Abstract
A novel cDNA EZI isolated as an oncostatin M- inducible gene encoded a protein containing 12 C2H2-type zinc fingers. EZI was found to transactivate the promoters that are also responsive to STAT3 and activated the acute phase response element (APRE) synergistically with STAT3. Co-immunoprecipitation demonstrated the association of EZI with STAT3, which was mediated by the N-terminal region (1–183) of EZI. The EZI mutant lacking this region showed reduced transcriptional activity, indicating that EZI and STAT3 function cooperatively through physical interaction. While EZI predominantly localized in the nucleus and enhanced the nuclear localization of STAT3, the EZI mutant lacking 11 zinc finger motifs failed to translocate into the nucleus and also inhibited nuclear localization of STAT3 as well as STAT3-mediated transactivation. These results indicate that EZI is a novel nuclear zinc finger protein that augments STAT3 activity by keeping it in the nucleus.
Keywords: nuclear localization/oncostatin M/STAT/zinc finger protein
Introduction
Zinc finger motifs are involved in protein binding to DNA as well as protein–protein interactions. Zinc finger proteins are classified basically into C2H2, C3H, C4 and ring finger types based on the amino acid sequences comprising the zinc finger domain. C2H2 zinc finger proteins such as EKLF are a major type among the zinc finger family, and their zinc finger domain consists of two cysteine and two histidine residues, i.e. CX2C-X12-HX3H, where X stands for any amino acid. In contrast, C3H zinc fingers, exemplified by TIS11, consist of three cysteines and one histidine, and C4 zinc fingers such as GATA-1 have four cysteines (Evans and Felsenfeld, 1989; Varnum et al., 1991). Because of these unique structures, a large number of zinc finger proteins have been found in the DNA sequence database, while most of their cellular functions still remain to be discovered. There are many zinc finger transcription factors that are known to play important roles in proliferation, differentiation and cell fate determination. For example, GATA-1 is an extensively characterized zinc finger protein and plays an essential role in erythroid differentiation by binding to the GATA sequence in several promoters, including the globin genes (Shivdasani and Orkin, 1996). GATA-1 not only binds to DNA but also interacts with other factors such as FOG, CBP/p300 and PU.1 through their zinc finger motifs (Tsang et al., 1997; Boyes et al., 1998; Rekhtman et al., 1999; Zhang,P. et al., 1999). Likewise, many zinc finger transcription factors bind their specific DNA sequence and also interact with various proteins.
Signal transducers and activators of transcription (STATs) are a family of transcription factors with an SH2 domain. Seven STATs have been identified and mediate signals for various cytokines (Darnell, 1997). STATs are recruited to the tyrosine-phosphorylated cytokine receptors via their SH2 domain and are phosphorylated by Jak kinase bound to the receptor. Tyrosine-phosphorylated STATs then form a homo- or a heterodimer and translocate to the nucleus where they bind to their target DNA sequences. However, many questions still remain unanswered as to the regulation of STATs following tyrosine phosphorylation. Although tyrosine phosphatases must be an important factor for regulation of STATs, such phosphatases are yet to be identified. Protein inhibitors of activated STAT (PIASs) associate with the phosphorylated STATs and inhibit their DNA binding, leading to repression of transactivation (Chung et al., 1997; Liu et al., 1998). There may be more such regulators in various steps of STAT’s action, such as translocation to the nucleus and proteolysis, the mechanisms of which remain largely unknown.
STAT3 was identified originally as a factor that induces acute phase proteins in hepatocytes in response to interleukin-6 (IL-6) or epidermal growth factor (EGF) (Akira et al., 1994; Raz et al., 1994; Wegenka et al., 1994; Zhong et al., 1994, Cantwell et al., 1998). STAT3 forms a homo- or heterodimer with STAT1 and regulates the expression of a number of genes including those for α2-macroglobulin, c-Fos, junB, interferon regulatory factor 1 (IRF-1) and angiotensinogen, genes whose promoters contain a STAT3-binding site (Fujitani et al., 1994; Harroch et al., 1994; Yuan et al., 1994; Campbell et al., 1995; Look et al., 1995; Mascareno et al., 1998). The acute phase response element (APRE) in the α2-macroglobulin promoter region containing the STAT3-binding site consists of 16 nucleotides and has been used extensively for assessing STAT3 activation (Hocke et al., 1992). Like many other transcription factors, STAT3 associates with CBP/p300 to form a transcriptional complex. Co-activator CBP/p300 integrates various signals by binding transcription factors. For example, leukemia inhibitory factor (LIF) and BMP are known to act synergistically on neural progenitor cells to induce astrocytes, and this synergistic action of LIF and BMP is explained by the cooperative action of LIF-activated STAT3 and BMP-activated SMAD1 through their binding to the co-activator (Nakashima et al., 1999). STAT3 was also shown to interact with c-Jun and they form a complex on the α2-macroglobulin promoter that contains both STAT3- and c-Jun-binding sites (Schaefer et al., 1995; Zhang,X. et al., 1999). PIAS3 binds to STAT3 and inhibits its activity, whereas a zinc finger protein Gfi-1 enhances STAT3 signaling by preventing the binding of PIAS3 to STAT3 (Chung et al., 1997; Rodel et al., 2000). Thus, there are several proteins that are involved in STAT3-mediated gene expression.
Oncostatin M (OSM) is a member of the IL-6 cytokine subfamily. The receptors for this family of cytokines share the common signal transducer, gp130, and induce activation of STAT3 and mitogen-activated protein (MAP) kinase pathways upon binding of the cognate ligand (Taga and Kishimoto, 1997). Previously we showed that OSM plays a role in development of hematopoietic stem cells in the embryonic aorta–gonad–mesonephros (AGM) region and in differentiation of fetal hepatocytes (Mukouyama et al., 1998; Kamiya et al., 1999). In an effort to elucidate the function of OSM in development of hematopoiesis, we searched for OSM-inducible genes in hematogenic endothelial cells derived from the AGM region. Among them, we found a novel zinc finger protein, EZI (endothelial cell-derived zinc finger protein), that contains 12 repeats of C2H2 zinc finger motifs. Here we show that EZI is a novel nuclear protein with the potential to transactivate genes in cooperation with STAT3.
Results
Structure and expression of EZI
By using the cDNA library subtraction method, we isolated OSM-inducible genes from a hematogenic endothelial cell line derived from the mouse AGM region. Among them, we found a cDNA encoding a novel zinc finger protein, which we named EZI. The deduced amino acid sequence of EZI consists of 594 amino acids that contain 12 repeats of the zinc finger motif in the middle part of the sequence (Figure 1). There are 161 amino acids at the N-terminus and 30 amino acids at the C-terminus besides the zinc finger cluster. No known protein motif was found in those regions. Generally, a zinc finger motif binds a zinc ion and forms a tertiary structure suitable for protein–protein interaction or binding to its specific DNA sequence. The zinc finger motifs in EZI are all classified into the C2H2 type. A typical member of this family is EKLF, one of the Kruppel-like transcription factors, which binds to the β-globin promoter and is essential for its expression (Nuez et al., 1995). While there are many such factors implicated in developmental processes, their targets are yet to be identified.
Fig. 1. The deduced amino acid sequence of EZI. The numbers on the right indicate the amino acid number from the first methionine. The asterisk shows the stop codon. The zinc finger motifs in the sequence are indicated by the underlines, and the cluster of basic amino acids, which may function as an NLS, is marked by a box. This sequence data has been submitted to the DDBJ/EMBL/GenBank databases under accession No. AB076746.
