 37.7 ± 24.2 | 33.6 ± 22.8 | 4.3 |
| F | G2 Terc–/–/DNA-PKcs–/– (98) | 9 | 1416 | 17.4 ± 12.1 | 27.3 ± 19.4 | 22.3 ± 15.7 | 7.1 |
Littermates: 95 and 98; 101 and 103. Student’s t-test comparing all telomeres analyzed for the different genotypes (>1000 telomere values for each mice) was used for statistical significance: A/B, P = 0.14; C/D, P = 1.8 × 10–81; E/F, P = 3.6 × 10–52.
It is important to note that DNA-PKcs deficiency per se did not result in significant telomere loss, in agreement with prior reports (Gilley et al., 2001; Goytisolo et al., 2001). As shown in Table III, splenocytes isolated from Terc+/+/DNA-PKcs–/– animals displayed an average telomere length that is not significantly different from that of Terc+/+/DNA-PKcs+/+ splenocytes, 45.7 ± 19.3 kb compared with 42.8 ± 18.8 kb, respectively (Student’s t-test, P = 0.14, not significant; Table III).
Both Terc–/–/DNA-PKcs–/– MEFs and splenocytes showed a slightly higher percentage of undetectable telomeres as determined by Q-FISH than those of the corresponding Terc–/–/DNA-PKcs+/+ controls, although the difference was not very striking (Tables II and III). It is tempting to speculate that simultaneous absence of DNA-PKcs and telomerase not only results in a faster rate of telomere shortening, but also decreases the average telomere length at which telomere dysfunction occurs, similar to what has been described recently for a dominant-negative mutant of TRF2 (Karlseder et al., 2002).
The data obtained from Q-FISH were confirmed by using terminal restriction fragment (TRF) analysis, which allows visualization of the size of TRFs that contain telomeres by using Southern blotting and pulsed-field electrophoresis (data not shown; Materials and methods). Using the TRF technique under native conditions, we have also determined the integrity of the G-strand overhang in all different genotypes (Figure 3C). For this, liver nuclei were isolated from the mice indicated and subjected to TRF analysis (Materials and methods). The intensity of the G-strand overhang signal is indicative of the G-strand overhang length. As reported before, neither single Terc or DNA-PKcs deficiency affected the integrity of the G-strand overhang, as indicated by normal G-strand signals (Hemann and Greider, 1999; Goytisolo et al., 2001) (Figure 3C). Importantly, the simultaneous absence of both telomerase and DNA-PKcs did not affect the intensity of the G-strand overhang signals, although the G-strand overhang signals appeared at lower molecular weight in agreement with shorter telomeres in these cells (Figure 3C). It is important to point out, however, that while the overall length of the overhangs appears unaffected, this does not rule out the possibility that individual overhangs may be either very short or even non-existent.
Taken together, these results indicate that while DNA-PKcs deficiency alone does not have an impact on telomere length, the simultaneous absence of both telomerase and DNA-PKcs exerts a synergic effect that results in an accelerated rate of telomere loss compared with that observed in the absence of telomerase activity alone. Employing two different cell types, we have estimated a 2-fold increase in the rate of telomere shortening per mouse generation in the Terc–/–/DNA-PKcs–/– mice compared with the Terc–/–/DNA-PKcs+/+ controls. These findings suggest that both activities functionally interact in maintaining telomere length in all of the different cell types studied here.
Short telomeres in Terc–/–/DNA-PKcs–/– cells do not trigger end-to-end chromosomal fusions
A hallmark of telomerase-deficient mice is progressive loss of telomeres, and the short termini have been shown to be responsible for end-to-end chromosomal fusions in various cell types studied with the exception of male germ cells, which, as we mentioned previously, undergo a massive cell death (Blasco et al., 1997; Lee et al., 1998; Hande et al., 1999; Herrera et al., 1999; Hemann et al., 2001a,b; Samper et al., 2001). These end-to-end fusions were proposed to be the result of NHEJ events (Goytisolo et al., 2000; Hemann et al., 2001b). Evidence for this came from the fact that Ku86 activity is necessary to mediate these fusions, thus demonstrating that the NHEJ machinery is involved in the process (Espejel et al., 2002).
To unravel whether the accelerated loss of telomeres described in Terc–/–/DNA-PKcs–/– mice correlates with increased end-to-end fusions, we have determined using Q-FISH the spontaneously arising chromosomal aberrations on 100 metaphases from each of the MEFs described above. As described in this study, G2 Terc–/–/DNA-PKcs–/– MEFs harbor an increased population of very short telomeres. In spite of this, we failed to detect a significant increase in end-to-end chromosomal fusions in G2 Terc–/–/DNA-PKcs–/– MEFs compared with the wild-type or to the G2 Terc–/–/DNA-PKcs+/+ MEFs (Table IV). Only one Robertsonian-like fusion was detected in a total of 200 metaphases from the two different G2 Terc–/–/DNA-PKcs–/– MEFs studied, and this fusion had detectable TTAGGG repeats at the fusion point, suggesting that this was not due to critical shortening of telomeres but was most likely the result of DNA-PKcs deficiency (see below for CO-FISH analysis) (Table IV) (Bailey et al., 1999; Goytisolo et al., 2001). These results suggest that short telomeres in the absence of DNA-PKcs activity do not lead to increased end-to-end fusions. A similar scenario has been shown for Ku86 deficiency (Espejel et al., 2002).