As EZI was isolated by subtraction of the cDNA libraries prepared from OSM-stimulated and unstimulated LO cells, the induction of EZI mRNA in LO cells was examined. Northern hybridization showed that the EZI gene was induced by 30 min after OSM stimulation, though the induction level was modest. The expression level of EZI returned to the basal level in 1–2 h after stimulation (Figure 2A). We also examined induction of EZI by IL-6 in M1 cells and by OSM in BaF3 cells transfected with the OSM receptor. However, no significant induction was observed (data not shown). To determine its tissue distribution, we performed northern blot analysis using total RNA derived from mouse adult tissues and cell lines and found that EZI was ubiquitously expressed in adult tissues and cell lines (Figure 2B). It was also shown that EZI was expressed in various mouse embryonic tissues, including liver, limbs, brain and the AGM region (Figure 2C).
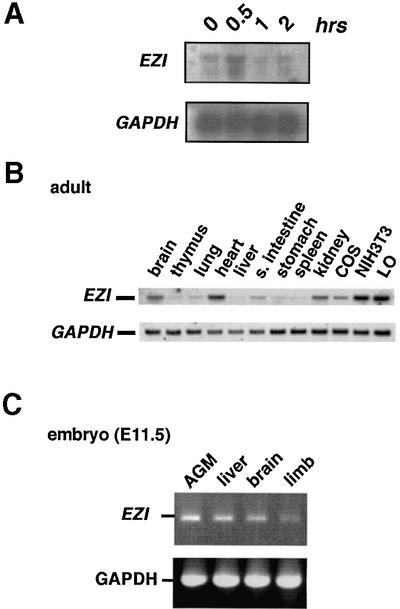
Fig. 2. Expression of EZI. (A) OSM-dependent LO cells were treated with OSM and harvested at 0, 0.5, 1 and 2 h after stimulation, and RNA was extracted for northern blotting. EZI mRNA was already present before stimulation and was induced slightly at 0.5 h after stimulation. (B) Expression of EZI in various adult tissues and cell lines was detected by northern blotting. (C) Expression of EZI in mouse embryonic tissues at E11.5 was examined by RT–PCR.
Cooperative transactivation by EZI and STAT3
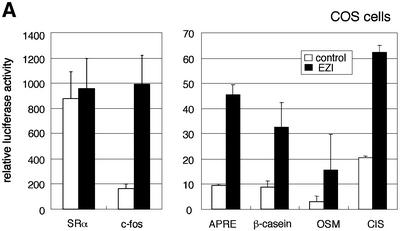
As a major function of zinc finger proteins is regulation of transcription, we asked whether EZI has such activity. The EZI expression vector was co-transfected with several promoter constructs linked to the luciferase gene in COS cells and it was revealed that EZI has the ability to transactivate several promoters. We tested the APRE, β-casein, OSM, CIS, c-fos and SRα promoters, and found that, with the exception of SRα, all these promoters were transactivated significantly by EZI (Figure 3A). A common feature of those promoter sequences activated by EZI is that they are known to be activated by STATs. STAT3 activates APRE most effectively, while STAT5 and also STAT3, to a lesser extent, activate β-casein, OSM and CIS promoters. As our APRE–luciferase construct consists of a tandem repeat of the α2-macroglobulin promoter-derived APRE containing a STAT3-binding site and was transactivated efficiently by EZI, we used this construct for further analysis.
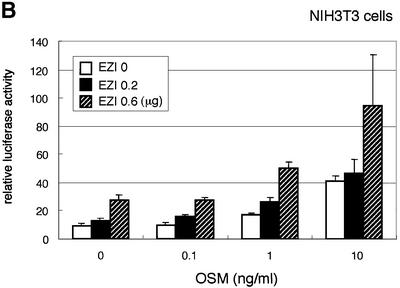
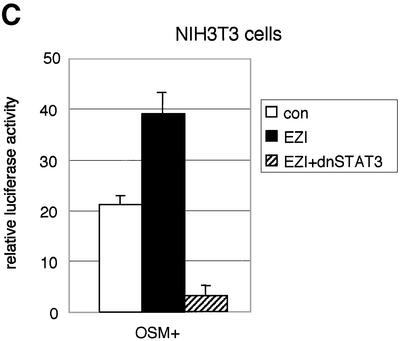
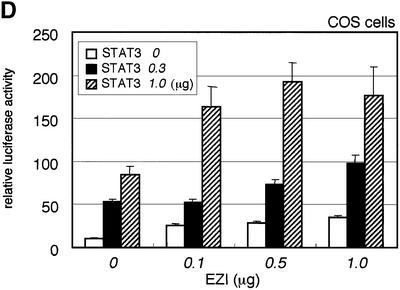
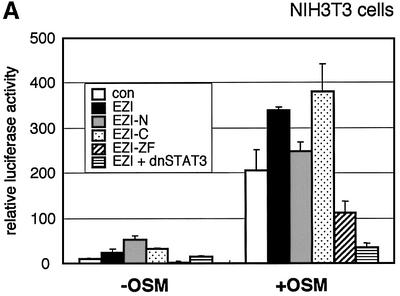
Fig. 3. EZI cooperatively activates APRE with STAT3. (A) COS cells were transfected with the EZI expression vector and the luciferase reporter constructs. Luciferase activity was measured 2 days after transfection. EZI activated several promoters that contain a STAT-binding site. (B) NIH-3T3 cells were co-transfected with the EZI expression vector and the APRE–luciferase construct, and stimulated with various concentrations of OSM as indicated. At 8 h after OSM stimulation, luciferase activity was measured. The EZI-dependent transactivation was enhanced by OSM stimulation. (C) NIH-3T3 cells were co-transfected with expression vectors for EZI, a dominant-negative form of STAT3 (dnSTAT3) and the APRE–luciferase reporter. Luciferase activity in cell lysates was measured as above. Transactivation by EZI was significantly suppressed by dnSTAT3. (D) Expression vectors of EZI and STAT3 were introduced into COS cells and the luciferase activity of APRE was measured. APRE was activated dose-dependently on EZI and STAT3, indicating that EZI functions cooperatively with STAT3.
We first transfected the EZI cDNA with the reporter construct in OSM-responsive NIH-3T3 cells because OSM activates intracellular signaling pathways including that of STAT3. The OSM-induced luciferase activity in NIH-3T3 cells was enhanced by expression of EZI (Figure 3B). Furthermore, expression of a dominant-negative form of STAT3 almost completely suppressed luciferase activity induced by EZI in NIH-3T3 cells (Figure 3C). Thus STAT3 plays a role in the transactivation by EZI, while additional factors may also be involved in the process. To verify the interaction between STAT3 and EZI, we expressed both of them in COS cells with the APRE–luciferase construct and found that EZI and STAT3 cooperatively activated APRE and increased the luciferase activity in a dose-dependent manner (Figure 3D).
Association of EZI with STAT3
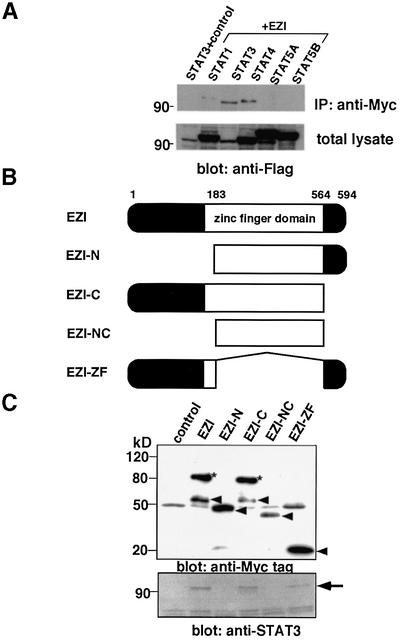
The above results suggest that EZI is able to activate transcription and its activity is enhanced by OSM through STAT3. We considered the possibility that EZI might associate with STAT3 to function cooperatively. We then co-expressed Myc-tagged EZI and Flag-tagged STAT3 in COS cells and immunoprecipitated them with anti-Myc tag antibody followed by western blotting with anti-Flag tag antibody, and found that EZI and STAT3 were co-immunoprecipitated (Figure 4A). We also examined if EZI interacts with other STATs, and found that STAT1 and STAT4 also interacted with EZI when they were co-expressed in COS cells. Although STAT5 cannot be observed clearly in Figure 4A, a faint band was detected occasionally (data not shown). As the expression level of STAT3 in the blot of the total lysate was much lower than that of the other STATs (Figure 4A, lower panel), binding of EZI to STAT3 is the strongest among the STATs we tested (Figure 4A).
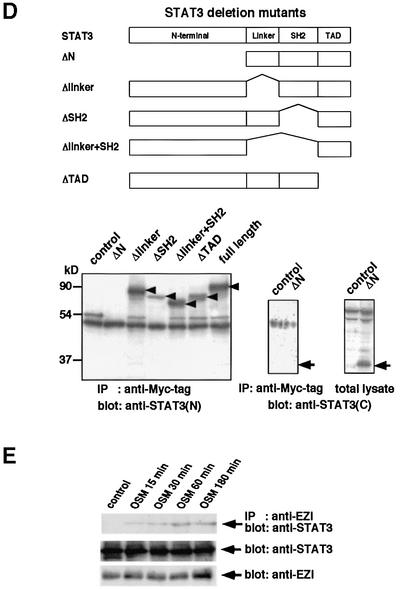
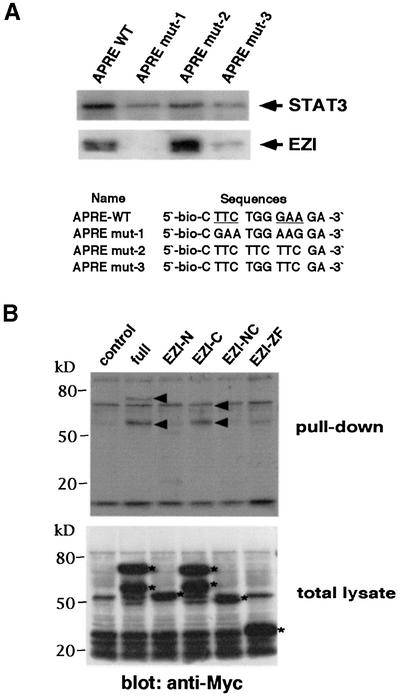
Fig. 4. Association of EZI with STAT3. (A) Expression vectors of Myc-tagged EZI and of Flag-tagged STATs were transfected into COS cells. COS cells transfected with Flag-tagged STAT3 and the empty vector were used as a control. Cells were collected 2 days after transfection and used for immunoprecipitation with anti-Myc tag antibody followed by western blotting with anti-Flag tag antibody. In order to evaluate the levels of STATs present in the total lysate, equal amounts of total lysates were blotted with anti-Flag tag antibody. As the expression level of STAT3 was the lowest in the blot of total lysate, STAT3 associated most efficiently with EZI. (B) Structures of EZI mutants. To determine the domain responsible for STAT3 binding of EZI, four kinds of deletion mutants were prepared. Myc tag was added to the N-terminus of these mutants. (C) EZI mutants were tested for the association with STAT3 by co-transfection into COS cells. Cell lysates were prepared and immunoprecipitated with anti-Myc tag antibody, followed by blotting with anti-Myc tag and anti-STAT3 antibody. The protein levels of EZI and its mutants were evaluated by anti-Myc tag blotting. Arrowheads indicate EZI and its mutants (upper panel). The asterisks indicate the high molecular weight species of EZI. STAT3 was co-immunoprecipitated by full-length EZI, EZI-C and EZI-ZF, which contained the N-terminal region, but not by EZI-N and EZI-NC (lower panel). (D) Deletion mutants of STAT3 were prepared and expressed in COS cells with Myc-tagged EZI. Cell lysates were immunoprecipitated with anti-Myc tag antibody and STAT3 was detected with anti-STAT3 (epitope; N- or C-terminus) antibodies. Arrowheads indicate the STAT3 mutants. As the STAT3 mutant lacking the N-terminal region could not be detected with anti-STAT3(N) antibody, anti-STAT3(C) antibody was used to detect ΔN-STAT3. While it was detected in the total lysate, ΔN-STAT3 was not present in the anti-Myc tag immunoprecipitate (lower right panel). (E) Binding of endogenous EZI and STAT3. NIH-3T3 cells cultured for 2 days in low serum (0.5% FCS) media. Cells were deprived of serum for 2 h and then stimulated with 10 ng/ml OSM. Cell lysates were immunoprecipitated with anti-EZI antibody and protein A. STAT3 and EZI present in the lysates were detected by anti-EZI and anti-STAT3(N) antibodies, respectively.
In order to find the domain in EZI which is responsible for STAT3 binding, we generated four kinds of EZI mutants, i.e. EZI-N lacking the N-terminal portion with 183 amino acids including one zinc finger motif; EZI-ZF lacking the central region including 11 zinc finger motifs, consisting of 381 amino acids; EZI-C lacking the short C-terminal portion with 30 amino acids; and EZI-NC lacking both the N- and C-terminal domains (Figure 4B). These deletion mutants with a Myc tag were co-expressed with STAT3 in COS cells and co-immunoprecipitation assays were performed. STAT3 was not co-precipitated with the EZI-N and EZI-NC mutants lacking the N-terminal domain, while the other mutants which still retained the N-terminal region were co-precipitated (Figure 4C). These results indicate that the N-terminal 183 amino acids of EZI are responsible for the association with STAT3. Furthermore, the larger molecular weight band at ∼90 kDa was always observed only when full-length EZI and EZI-C were expressed (Figure 4C, upper panel), suggesting that some modification occurs in the N-terminal region of EZI including some of the zinc finger motifs.
We also made a series of STAT3 mutants and co-expressed them with full-length EZI (Figure 4D). A co-immunoprecipitation experiment revealed that a mutant lacking either the linker domain, SH2 domain or transactivation domain (TAD) was able to bind EZI, but a mutant lacking the N-terminal 505 amino acids failed to bind EZI (Figure 4D). This indicates that EZI binds to the N-terminal 505 amino acids of STAT3.
These results strongly suggest that EZI physically interacts with STAT3. However, as these results were obtained by transfecting cDNAs into COS7 cells, it remained possible that the interaction was due to overexpression. To exclude this possibility, we examined the interaction between endogenous EZI and STAT3 in NIH-3T3 cells by immunoprecipitation. We immunoprecipitated proteins with anti-EZI peptide antibodies and blotted them with anti-STAT3 antibody. As shown in Figure 4E, anti-EZI antibodies immunoprecipitated STAT3 when NIH-3T3 cells were stimulated with OSM, indicating that the binding of EZI to STAT3 occurs at physiological expression levels by OSM stimulation.