Table IV. Cytogenetic analysis by Q-FISH of spontaneous chromosomal aberrations in primary passage 1 MEFs.
| Genotype (MEF) | Metaphase number | RT-likea (TTAGGGb) | Dica |
|---|---|---|---|
| Wild-type (6) | 100 | 0 | 0 |
| Wild-type (7) | 100 | 0 | 0.01 |
| G2 Terc–/–/DNA-PKcs+/+ (4) | 100 | 0 | 0 |
| G2 Terc–/–/DNA-PKcs+/+ (5) | 100 | 0 | 0 |
| G2 Terc–/–/DNA-PKcs–/– (3) | 100 | 0 | 0 |
| G2 Terc–/–/DNA-PKcs–/– (4) | 100 | 0.01 | 0 |
aFrequency per metaphase; RT-like, Robertsonian-like fusion involving the p-arms; Dic, dicentric chromosome fusion involving the q-arms.
bTTAGGG, fusion that contains TTAGGG signal at the fusion point as determined by Q-FISH.
This finding, together with the previous observation that short telomeres in the context of DNA-PKcs deficiency do not trigger apoptosis of the male germ line (see above), have important implications for cancer and aging, since they suggest that short telomeres in a NHEJ-deficient context would not trigger chromosome end-to-end fusions or apoptosis.
End-to-end fusions in Terc–/–/DNA-PKcs–/– cells, not due to telomere shortening, are the result of dysfunctional leading-strand telomeres
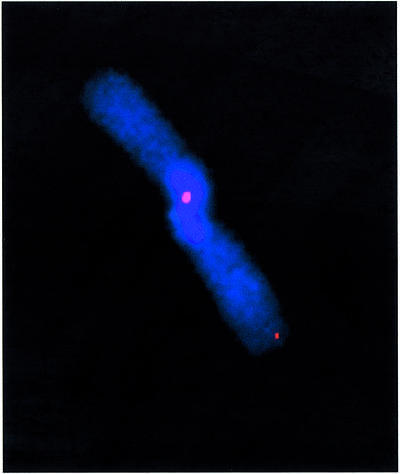
To investigate the nature of the end-to-end chromosome fusions present in the Terc–/–/DNA-PKcs–/– cells, we used chromosome orientation FISH (CO-FISH), which allows discrimination between telomeres produced by leading- versus lagging-strand DNA synthesis (see Materials and methods). It has been demonstrated recently that in a number of mouse DNA-PKcs-deficient, telomerase-competent backgrounds, it is the leading-strand telomere that is preferentially involved in these fusions (Bailey et al., 2001). We found five chromatid-type telomere fusions in a total of 200 G2 Terc–/–/DNA-PKcs–/– metaphases scored compared with zero chromatid-type fusions in 200 metaphases from the G2 Terc–/–/DNA-PKcs+/+ controls. All the fusions retained telomere sequence at the fusion point and were the result of leading-strand telomere fusion (see example in Figure 4). This difference is statistically significant (P < 0.02) and further supports a role of DNA-PKcs in leading-strand processing (Bailey et al., 2001). We conclude, therefore, that in DNA-PKcs-deficient cells, and regardless of telomerase status, it is the leading-strand telomere that becomes uncapped and is subject to end-to-end fusions.
Fig. 4. Representative CO-FISH image of a chromatid-type dicentric leading-strand telomere fusion present in G2 Terc–/–/DNA-PKcs–/– cells. Blue, DAPI; red, TTAGGG signal.
Discussion
Pathways leading to telomere dysfunction in mammalian cells
There are at least two major pathways by which a telomere can become uncapped or dysfunctional and lead to end-to-end chromosomal fusions. The first one involves progressive loss of telomeric sequences that occurs in the absence of telomerase due to the end-replication problem. Once a critically shortened telomere length has been reached, not enough telomeric sequence remains to function properly (e.g. preventing the formation of a T-loop), and this unsequestered chromosome end is detected as damaged DNA and processed as such by the DNA repair machinery. Compelling evidence for this came from the study of mice deficient in the RNA component of telomerase, Terc–/– (Blasco et al., 1997). In particular, the study of these mice has demonstrated that telomere loss to a critically shortened length has two preferential outcomes: (i) decreased viability due to decreased proliferation and/or increased apoptosis; (ii) increased end-to-end chromosome fusions, both of which impact on the aging and cancer patterns of these mice (Blasco et al., 1997; Lee et al., 1998; Chin et al., 1999; Herrera et al., 1999; Rudolph et al., 1999; González-Suárez et al., 2000). The decreased proliferation and increased apoptosis, but not the end-to-end fusions, produced by critically short telomeres can be rescued in a p53 mutant background, indicating that p53 is one of the major mediators of cell cycle arrest and apoptosis triggered by short telomeres (Chin et al., 1999).