Nuclear localization and DNA binding of EZI
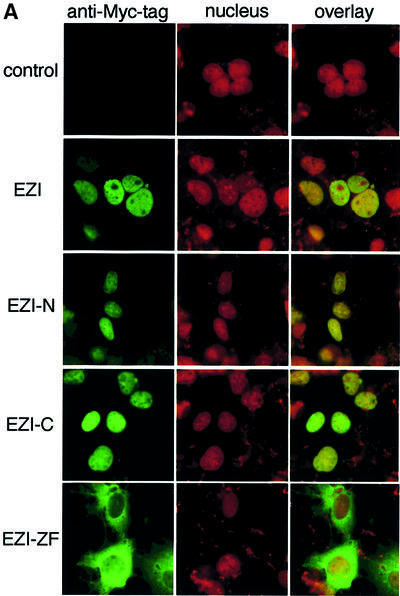
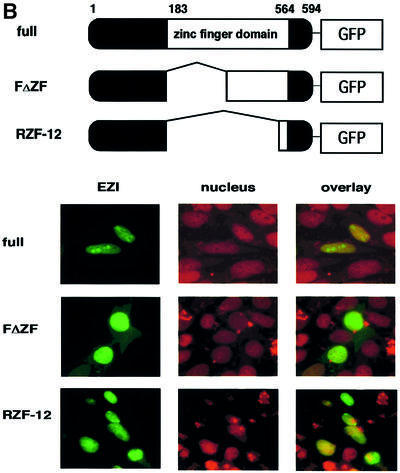
As luciferase reporter assays indicated that EZI activated transcription, it was expected that EZI localized to the nucleus. To test this possibility, EZI tagged with the Myc peptide was transiently expressed in COS cells. Cells were stained with anti-Myc tag antibody followed by fluorescein isothiocyanate (FITC)-labeled anti-mouse antibody, and intracellular localization of EZI was examined by fluorescence microscopy. The nucleus was counterstained with propidium iodide (PI). The staining pattern of EZI was completely merged with that of PI, indicating that EZI is localized predominantly in the nucleus (Figure 5A). Although EZI seemed to be present in the nucleus constitutively, EZI does not contain a typical nuclear localization signal (NLS). To delineate the domain of EZI responsible for nuclear localization, deletion mutants were expressed in COS cells. All the deletion mutants, except for EZI-ZF, localized in the nucleus in a manner similar to wild-type EZI. On the other hand, EZI-ZF that lacked 11 zinc fingers was almost completely excluded from the nucleus, indicating that zinc fingers are necessary for the nuclear localization of EZI. Although there is an NLS-like sequence KRFRKK in the fifth zinc finger, the FΔZF mutant that lacked the first seven zinc fingers localized predominantly in the nucleus, indicating that the NLS-like sequence is not necessary for nuclear localization. Furthermore the C-terminal zinc finger motif (twelfth zinc finger) was found to be sufficient for nuclear localization as the fusion protein of this domain and green fluorescent protein (GFP) localized predominantly in the nucleus (Figure 5B).
Fig. 5. Subcellular localization of EZI. (A) EZI and EZI mutants were expressed in COS cells. Cells were fixed and stained with anti-Myc tag antibody followed by FITC-labeled anti-mouse IgG antibody (green signal). Nuclei were counterstained with propidium iodide (red signal). Full-length EZI and most of the mutants were localized to the nucleus, while EZI-ZF lacking the zinc finger motifs was almost completely excluded from the nucleus and remained in the cytoplasm. (B) Nuclear localization of EZ1 by the C-terminal zinc finger. NIH-3T3 cells were transiently transfected with expression vectors for EZ1–GFP fusion protein. The green fluorescence signal from the GFP fusion construct was monitored using an inverted fluorescence microscope. Nuclei were counterstained with propidium iodide (red signal).
DNA binding of EZI
To test the ability of EZI to bind DNA, we employed the protein pull-down method using double-stranded biotinylated oligonucleotides and magnetic beads. As EZI effectively activated the APRE–luciferase construct, APRE was used for DNA binding experiments. Cell extract of COS cells transfected with the expression vectors of Myc-tagged EZI or that of STAT3 was incubated with APRE-conjugated beads, and proteins bound to the beads were analyzed by western blotting with either anti-Myc tag or anti-STAT3 antibody. The results indicated that both EZI and STAT3 bound the APRE oligonucleotide (Figure 6A). In order to delineate the EZI-binding site, we generated APRE mutants (Figure 6A). STAT3 is known to recognize TTCXXX GAA, and substitution of TTC by GAA (mutant 1) resulted in a severe reduction of STAT3 binding and complete elimination of EZI binding. In contrast, TTCTTCTTC (mutant 2) bound more EZI than the original APRE, whereas STAT3 binding was significantly reduced. TTCTGGTCC (mutant 3) showed very weak binding to STAT3 as well as EZI. These results suggested that EZI and STAT3 recognize distinct DNA motifs, though they may not be completely segregated.
Fig. 6. DNA binding of EZI. (A) Binding of EZI to the APRE sequence. EZI was transiently expressed in COS cells. Two days after transfection, the cells were lysed and nuclear extracts were incubated with APRE-conjugated magnetic beads. The proteins associated with the beads were eluted and subjected to western blotting with anti-Myc and anti-STAT3 antibody. The APRE mutant, with a mutation in the STAT3-binding site, showed increased binding to EZI, but STAT3 binding was decreased. The STAT3 recognition site is underlined. (B) DNA binding activity of EZI mutants. Nuclear extracts of COS cells expressing EZI mutants were incubated with the APRE oligonucleotide as above, and EZI protein bound to beads was detected with anti-Myc tag antibody. The arrowheads in the upper panel indicate the proteins pulled-down with the APRE sequence, while the asterisks in the lower panel show the expression pattern of EZI and EZI mutants in the nuclear extract. The higher molecular weight EZI-C was at the same location as the non-specific band in this experiment.
The possibility that EZI directly binds DNA was supported further by the assays using EZI mutants. While full-length EZI as well as EZI-C were able to bind the APRE, EZI-N, EZI-NC and EZI-ZF failed to bind (Figure 6B), indicating that the N-terminal as well as zinc finger domains of EZI are necessary for the DNA binding. As EZI-ZF was able to bind STAT3 (Figure 4C) but failed to bind the APRE sequence, it is unlikely that the binding of EZI to APRE is mediated by STAT3.
A role for EZI in STAT3 activation
As our results indicated that EZI is a nuclear protein that binds DNA, we examined whether EZI contains a transactivation domain. We made a plasmid construct, in which EZI was fused to the GAL4 DNA-binding domain, and co-transfected it with the GAL4 reporter plasmid in COS7 cells. However, the EZI–GAL4 fusion protein failed to transactivate the reporter gene (data not shown). We then examined if EZI is also involved in STAT3-mediated gene expression by expressing EZI mutants in NIH-3T3 cells. Expression of full-length EZI and EZI-C slightly enhanced the OSM-induced expression of luciferase by ARPE, while expression of EZI-N showed no significant effect. In contrast, expression of EZI-ZF suppressed OSM-induced expression of luciferase to a level close to that produced by expression of a dominant-negative STAT3, suggesting that EZI-ZF functions as a dominant-negative molecule for STAT3-mediated gene expression (Figure 7A).