A second major pathway for telomere dysfunction is the loss of a telomeric capping structure. In this case, even though telomeres are sufficiently long, and telomeric sequences are visible by Q-FISH at the point of fusion between the misjoined chromosome ends, the telomere end is unable to form the correct higher order structure (i.e. T-loops), thus leading to fusions. These conclusions are based on the analysis of cells and mice defective for telomere-binding proteins, such as TRF2, Ku86 and DNA-PKcs (van Steensel et al., 1998; Bailey et al., 1999; Bianchi and de Lange, 1999; Hsu et al., 1999, 2000; Samper et., 2000; Zhu et al., 2000; d’Adda di Fagagna et al., 2001; Gilley et al., 2001; Goytisolo et al., 2001). A common phenotype associated with mutations in these proteins is increased end-to-end chromosome fusions, but without telomere shortening. In the case of DNA-PKcs and TRF2 mutation, the telomere fusions always involve telomeres produced by leading-strand synthesis, suggesting that the role of these proteins in telomere capping is to process the initially blunt-ended leading-strand telomere immediately following replication (Bailey et al., 2001).
Role of DNA-PKcs in telomere length maintenance
Here, we have generated a new mouse model, which lacks both DNA-PKcs and telomerase, Terc–/–/DNA-PKcs–/– mice. A new unexpected role of DNA-PKcs at the telomere (besides its role in generating the proper capping structure) has been unmasked by studying this new murine model. We show that DNA-PKcs deficiency in the context of telomerase deficiency leads to a faster rate of telomere loss than that of the single knock-out controls in four different cell types studied. We have estimated that telomeres are lost at a rate that is 2-fold higher than that of the single telomerase knockouts. As a consequence of the faster rate of telomere loss, Terc–/–/DNA-PKcs–/– mice showed early appearance of infertility at the first generation (G1). The G1 Terc–/–/DNA-PKcs–/– mice showed severe germ-line depletion, reduced testis size and decreased proliferation of germ cells at the first generation, while these phenotypes only appear from the third generation (G3) onwards in the single telomerase knockout mice in the same genetic background (Herrera et al., 1999).
The fact that single DNA-PKcs-deficient mice, which are telomerase-proficient, have been shown to have normal length telomeres (Gilley et al., 2001; Goytisolo et al., 2001), argues in favor of a role for telomerase in stabilizing/elongating the uncapped telomeres present in DNA-PKcs-deficient cells, which otherwise could suffer a faster rate of degradation (see below).
DNA-PKcs mediates both end-to-end fusions and apoptosis, but not cell cycle arrest, produced by critically short telomeres
Interestingly, in contrast to G3 Terc–/– mice, which are wild type for DNA-PKcs, G1 Terc–/–/DNA-PKcs–/– doubly mutant mice did not show increased apoptosis of male germ cells, indicating that DNA-PKcs activity mediates the male early germ cell apoptosis triggered by critically short telomeres. As mentioned above, however, G1 Terc–/–/DNA-PKcs–/– testis still showed a marked decrease in cell proliferation, indicating that this outcome of telomere dysfunction is not mediated by DNA-PKcs activity, in agreement with a previous report (Wang et al., 2000). Importantly, the absence of DNA-PKcs in Terc–/–/DNA-PKcs–/– MEFs also prevented the end-to-end fusions involving short telomeres. These findings agree with those described previously by us for Terc/Ku86-deficient cells (Espejel et al., 2002). DNA-bound Ku86 serves to efficiently target DNA-PKcs to the ends and enhance its kinase activity (reviewed in Smith and Jackson, 1999). Therefore, it is not surprising that both Ku86 and DNA-PKcs are required for the efficient detection and signaling of shortened telomeres as ‘damaged’ DNA. These findings could also suggest that NHEJ activities, such as Ku86 and DNA-PKcs, are upstream of p53 in detecting and processing ‘damaged’ telomeres. Although this model is not supported by previous data showing that p53 is not phosphorylated by DNA-PKcs in vivo (Jimenez et al., 1999), it is still possible that, in the particular case of apoptosis mediated by telomere dysfunction, DNA-PKcs could be upstream of p53. In this regard, it has been reported recently that DNA damage-induced apoptosis requires DNA-PK activity and is mediated by the latent population of p53 (Woo et al., 2002).