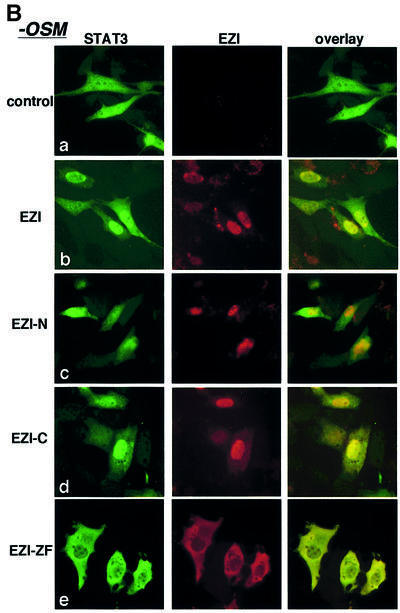
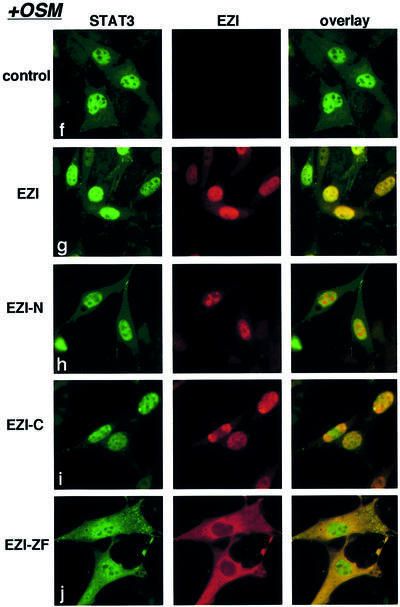
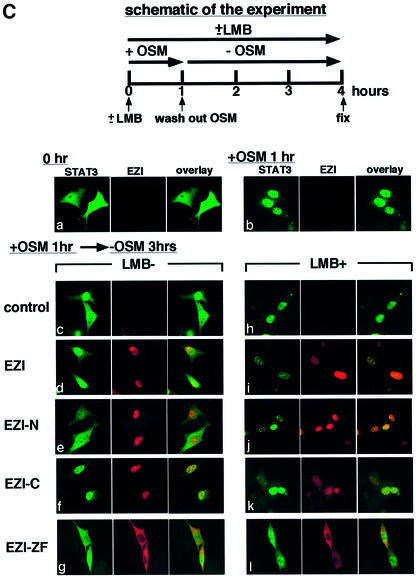
Fig. 7. Roles of EZI for the nuclear localization of STAT3. (A) Expression vectors of EZI or its mutants were transfected into NIH-3T3 cells with the APRE–luciferase reporter. Stimulation with OSM (30 h) produced a significant increase in the luciferase activity, and full-length EZI and EZI-C further enhanced the activity. The EZI-N mutant lacking the ability to bind STAT3 did not enhance the luciferase activity. The EZI-ZF mutant, which failed to localize to the nucleus, inhibited OSM-induced transcriptional activity. (B) Co-localization of EZI and STAT3. Expression vectors of EZI and STAT3 were transiently transfected into NIH-3T3 cells and stimulated with 10 ng/ml of OSM (+OSM) or left unstimulated (–OSM). Cells were fixed and stained by incubation with anti-STAT3 antibody and anti-Myc tag antibody, followed by FITC-labeled (green signal) and PE-labeled (red signal) secondary antibodies, respectively. EZI localized in the nucleus, while STAT3 distributed to both the cytoplasm and nucleus in the unstimulated condition (a–d). STAT3 was mostly excluded from the nucleus when co-expressed with EZI-ZF (e). EZI-ZF also suppressed the nuclear localization of STAT3 when stimulated with OSM (j). (C) Nuclear retention of STAT3 by EZI. NIH-3T3 cells transfected with EZI and STAT3 cDNAs (a) were first stimulated with OSM for 1 h (b) in the presence (10 ng/ml) or absence of LMB. Then, OSM was removed and the cells were incubated for an additional 3 h in the presence (h) or absence of LMB (c). Treatment with LMB suppressed the nuclear export of STAT3 (h–l). On the other hand, expression of EZI (d) and EZI-C (f) led to greater accumulation of STAT3 in the nucleus than vector control (c) and EZI-N (e) in the absence of LMB.
In order to find how EZI-ZF inhibits STAT3 activity, we analyzed localization of STAT3 in NIH-3T3 cells expressing EZI or its mutants. Microscopic observation indicated that EZI was localized predominantly in the nucleus, while STAT3 was found in both the nucleus and cytoplasm without OSM stimulation (Figure 7B, a–e). Interestingly, expression of the full-length EZI as well as EZI-C increased the nuclear STAT3 to some extent without stimulation, whereas the level of nuclear STAT3 was lower in cells expressing EZI-N (Figure 7B, b, c and d). Significantly, STAT3 was almost completely excluded from the nucleus by expression of EZI-ZF (Figure 7B, e).
These results were obtained in the absence of OSM, and stimulation with OSM further highlighted the differential effect of these mutants on STAT3 localization. OSM stimulation resulted in the accumulation of most of STAT3 in the nucleus in control cells as well as in cells expressing the full-length EZI, EZI-N and EZI-C (Figure 7B, f–i). However, even in the presence of OSM, a significant portion of STAT3 remained in the cytoplasm when EZI-ZF was expressed (Figure 7B, j). A difference in the localization of STAT3 was noticed in cells expressing EZI-N in the presence or absence of OSM, i.e. without OSM stimulation, nuclear accumulation of STAT3 in cells expressing EZI-N was much less than that in cells expressing the full-length EZI or EZI-C (Figure 7B, c). However, upon OSM stimulation, no such difference was observed (Figure 7B, h). While EZI-N was localized to the nucleus, it failed to bind STAT3 (Figure 4C). Thus, it is possible that the ability of EZI-N to retain STAT3 in the nucleus is minimal, if any. In contrast to STAT3, distribution of STAT1, STAT4 and STAT5 was unaffected by expressing any of the EZI mutants regardless of OSM stimulation (data not shown).
Leptomycin B (LMB) is known to inhibit protein export from the nucleus, and the addition of LMB resulted in the accumulation of STAT3 in the nucleus, indicating that the export of STAT3 from the nucleus is mediated by an LMB-sensitive mechanism (Figure 7C, h–l). Expression of full-length EZI and EZI-C also resulted in accumulation of STAT3 as well as EZI in the nucleus, though the nuclear accumulation of STAT3 was not as complete as that caused by the addition of LMB (Figure 7C, d and f). As EZI was localized predominantly in the nucleus and bound STAT3, the accumulation of STAT3 in the nucleus of cells expressing EZI appears to be mediated by the binding of STAT3 to EZI. In contrast, EZI-ZF stayed in the cytoplasm and nuclear localization of STAT3 was blocked in cells expressing EZI-ZF (Figure 7C, g and l). Moreover, the distribution of STAT3 in cells expressing EZI-ZF was unaffected by LMB (Figure 7C, l). These results indicate that nuclear localization of STAT3 is inhibited by EZI-ZF, which binds STAT3 but is unable to enter the nucleus.
Discussion
EZI is a novel zinc finger protein that was identified as an OSM-inducible gene product, although it is expressed rather ubiquitously. EZI is localized predominantly in the nucleus by its zinc finger motifs and transactivates genes in cooperation with STAT3. EZI binds STAT3 and enhances its nuclear localization. As the deletion mutant of EZI lacking 11 zinc fingers is capable of binding STAT3 but fails to translocate to the nucleus, it inhibits nuclear localization of STAT3 as well as STAT3-mediated transactivation. Based on these results, it is concluded that EZI is a novel nuclear zinc finger protein that augments STAT3 activity by retaining it in the nucleus.
Expression and structure of EZI
While EZI cDNA was cloned as an OSM-inducible gene, induction by OSM in LO cells was modest. As expression of EZI in AGM cells increased during in vitro culture in the presence of OSM (data not shown), it might be a factor that induces EZI expression, although OSM may not be the only factor required for the expression of EZI in the AGM region. As EZI was found to be rather ubiquitous in adult tissues and also in embryonic tissues at E11.5, it is suggested that EZI plays a role in a general cellular function.
EZI contains 12 repeats of a zinc finger motif in the central part of the molecule, but has no other motifs. The zinc finger motifs in EZI are the C2H2 type that consist of 21 amino acid residues with two cysteines and two histidines. Kruppel-like factors are a representative family of C2H2 type zinc finger proteins. They have three zinc finger repeats at their C-terminus and bind DNA through those repeats (Bieker, 2001). As zinc finger motifs of EZI exhibit homology to those of Kruppel-like protein family members, it is possible that the zinc finger motifs of EZI are involved in DNA binding. This possibility is supported by the deletion analysis; the zinc finger region of EZI is required for nuclear localization and DNA binding, but not for association with STAT3. The 183 amino acids in the N-terminal region of EZI are responsible for binding to STAT3. On the other hand, the deletion analysis of STAT3 indicates that the region including the N-terminal 505 amino acids in STAT3 is responsible for EZI binding. As EZI also binds STAT1, STAT4 and STAT5 weakly, there might be a conserved structure among these STATs for binding to EZI.
Nuclear localization of EZI
A unique feature of EZI is its predominant nuclear localization. Protein transport into the nucleus generally is mediated by the shuttle protein, importin, which recognizes the NLS in various nuclear proteins (Nigg, 1997). A typical NLS, originally found in the SV40 T antigen, consists of seven amino acids, PKKKRKV, including a cluster of basic amino acids. In addition to such NLSs, there is a report that zinc finger domains of C2H2-type zinc finger proteins such as EKLF and zif268 were sufficient for nuclear localization (Matheny et al., 1994; Shields and Yang, 1997). Although there is a basic amino acid cluster, KRFRKK, between amino acids 278 and 284 in the fifth zinc finger domain of EZI, deletion of the first seven zinc fingers did not affect nuclear localization, indicating that the C-terminal zinc finger motif (twelfth zinc finger) but not this NLS-like motif is involved in nuclear localization.
Transactivation and possible roles for EZI in STAT3-mediated gene expression
By using the luciferase reporter system, EZI was shown to transactivate the APRE, β-casein, CIS, c-fos and OSM promoters, but not the SRα promoter. Those promoters activated by EZI are also regulated by STAT proteins, STAT3 and STAT5 (Takebe et al., 1988; Wakao et al., 1994; Campbell et al., 1995; Yoshimura et al., 1995; Matsumoto et al., 1997). In fact, expression of a dominant-negative form of STAT3 effectively inhibited the EZI-induced expression of luciferase from the APRE, suggesting that STATs are involved in the transactivation by EZI. Protein pull-down assays using APRE showed that EZI and STAT3 bind APRE. EZI-ZF was capable of binding STAT3 but failed to bind APRE, suggesting that EZI binds DNA independently from STAT3. EZI was shown to bind STAT3 by co-immunoprecipitation, and STAT3 was shown previously to bind p300. Coomassie Blue staining of anti-Myc immunoprecipitates from transfectants of Myc-tagged EZI showed a co-precipitated protein of ∼90 kDa, which is likely to be STAT3. However, no other protein was found (data not shown), suggesting that EZI associates with STAT3 directly.
Both EZI and STAT3 bind the APRE wild-type DNA consisting of 16 nucleotides, 5′-CTTCTGGGAAGA-3′. However, the APRE mutants exhibited altered binding profiles. Although all these mutants exhibited reduced binding to STAT3, EZI binding to these mutants was distinct from STAT3 binding. While mutants 1 and 3 lost EZI binding almost completely, mutant 2 bound more EZI than the original APRE. Despite the lack of the authentic STAT3 recognition site, TTCXXXGAA, in mutant 2 (CTTCTTCTTCGA), it bound significantly more STAT3 than other mutants. As this mutant bound more EZI, it is possible that EZI binding to this sequence allows STAT3 to bind. Taking these results together, it is suggested that EZI and STAT3 recognize distinct DNA motifs and that binding of EZI to DNA may help the binding of STAT3 to DNA either by inducing a conformational change in DNA suitable for STAT3 binding or by stabilizing the binding of STAT3 to DNA. As EZI physically interacts with STAT3, it is likely that EZI and STAT3 form a complex with APRE.
EZI exhibits characteristics of transcription factors, i.e. transactivation, nuclear localization and DNA binding. An interesting finding in this study is that EZI cooperatively transactivates with STAT3, possibly through forming a complex on the APRE. While the precise mechanism of how EZI and STAT3 cooperatively induce gene expression awaits further investigation, our results clearly demonstrate that EZI keeps STAT3 in the nucleus. EZI-N stayed in the nucleus but failed to retain STAT3 in the nucleus because it lacks the N-terminal domain responsible for STAT3 binding. In contrast, EZI-ZF stayed in the cytoplasm and blocked the accumulation of STAT3 in the nucleus even in the presence of LMB. These results indicate that EZI keeps STAT3 in the nucleus. It is known that CRM1 binds the nuclear export signal (NES) directly and actively transports NES-containing proteins from the nucleus to the cytoplasm in cooperation with Ran-GTP. CRM1 was shown previously to export STAT1 from the nucleus when STAT1 was detached from DNA (McBride et al., 2000). As discussed above, one possible mechanism for EZI to enhance STAT3-mediated transcription is to form a stable complex with STAT3 and DNA, resulting in prolonged activation of STAT3. In addition, as EZI binds STAT3 that is not bound to DNA, EZI may also retain free STAT3 in the nucleus as a reservoir.
Recently, significant progress has been made in the elucidation of negative regulation of cytokine signaling. Negative regulators such as SOCS play an important role in cellular function by limiting the cytokine signals. However, continuous cytokine signaling is also necessary in some instances. In the primary culture system of the AGM region, production of hematopoietic cells requires continuous stimulation by OSM (Mukouyama et al., 1998). Likewise, continuous OSM stimulation is necessary for fetal hepatocytes to differentiate in vitro (Kamiya et al., 1999). Transient cytokine stimulation may be sufficient for some cases; however, continuous cytokine stimulation is also required for other cases such as the AGM region and fetal liver cells, and EZI may be a factor that plays a role in the duration of cytokine signaling by keeping STAT3 in the activated state.