Differential roles of DNA-PKcs and Ku86 in telomere length maintenance
Although cells derived from Ku86–/– and DNA-PKcs–/– animals share a common phenotype of enhanced end-to-end fusion, these proteins seem to play differential roles at telomeres. In particular, the effect of DNA-PKcs deficiency in accelerating telomere shortening in the absence of telomerase is contrary to that described previously for mice lacking both Ku86 and telomerase (Espejel et al., 2002), pointing to differential roles of these proteins in telomere length maintenance. Indeed, Ku86 does not interact functionally with telomerase to maintain telomere length, but instead acts as a negative regulator, and has been proposed to block the access of the enzyme to the terminus (Espejel et al., 2002). A possible explanation for this difference is that the presence of Ku86 in doubly mutant Terc/DNA-PKcs, but not in mutant Terc/Ku86 cells, may be responsible for the accelerated rate of telomere loss; for instance, Ku86 may mediate increased telomere degradation by 3′–5′ exonuclease activities at the telomere, such as that of the Mre11/Rad50/Nbs1 complex or that of Werner protein (WRN), that has been shown to be stimulated by Ku86 (Trujillo et al., 1998; Li and Comai, 2000; Orren et al., 2001). WRN is particularly attractive as an explanation of accelerated telomere loss in Terc–/–/DNA-PKcs–/– cells, since its exonuclease activity is not only stimulated by Ku86, but is also negatively regulated by DNA-PK-mediated phosphorylation (Karmakar et al., 2002). In addition, Ku has been shown to interact with TRF1 and TRF2, which in turn could influence telomere structure and length (reviewed in de Lange, 2002).
Two essential roles for DNA-PKcs in maintenance of functional telomeres
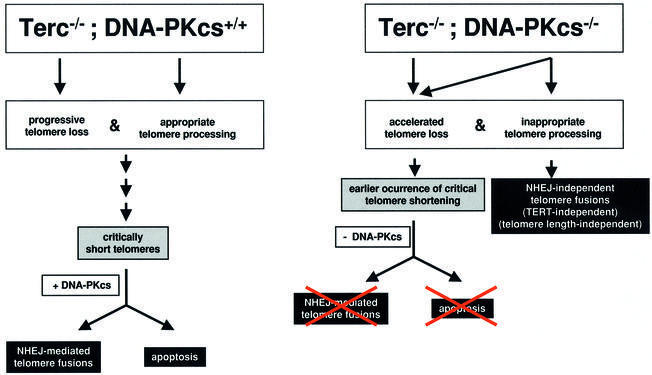
The results presented here indicate that DNA-PKcs activity is central to the two major known pathways leading to loss of telomere function in mammalian cells (see above; Figure 5). On the one hand, DNA-PKcs interacts with telomerase to maintain telomere length, as just discussed. On the other hand, as also suggested here, as well as elsewhere (Bailey et al., 2001), DNA-PKcs is involved in generating and/or maintaining the proper terminal structure by its proposed role in telomere processing. In the first instance, in the absence of telomerase and as telomeres suffer a faster rate of shortening, DNA-PKcs deficiency will result in defective signaling of short telomeres as ‘damaged’ DNA, rescuing apoptosis and end-to-end chromosome fusions triggered by them (Figure 5). In the second instance, DNA-PKcs deficiency will result in loss of proper telomere capping/processing, leading to telomere fusions that maintain telomeric signals at the fusion point and involve telomeres produced by leading-strand synthesis (Bailey et al., 2001; Figure 5). Since these telomere fusions are not the result of shortening, their frequency would not be expected to increase in the absence of telomerase (unless the physical/structural presence of telomerase aids in capping). In agreement with this notion, the occurrence of these fusions did not increase further as a consequence of telomerase deficiency, suggesting that telomerase is not needed for the formation of the final leading-strand telomere capping structure in mouse cells.
Fig. 5. Working model showing that DNA-PKcs is essential for both telomere length maintenance (as shown here for Terc–/–/DNA-PKcs–/– mice) and for proper telomere processing (as shown here, as well as elsewhere; Bailey et al., 2001). In addition, the results shown here also demonstrate that DNA-PKcs mediates both end-to-end fusion and apoptosis produced by critically short telomeres. In the absence of both Terc and DNA-PKcs, telomeres suffer a faster rate of shortening; however, these prematurely shortened telomeres do not result in end-to-end fusions or apoptosis.
In summary, the results presented here indicate that DNA-PKcs interacts with telomerase to maintain telomere length, as well as signaling short telomeres as DNA damage, triggering apoptosis and facilitating end-to-end fusion of short telomeres, and that DNA-PKcs, but not telomerase, is involved in generating and/or maintain ing the proper capping structure at the leading-strand telomeres.