Materials and methods
Cell culture and media
LO is an OSM-dependent endothelial-like cell line derived from the mouse AGM region. LO cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 15% fetal bovine serum (FBS), 10 ng/ml OSM, 1 mM sodium pyruvate and antibiotics. NIH-3T3 cells were maintained in DMEM with 10% calf serum (CS) and antibiotics. COS7 cells were cultured in DMEM supplemented with 10% FBS and antibiotics.
Isolation of full-length EZI cDNAs
We prepared cDNA libraries of LO cells stimulated or not with OSM. Subtraction of cDNA libraries was performed as described previously (Nakayama et al., 1999). The subtracted cDNAs were sequenced randomly and a cDNA fragment encoding a novel zinc finger protein EZI was obtained. The partial cDNA fragment of EZI was labeled with digoxigenin (DIG) by PCR amplification and was used to isolate the full-length cDNA clones from a randomly primed LO cDNA library. The sequences of the isolated clone were determined by an automated DNA sequencer (Perkin Elmer).
Northern blotting
LO cells were deprived of OSM and serum for 24 h and cells were harvested at 0, 0.5, 1 and 2 h after OSM stimulation. Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. A 10 µg aliquot of total RNA for each sample was electrophoresed on a 1.2% agarose gel containing 2.4% formaldehyde. RNA was then transferred to a positively charged nylon membrane and hybridized with DIG-labeled EZI probe in high-SDS buffer (50% formamide, 5× SSC, 0.05 M phosphate buffer pH 7.0, 2% DIG blocking reagent, 0.1% lauroylsarcosine) at 45°C for 12 h. After washing the filter to remove the non-specifically bound probe, the filter was incubated with alkaline phosphatase-conjugated anti-DIG antibody. Bound alkaline phosphatase was detected by a chemiluminescent substrate according to the manufacturer’s instructions (Roche Diagnostics).
RT–PCR analyses
Expression of EZI was tested by RT–PCR using RNA derived from various tissues. The sequence between nucleotides 517 and 1052 of the EZI cDNA was amplified by the forward primer 5′-GCACTC CTGCTCAGGGCA-3′ and the reverse primer 5′-CTTGTCGC ACTCTGAGCA-3′. Twenty-five cycles of amplification were performed under the following conditions: denaturation at 94°C for 30 s, annealing at 53°C for 30 s and elongation at 72°C for 1 min.
Immunocytochemistry
COS cells were transiently transfected with the Myc-tagged EZI expression vector by the lipofection method using lipofectamine (Invitrogen) and fixed with 4% paraformaldehyde 2 days later. These cells were stained with anti-Myc tag antibody (1 µg/ml) (Santa Cruz) followed by incubation with FITC-conjugated anti-mouse IgG antibody (5 µg/ml). Nuclei were stained by incubating cells in 10 µg/ml of PI solution. NIH-3T3 cells were transiently transfected with the Myc-tagged EZI expression vector and expression vectors for STATs by the lipofection method. Cells were fixed and double-stained with anti-Myc-tag antibody followed by phycoerythrin (PE)-conjugated anti-mouse IgG antibody and anti-STATs (1 µg/ml) (Santa Cruz) antibody, followed by FITC-conjugated anti-rabbit IgG antibody (5 µg/ml). Stained cells were observed with a fluorescent microscope (Nikon).
Luciferase assay
The EZI cDNA was inserted into the pME18S expression vector, and COS cells (3 × 105) were transiently transfected with the EZI cDNA (600 ng) and several promoter–luciferase constructs (600 ng) by the lipofection method using lipofectamine. COS cells were harvested 2 days after the transfection and lysed in 300 µl of passive lysis buffer (Promega) in each well of a 12-well culture plate. A 10 µl aliquot of cell lysate was subjected to a luciferase assay by using the dual promoter luciferase kit (Promega). Luciferase activity was measured by Microlumat (Perkin Elmer). Relative luciferase activity was calculated by dividing the value of the firefly luciferase activity with that of Renilla control luciferase activity.
Immunoprecipitation and immunoblotting
COS cells (5 × 106) were transfected with 5 µg of expression vectors for Myc-tagged EZI and STATs by the lipofection method. Transfected COS cells were lysed in protein lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.1% Triton, 1 mM sodium vanadate, 0.1 µg/ml phenylmethyl sulfonyl fluoride and 1 µg/ml leupeptin). A 2 µl aliquot of anti-Myc-tag antibody was added to the lysate, followed by the addition of protein A (Pierce). The immune complexes were subjected to SDS–PAGE on an 8% acrylamide gel and transferred to a PVDF membrane (Millipore). The filter was incubated with anti-Myc tag antibody or anti-STAT3 antibody (Transduction Laboratory), followed by incubation with the secondary antibody conjugated to horseradish peroxidase. The immune complex was detected by the enhanced chemiluminescence system (AP-Bioteh). Anti-EZI antibodies used for immunoprecipitation were prepared by immunizing rabbits with the C-terminal EZI peptide NPSEVVPPPIFF.
Protein pull-down assays by biotinylated oligonucleotides
Protein pull-down assays using biotinylated oligonucleotides were employed to test the ability of the protein to associate with specific DNA sequences (Ebert and Bunn, 1998; Buzek et al., 2002). COS cells were transiently transfected with an expression vector for Myc-tagged EZI or STAT3 and lysed in lysis buffer (20 mM HEPES pH 7.9, 0.1% NP-40, 75 mM NaCl, 100 mM KCl, 0.2 mM EDTA, 10% glycerol). The oligonucleotides used for the pull-down assays were α2-macroglobulin APRE-WT (5′-CTTCTGGGAAGA-3′) and its mutants (Takeda et al., 1994). The cell lysate was incubated for 12 h with streptavidin-conjugated Dynabeads (Dynal, M280) that bound biotinylated APRE double-stranded oligonucleotides in 1× binding buffer (10 mM Tris–HCl pH 7.5, 50 mM KCl, 1 mM MgCl2, 1 mM EDTA, 5.5 mM dithiothreitol, 5% glycerol, 0.03% NP-40). The nucleotide–protein complex was washed twice with 0.5× binding buffer and subjected to SDS–PAGE. Western blotting was performed with anti-Myc tag or anti-STAT3 antibody.
Accession number
The sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession No. AB076746.
Acknowledgments
Acknowledgements
We thank Drs Minoru Tanaka and Keisuke Sekine for helpful discussions. K.N. was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists. This work was supported in part by Grants-in-Aid for Scientific Research and Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports and Technology, the Japanese Government.