Implications of DNA-PKcs mutation for human cancer and aging
These findings have potentially important implications for both cancer and organismal aging. Human somatic cells do not possess active telomerase, so if DNA-PKcs activity is lost (via naturally occurring polymorphisms or muta tion) an accelerated rate of telomere dysfunction will occur. This model suggests two predictable outcomes: (i) increased telomere shortening and eventual loss of telomeres will occur, resulting in accelerated aging (loss of viability and decreased proliferation) in the absence of increased fusions of telomere-exhausted chromosomes and apoptosis, and (ii) loss of the telomere capping structure will occur, resulting in the fusion of telomeres produced by leading-strand synthesis. Both outcomes could lead to an increased risk of cancer, as well as to premature aging phenotypes. Long-term studies are underway to determine the impact of simultaneous absence of telomerase and DNA-PKcs on the normal aging and tumor incidence of these mice.
Materials and methods
Mice and cells
To obtain successive generations of the double Terc–/–/DNA-PKcs–/– mutant mice and MEFs, as well as the corresponding Terc–/–/DNA-PKcs+/+ controls, Terc–/–/DNA-PKcs+/– mice from each generation were intercrossed. For this, double heterozygous Terc+/–/DNA-PKcs+/– mice were first derived from crossing Terc+/– and DNA-PKcs+/– mice in a C57Bl6 background (Taccioli et al., 1998; Herrera et al., 1999). Terc+/–/DNA-PKcs+/– intercrosses produced the first generation (G1) of double mutant mice. Successive generations of Terc–/–/DNA-PKcs–/– mice, as well as the corresponding Terc–/–/DNA-PKcs+/+ littermate controls, were obtained in parallel by serial intercrossing of Terc–/–/DNA-PKcs+/– mice until the fourth generation (G4), at which point G4 Terc–/–/DNA-PKcs+/– became infertile and no further generations could be obtained, as described previously for single Terc–/– mice in a C57Bl6 background (Herrera et al., 1999).
For all studies, primary MEFs (passage 1) were used. MEFs were prepared from day 13.5 embryos as described previously (Blasco et al., 1997). First-passage MEFs corresponded to approximately two population doublings (PDL 2). Mice used for all studies were 8–12 weeks old.
Mouse handling
All mice were housed at our barrier area in Madrid, where pathogen-free procedures are employed in all mouse rooms. Quarterly health monitoring reports have been negative for all pathogens in accordance with Federation of European Laboratory Animal Science Associations (FELASA) recommendations.
Cell culture
Low-passage primary MEFs were incubated at 37°C in 5% CO2 and cultured in β-MEM supplemented with 15% fetal bovine serum and antibiotics. Cultures were grown to confluence in 75 cm2 flasks, then returned to exponential growth by subculturing into fresh medium (either with or without BrdU; see below) and incubated at 37°C for an additional 24 h. Colcemid (0.2 µg/ml) was added during the last 4 h to accumulate mitotic cells. Cultures were trypsinized and cells suspended in 75 mM KCl at 37°C for 15 min before fixing in 3:1 methanol/acetic acid. Fixed cells were dropped onto cold, wet, glass microscope slides.
CO-FISH
CO-FISH has been described in detail previously and was used here with some modification (Bailey et al., 2001). Briefly, confluent cell monolayers were subcultured into medium containing 5′-bromo-2′-deoxyuridine (BrdU; Sigma, St Louis, MO) at a total final concentration of 1 × 10–5 M then incubated at 37°C for an additional 24 h. Colcemid (0.2 µg/ml) was added during the final 4 h to accumulate mitotic cells. Cells, a majority of which are now singly substituted with BrdU, were harvested and metaphase spreads prepared on microscope slides as described above. Prior to hybridization of the single-stranded (TTAGGG)7 telomere probe, slides were treated with 0.5 mg/ml RNase A for 10 min at 37°C, then stained with 0.5 µg/ml Hoechst 33258 (Sigma) in 2× SSC for 15 min at room temperature. Slides were then exposed to 365 nm UV light (Stratalinker 1800 UV irradiator) for 25–30 min. Enzymic digestion of the BrdU-substituted DNA strands with 3 U/µl exonuclease III (Promega) in buffer supplied by the manufacturer (50 mM Tris–HCl, 5 mM MgCl2 and 5 mM DTT pH 8.0) was allowed to proceed for 10 min at room temperature. An additional denaturation in 70% formamide, 2× SSC at 70°C for 1 min was performed, followed by dehydration in a cold ethanol series (70, 85 and 100%). Probe hybridization and analysis was identical to that described for FISH experiments. This strategy facilitates identification of the telomere produced by leading-strand DNA synthesis and is described in detail elsewhere (Bailey et al., 2001).