References
- Akira S. et al. (1994) Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell, 77, 63–71. [DOI] [PubMed] [Google Scholar]
- Bieker J.J. (2001) Kruppel-like factors: three fingers in many pies. J. Biol. Chem., 276, 34355–34358. [DOI] [PubMed] [Google Scholar]
- Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- Buzek J., Latonen,L., Kurki,S., Peltonen,K. and Laiho,M. (2002) Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res., 30, 2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G.S., Meyer,D.J., Raz,R., Levy,D.E., Schwartz,J. and Carter-Su,C. (1995) Activation of acute phase response factor (APRF)/Stat3 transcription factor by growth hormone. J. Biol. Chem., 270, 3974–3979. [DOI] [PubMed] [Google Scholar]
- Cantwell C.A., Sterneck,E. and Johnson,P.F. (1998) Interleukin-6-specific activation of the C/EBPδ gene in hepatocytes is mediated by Stat3 and Sp1. Mol. Cell. Biol., 18, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.D., Liao,J., Liu,B., Rao,X., Jay,P., Berta,P. and Shuai,K. (1997) Specific inhibition of Stat3 signal transduction by PIAS3. Science, 278, 1803–1805. [DOI] [PubMed] [Google Scholar]
- Darnell J.E. Jr (1997) STATs and gene regulation. Science, 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- Ebert B.L. and Bunn,H.F. (1998) Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor and p300/CREB binding protein. Mol. Cell. Biol., 18, 4089–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T. and Felsenfeld,G. (1989) The erythroid-specific transcription factor Eryf1: a new finger protein. Cell, 58, 877–885. [DOI] [PubMed] [Google Scholar]
- Fujitani Y., Nakajima,K., Kojima,H., Nakae,K., Takeda,T. and Hirano,T. (1994) Transcriptional activation of the IL-6 response element in the junB promoter is mediated by multiple Stat family proteins. Biochem. Biophys. Res. Commun., 202, 1181–1187. [DOI] [PubMed] [Google Scholar]
- Harroch S., Revel,M. and Chebath,J. (1994) Induction by interleukin-6 of interferon regulatory factor 1 (IRF-1) gene expression through the palindromic interferon response element pIRE and cell type-dependent control of IRF-1 binding to DNA. EMBO J., 13, 1942–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocke G.M., Barry,D. and Fey,G.H. (1992) Synergistic action of interleukin-6 and glucocorticoids is mediated by the interleukin-6 response element of the rat α2 macroglobulin gene. Mol. Cell. Biol., 12, 2282–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A. et al. (1999) Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J., 18, 2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liao,J., Rao,X., Kushner,S.A., Chung,C.D., Chang,D.D. and Shuai,K. (1998) Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl Acad. Sci. USA, 95, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look D.C., Pelletier,M.R., Tidwell,R.M., Roswit,W.T. and Holtzman,M.J. (1995) Stat1 depends on transcriptional synergy with Sp1. J. Biol. Chem., 270, 30264–30267. [DOI] [PubMed] [Google Scholar]
- Mascareno E., Dhar,M. and Siddiqui,M.A. (1998) Signal transduction and activator of transcription (STAT) protein-dependent activation of angiotensinogen promoter: a cellular signal for hypertrophy in cardiac muscle. Proc. Natl Acad. Sci. USA, 95, 5590–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny C., Day,M.L. and Milbrandt,J. (1994) The nuclear localization signal of NGFI-A is located within the zinc finger DNA binding domain. J. Biol. Chem., 269, 8176–8181. [PubMed] [Google Scholar]
- Matsumoto A., Masuhara,M., Mitsui,K., Yokouchi,M., Ohtsubo,M., Misawa,H., Miyajima,A. and Yoshimura,A. (1997) CIS, a cytokine inducible SH2 protein, is a target of the JAK–STAT5 pathway and modulates STAT5 activation. Blood, 89, 3148–3154. [PubMed] [Google Scholar]
- McBride K.M., McDonald,C. and Reich,N.C. (2000) Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J., 19, 6196–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukouyama Y. et al. (1998) In vitro expansion of murine multipotential hematopoietic progenitors from the embryonic aorta–gonad– mesonephros region. Immunity, 8, 105–114. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yanagisawa,M., Arakawa,H., Kimura,N., Hisatsune,T., Kawabata,M., Miyazono,K. and Taga,T. (1999) Synergistic signaling in fetal brain by STAT3–Smad1 complex bridged by p300. Science, 284, 479–482. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Hara,T., Hibi,M., Hirano,T. and Miyajima,A. (1999) A novel oncostatin M-inducible gene OIG37 forms a gene family with MyD118 and GADD45 and negatively regulates cell growth. J. Biol. Chem., 274, 24766–24772. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. (1997) Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature, 386, 779–787. [DOI] [PubMed] [Google Scholar]
- Nuez B., Michalovich,D., Bygrave,A., Ploemacher,R. and Grosveld,F. (1995) Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature, 375, 316–318. [DOI] [PubMed] [Google Scholar]
- Raz R., Durbin,J.E. and Levy,D.E. (1994) Acute phase response factor and additional members of the interferon-stimulated gene factor 3 family integrate diverse signals from cytokines, interferons and growth factors. J. Biol. Chem., 269, 24391–24395. [PubMed] [Google Scholar]
- Rekhtman N., Radparvar,F., Evans,T. and Skoultchi,A.I. (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev., 13, 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodel B. et al. (2000) The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J., 19, 5845–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer T.S., Sanders,L.K. and Nathans,D. (1995) Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc. Natl Acad. Sci. USA, 92, 9097–9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields J.M. and Yang,V.W. (1997) Two potent nuclear localization signals in the gut-enriched Kruppel-like factor define a subfamily of closely related Kruppel proteins. J. Biol. Chem., 272, 18504–18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani R.A. and Orkin,S.H. (1996) The transcriptional control of hematopoiesis. Blood, 87, 4025–4039. [PubMed] [Google Scholar]
- Stocklin E., Wissler,M., Gouilleux,F. and Groner,B. (1996) Functional interactions between Stat5 and the glucocorticoid receptor. Nature, 383, 726–728. [DOI] [PubMed] [Google Scholar]
- Taga T. and Kishimoto,T. (1997) Gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol., 15, 797–819. [DOI] [PubMed] [Google Scholar]
- Takebe Y., Seiki,M., Fujisawa,J., Hoy,P., Yokota,K., Arai,K., Yoshida,M. and Arai,N. (1988) SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol., 8, 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Nakajima,K., Kojima,H. and Hirano,T. (1994) E1A repression of IL-6-induced gene activation by blocking the assembly of IL-6 response element binding complexes. J. Immunol., 153, 4573–4582. [PubMed] [Google Scholar]
- Tsang A.P., Visvader,J.E., Turner,C.A., Fujiwara,Y., Yu,C., Weiss,M.J., Crossley,M. and Orkin,S.H. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell, 90, 109–119. [DOI] [PubMed] [Google Scholar]
- Varnum B.C., Ma,Q.F., Chi,T.H., Fletcher,B. and Herschman,H.R. (1991) The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys–His repeat. Mol. Cell. Biol., 11, 1754–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao H., Gouilleux,F. and Groner,B. (1994) Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J., 13, 2182–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegenka U.M. et al. (1994) The interleukin-6-activated acute-phase response factor is antigenically and functionally related to members of the signal transducer and activator of transcription (STAT) family. Mol. Cell. Biol., 14, 3186–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A., Ohkubo,T., Kiguchi,T., Jenkins,N.A., Gilbert,D.J., Copeland,N.G., Hara,T. and Miyajima,A. (1995) A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J., 14, 2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Wegenka,U.M., Lutticken,C., Buschmann,J., Decker,T., Schindler,C., Heinrich,P.C. and Horn,F. (1994) The signalling pathways of interleukin-6 and γ interferon converge by the activation of different transcription factors which bind to common responsive DNA elements. Mol. Cell. Biol., 14, 1657–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Behre,G., Pan,J., Iwama,A., Wara-Aswapati,N., Radomska,H.S., Auron,P.E., Tenen,D.G. and Sun,Z. (1999) Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl Acad. Sci. USA, 96, 8705–8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wrzeszczynska,M.H., Horvath,C.M. and Darnell,J.E.,Jr (1999) Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol., 19, 7138–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Wen,Z. and Darnell,J.E.,Jr (1994) Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science, 264, 95–98. [DOI] [PubMed] [Google Scholar]