Q-FISH
Q-FISH on testis sections. Testis sections from four different mice of each genotype were hybridized with a PNA-tel probe and telomere length was determined as described previously (González-Suárez et al., 2000). For average telomere fluorescence calculations, >100 meiotic nuclei from each mouse were captured at 100× magnification and the telomere fluorescence was integrated using spot IOD analysis in the TFL-TELO program.
Q-FISH on metaphasic chromosomes. First passage MEFs or fresh splenocytes were prepared for Q-FISH and hybridized as described previously (Samper et al., 2000, 2001). To correct for lamp intensity and alignment, images from fluorescent beads (Molecular probes, USA) were analyzed using the TFL-Telo program. Telomere fluorescence values were extrapolated from the telomere fluorescence of LY-R (R cells) and LY-S (S cells) lymphoma cell lines of known lengths of 80 and 10 kb, respectively (McIlrath et al., 2001). There was a linear correlation (r2 = 0.999) between the fluorescence intensity of the R and S telomeres with a slope of 38.6. The calibration-corrected telomere fluorescence intensity (ccTFI) was calculated as described previously (Herrera et al., 1999).
Images were captured using Leica Q-FISH software at 400 ms integration time in a linear acquisition mode to prevent over-saturation of fluorescence intensity and recorded using a COHU CCD camera on a Leica Leitz DMRB fluorescence microscope.
TFL-Telo software (gift from Dr Peter Lansdorp, Vancouver), was used to quantify the fluorescence intensity of telomeres from at least 10 metaphases of each data point. The images of metaphases from different MEF cultures were captured on the same day, in parallel and scored blind.
TRF analysis
Fresh hepatocyte nuclei from the mice indicated were prepared as decribed previously (Kipling and Cooke, 1990), and TRF analysis was performed as described in Blasco et al. (1997).
G-strand overhang assay
Hepatocyte nuclei prepared as described previously (Kipling and Cooke, 1990) were included in agarose plugs following instructions provided by the manufacturer (Bio-Rad). After overnight digestion in LDS buffer (1% LDS, 100 mM EDTA pH 8.0 and 10 mM Tris pH 8.0), the plugs were digested with 0, 100 or 300 U of mung bean nuclease for 15 min; or with 100 U at 20 or 40 min, as indicated. Then the plugs were digested with MboI overnight and run on pulse-field electrophoresis gels as described previously (Blasco et al., 1997). The in-gel hybridizations in native and denaturing conditions were carried out as described previously (Samper et al., 2000). Quantification of the G-strand overhang radioactive signals was carried out using a STORM 860 PhosphorImager (Molecular Dynamics), using the software provided by the manufacturer. These values were corrected by the TRF signal in denaturing gel conditions (not shown). G-strand radioactive signals were normalized to that of control G1 Terc–/–/DNA-PKcs+/+ cells, which were considered to be 100%.
Scoring of chromosomal abnormalities by Q-FISH
The indicated numbers of metaphases from each MEF culture were scored for chromosomal aberrations by superimposing the telomere image on the DAPI chromosome image in the TFL-telo software (gift from Dr Peter Lansdorp). End-to-end fusions can be two chromosomes fused by their p-arms (Robertsonian-like fusions) or two chromosomes fused by their q-arms (dicentrics).
Statistical analysis
Statistical calculation was done using Microsoft Excel. For statistical significance, Student’s t-test values were calculated.
Histological analysis
Testis from age-matched mice were either fixed in 10% buffered formalin phosphate, dehydrated and paraffin-embedded, or snap frozen in tissue-freezing medium (Leica Instruments, Nussloch, Germany). For hematoxylin staining, 4 µm paraffin-embedded sections were used.
Germ cell apoptosis by the TUNEL assay
Whole testes were fixed in buffered 5% formaldehyde for 2 h and paraffin embedded following standard methods. For determination of the apoptotic index, 4 µm testicular sections were deparaffinized and rehydrated through graded alcohols. After quenching endogenous peroxidase by incubation in 0.3% H2O2 in methanol, tissues were permeabilized in 0.5% Triton-X/TBS, incubated in TUNEL reaction mix (In Situ Cell Death Detection, POD; Roche, Manhnheim, Germany) for 1 h at 37° C, washed, incubated in Converter-POD for 30 min at 37°C and developed with 3′-diaminobenzidine (Sigma Fast; Sigma, Steinheim, Germany). Slides were counter-stained with hematoxylin (Mayer’s hematoxylin solution; Sigma Diagnostics, St Louis, MO) and mounted with Eukitt (Panreac, Barcelona, Spain). Positive DNase I-digested and negative TdT-free controls were included. For quantification of apoptosis, the number of seminiferous tubules and the number of TUNEL+ cells were counted in at least four non-contiguous cross-sections (see Table I for total numbers of tubules scored). Results are expressed as TUNEL+ cells/100 meiotic tubules.
Germ cell proliferation by BrdU incorporation
Male mice of the indicated genotypes were given intraperitoneal injections of BrdU (Sigma), 100 µg/g of body weight in 200 µl of PBS, 1 h before euthanasia. Paraffin-embedded 4 µm testis sections were cut onto Superfrost/Plus slides (Fisher Scientific, Pittsburgh, PA), deparaffinized, hydrated through graded alcohols and pretreated with 3% H2O2 in 70% methanol to block endogenous peroxidases. After digestion with 0.02% pepsin for 15 min at 37°C, DNA was denatured in 2 NHCl for 30 min and neutralized in 0.1 M sodium borate pH 8.5 for 10 min. Slides were then permeabilized in 0.3% Triton X-100/PBS, blocked in BSA 10%/PBS for 1 h at 37°C and incubated with murine anti-BrdU antibody (Becton Dickinson, San Jose, CA) at a 1:10 dilution at 4°C overnight. For signal amplification, a secondary biotinylated anti-mouse IgG and the StreptABComplex/HRP (Dako A/S, Denmark) were used, following the manufacturer’s instructions. Slides were counter-stained in Mayer’s hematoxylin and mounted with Eukitt. The number of BrdU+ germ cells was counted in 100 meiotic tubules from at least two non-contiguous testicular sections.
Acknowledgments
Acknowledgements
We thank R.Serrano and E.Santos for mouse care and genotyping, and M.Serrano and E.Goodwin for critical reading of the manuscript and helpful discussions. S.E. is a pre-doctoral fellow of the European Union (EU), S.F. is a pre-doctoral fellow of the Fondo de Investigaciones Sanitarias (FIS). Research at the laboratory of M.A.B. is funded by grants PM97-0133 from the MCYT, 08.1/0030/98 from CAM, and by grants EURATOM/991/0201, FIGH-CT-1999-00002 and FIS5-1999-00055 from the European Union, and by the DIO. The DIO was founded and is supported by the Spanish Research Council (CSIC) and by Pharmacia. G.E.T is a scholar of the Leukemia and Lymphoma Society. The G.E.T laboratory is supported by the National Institutes of Health (NIH) CA76409, and the Aids for Cancer Research Foundation. The S.M.B. laboratory is supported by grants from DOE (ER632339) and NIH (CA43322).
References
- Bailey S.M., Meyne,J., Chen,D.J., Kurimasa,A., Li,G.C., Lehnert,B.E. and Goodwin,E.H. (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl Acad. Sci. USA, 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S.M., Conforth,M.N., Kurimasa,A., Chen,D.J. and Goodwin,E.H. (2001) Strand-specific postreplicative processing of mammalian telomeres. Science, 293, 2462–2465. [DOI] [PubMed] [Google Scholar]
- Bianchi A. and de Lange,T. (1999) Ku binds telomeric DNA in vitro. J. Biol. Chem., 274, 21223–21227. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee,H.-W., Hande,P., Samper,E., Lansdorp,P., DePinho,R. and Greider,C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- Chan S.W.-L. and Blackburn,E.H. (2002) New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene, 21, 553–563. [DOI] [PubMed] [Google Scholar]
- Chin L., Artandi,S.E., Shen,Q., Tam,A., Lee,S.L., Gottlieb,G.J., Greider,C.W. and DePinho,R.A. (1999) p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell, 97, 527–538. [DOI] [PubMed] [Google Scholar]
- Collins K. and Mitchell,J.R. (2002) Telomerase in the human organism. Oncogene, 21, 564–579. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Hande,M.P., Tong,W.M., Roth,D., Lansdorp,P.M., Wang,Z.Q. and Jackson,S.P. (2001) Effects of DNA non-homologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol., 11, 1192–1196. [DOI] [PubMed] [Google Scholar]
- de Lange T. (2002) Protection of mammalian telomeres. Oncogene, 21, 532–540. [DOI] [PubMed] [Google Scholar]
- Espejel S. and Blasco,M.A. (2002) Identification of telomere-dependent ‘senescence-like’ arrest in mouse embryonic fibroblasts. Exp. Cell Res., 276, 242–248. [DOI] [PubMed] [Google Scholar]
- Espejel S., Franco,S., Rodríguez-Perales,S., Bouffler,S.D., Cigudosa,J.C. and Blasco,M.A. (2002) Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J., 21, 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D., Tanaka,H., Hande,M.P., Kurimasa,A., Li,G.C., Oshimura,M. and Chen,D.J. (2001) DNA-PKcs is critical for telomere capping. Proc. Natl Acad. Sci. USA, 98, 15084–15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Suárez E., Samper,E., Flores,J.M. and Blasco,M.A. (2000) Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet., 26, 114–117. [DOI] [PubMed] [Google Scholar]
- Goytisolo F.A. and Blasco,M.A. (2002) Many ways to telomere dysfunction: in vivo studies using mouse models. Oncogene, 21, 584–591. [DOI] [PubMed] [Google Scholar]
- Goytisolo F., Samper,E., Martín-Caballero,J., Finnon,P., Herrera,E., Flores,J.M., Bouffler,S.D. and Blasco,M.A. (2000) Short telomeres result in organismal hypersensitivity to ionizing radiation in mammals. J. Exp. Med., 192, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytisolo F., Samper,E., Edmonson,S., Taccioli,G.E. and Blasco,M.A. (2001) Absence of DNA-PKcs in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol., 21, 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Hande P., Samper,E., Lansdorp,P. and Blasco,M.A (1999) Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol., 144, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- Hemann M.T and Greider,C.W. (1999) G-strand overhangs on telomeres in telomerase deficient mouse cells. Nucleic Acids Res., 27, 3964–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Rudolph,K.L., Strong,M.A., De Pinho,R.A., Chin,L. and Greider,C.W. (2001a) Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol. Biol. Cell, 12, 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Strong,M.A., Hao,L.Y. and Greider,C.W. (2001b) The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell, 107, 67–77. [DOI] [PubMed] [Google Scholar]
- Herrera E., Samper,E., Martín-Caballero,J., Flores,J.M., Lee,H.-W. and Blasco,M.A. (1999) Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J., 18, 2950–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-L., Gilley,D., Backburn,E. and Chen,D.J. (1999) Ku is associated with the telomere in mammals. Proc. Natl Acad. Sci. USA, 96, 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-L. et al. (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev., 14, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G.S. et al. (1999) DNA-dependent protein kinase is not required for the p53-dependent response to DNA damage. Nature, 400, 81–83. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Smogorzewska,A. and de Lange,T. (2002) Senescence induced by altered telomere state, not telomere loss. Science, 295, 2446–2449. [DOI] [PubMed] [Google Scholar]
- Karmakar, P., Piotrowski,J., Brosh,R.M.,Jr, Sommers,J.A., Miller,S.P., Cheng,W.H., Snowden,C.M., Ramsden,D.A. and Bohr,V.A. (2002) Werner protein is a target of DNA-PK in vivo and in vitro and its catalytic activities are regulated by phosphorylation. J. Biol. Chem., 277, 18291–18302. [DOI] [PubMed] [Google Scholar]
- Kipling D. and Cooke,H.J. (1990) Hypervariable ultra-long telomeres in mice. Nature, 347, 400–402. [DOI] [PubMed] [Google Scholar]
- Lee H.-W. et al. (1998) Essential role of mouse telomerase in highly proliferative organs. Nature, 392, 569–574. [DOI] [PubMed] [Google Scholar]
- Li B. and Comai,L. (2000) Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J. Biol. Chem., 275, 28349–28352. [DOI] [PubMed] [Google Scholar]
- McIlrath J. et al. (2001) Telomere length anormalities in mammalian radiosensitive cells. Cancer Res., 61, 912–915. [PubMed] [Google Scholar]
- Orren D.K., Machwe,A., Karmakar,P., Piotrowski,J., Cooper,M.P. and Bohr,V.A. (2001) A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res., 29, 1926–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph K.L., Chang,S., Lee,H.W., Blasco,M., Gottlieb,G.J., Greider,C. and DePinho,R.A. (1999) Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell, 96, 701–712. [DOI] [PubMed] [Google Scholar]
- Samper E., Goytisolo,F., Slijepcevic,P., van Buul,P. and Blasco,M.A. (2000) Mammalian Ku86 prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO rep., 1, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E., Flores,J.M. and Blasco,M.A. (2001) Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc–/– mice with short telomeres. EMBO rep., 2, 800–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.C.M. and Jackson,S.P. (1999) The DNA-dependent protein kinase. Genes Dev., 13, 916–934. [DOI] [PubMed] [Google Scholar]
- Taccioli G.E. et al. (1998) Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immuno deficiency and radiosensitivity. Immunity, 9, 355–366. [DOI] [PubMed] [Google Scholar]
- Trujillo K.M., Yuan,S.S., Lee,E.Y. and Sung,P. (1998) Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem., 273, 21447–21450. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Wang S. et al. (2000) The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc. Natl Acad. Sci. USA, 97, 1584–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo R.A., Jack,M.T., Xu,Y., Burma,S., Chen,D.J. and Lee,P.W. (2002) DNA damage-induced apoptosis requires the DNA-dependent protein kinase, and is mediated by the latent population of p53. EMBO J., 21, 3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.D., Kuster,B., Mann,M., Petrini,J.H. and de Lange,T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet., 25, 347–352. [DOI] [PubMed] [Google Scholar]
- Zijlmans J.M., Martens,U.M., Poon,S., Raap,A.K., Tanke,H.J., Ward,R.K. and Lansdorp,P.M. (1997) Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl Acad. Sci. USA, 94, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]